Molecular Targets for the Diagnosis and Treatment of Pancreatic Cancer
Abstract
1. Introduction
2. Risk Factors for PC
3. PC Biomarkers
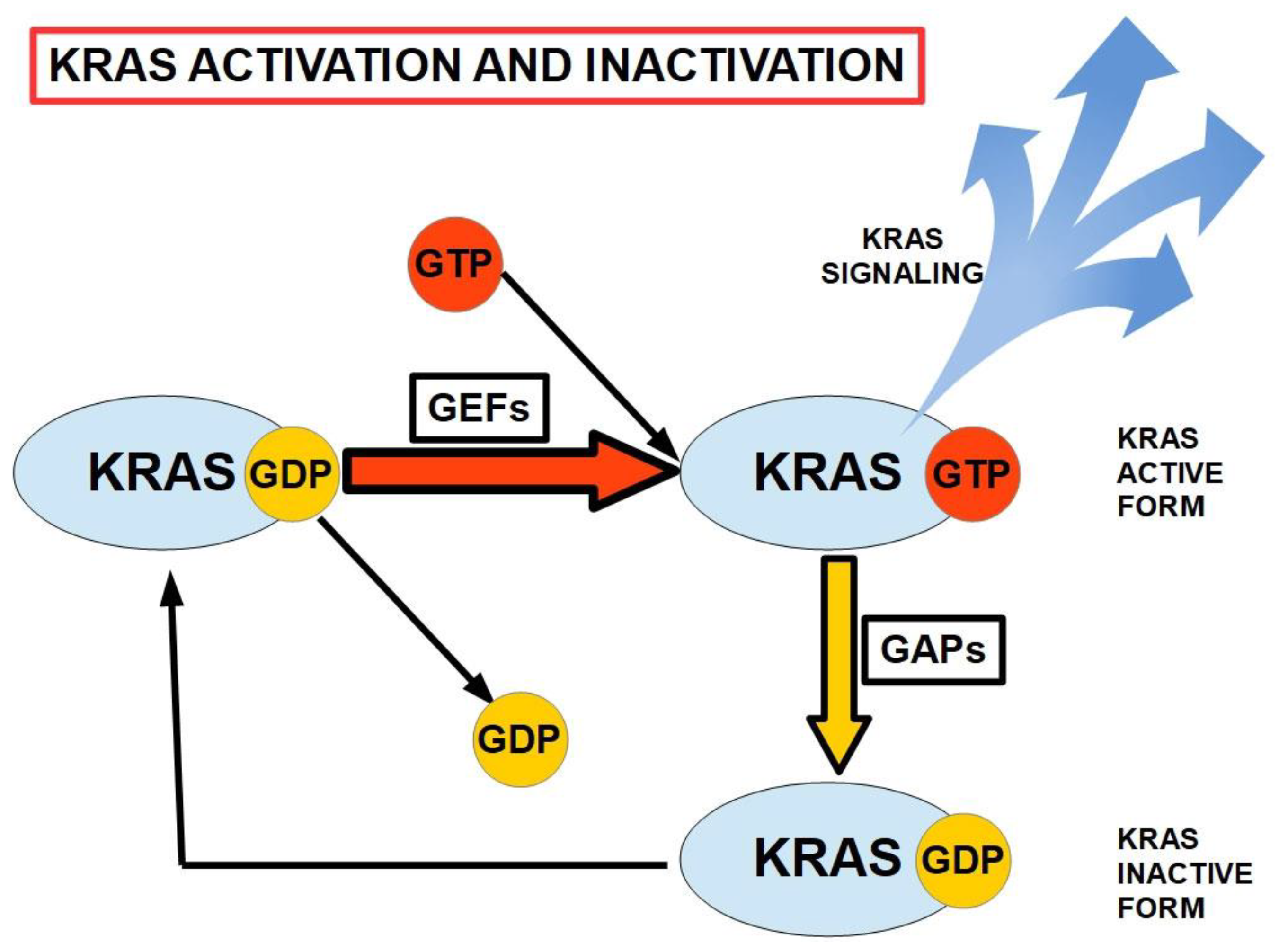
4. Genetic Factors in Pancreatic Cancer
- Kirsten rat sarcoma (KRAS).
- Cyclin-dependent kinase inhibitor 2 (CDKN2A/p16).
- ○
- aka MTS1 or multiple tumor suppressor 1.
- Tumor suppressor protein 53 (TP53).
- Small Mothers against Decapentaplegic homolog 4 (MADH4/DPC4/SMAD4).
5. Immunogenicity and Antigenicity of PC
6. MicroRNAs in PC
7. miRNA in Diagnosis of PC
8. Treatments for Pancreatic Cancer
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, J.X.; Zhao, C.F.; Chen, W.B.; Liu, Q.C.; Li, Q.W.; Lin, Y.Y.; Gao, F. Pancreatic cancer: A review of epidemiology, trend, and risk factors. World J. Gastroenterol. 2021, 27, 4298–4321. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Goral, V. Pancreatic cancer: Pathogenesis and diagnosis. Asian Pac. J. Cancer Prev. 2015, 16, 5619–5624. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.P.; Oldfield, L.; Ney, A.; Hart, P.A.; Keane, M.G.; Pandol, S.J.; Li, D.; Greenhalf, W.; Jeon, C.Y.; Koay, E.J.; et al. Early detection of pancreatic cancer. Lancet Gastroenterol. Hepatol. 2020, 5, 698–710. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shi, J.; Huang, J.; Cheng, M.; Geng, S.; Yu, W.; Chen, N.; Chen, C.; Wang, Z. Case-Control Trials on Risk Factors for Pancreatic Cancer: A Systematic Review and Meta-Analysis. Iran. J. Public Health 2023, 52, 1578–1588. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, Z.; Liu, W. Pancreatic Cancer: A Review of Risk Factors, Diagnosis, and Treatment. Technol. Cancer Res. Treat. 2020, 19, 1533033820962117. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Henriksen, A.; Dyhl-Polk, A.; Chen, I.; Nielsen, D. Checkpoint inhibitors in pancreatic cancer. Cancer Treat. Rev. 2019, 78, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.; Brown, S.R.; Mettu, N.B.; Miller, P.C.; Smyth, E.C.; Nixon, A.B. Incorporating Molecular Data Into Treatment Decision Making in Gastroesophageal and Pancreaticobiliary Cancers: Timing and Strategies. Am. Soc. Clin. Oncol. Educ. Book. 2024, 44, e433640. [Google Scholar] [CrossRef]
- Buscail, L.; Bournet, B.; Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Lanfranca, M.P.; Thompson, J.K.; Bednar, F.; Halbrook, C.; Lyssiotis, C.; Levi, B.; Frankel, T.L. Metabolism and epigenetics of pancreatic cancer stem cells. Semin. Cancer Biol. 2019, 57, 19–26. [Google Scholar] [CrossRef]
- Grady, W.M.; Yu, M.; Markowitz, S.D. Epigenetic Alterations in the Gastrointestinal Tract: Current and Emerging Use for Biomarkers of Cancer. Gastroenterology 2021, 160, 690–709. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Low, S.K.; Kuchiba, A.; Zembutsu, H.; Saito, A.; Takahashi, A.; Kubo, M.; Daigo, Y.; Kamatani, N.; Chiku, S.; Totsuka, H.; et al. Genome-wide association study of pancreatic cancer in Japanese population. PLoS ONE 2010, 5, e11824. [Google Scholar] [CrossRef]
- Wu, C.; Miao, X.; Huang, L.; Che, X.; Jiang, G.; Yu, D.; Yang, X.; Cao, G.; Hu, Z.; Zhou, Y.; et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat. Genet. 2011, 44, 62–66. [Google Scholar] [CrossRef]
- Wolpin, B.M.; Kraft, P.; Gross, M.; Helzlsouer, K.; Bueno-de-Mesquita, H.B.; Steplowski, E.; Stolzenberg-Solomon, R.Z.; Arslan, A.A.; Jacobs, E.J.; LaCroix, A.; et al. Pancreatic cancer risk and ABO blood group alleles: Results from the pancreatic cancer cohort consortium. Cancer Res. 2010, 70, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G.M.; Amundadottir, L.; Fuchs, C.S.; Kraft, P.; Stolzenberg-Solomon, R.Z.; Jacobs, K.B.; Arslan, A.A.; Bueno-de-Mesquita, H.B.; Gallinger, S.; Gross, M.; et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010, 42, 224–228. [Google Scholar] [CrossRef]
- Midha, S.; Chawla, S.; Garg, P.K. Modifiable and non-modifiable risk factors for pancreatic cancer: A review. Cancer Lett. 2016, 381, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.D.; Hruban, R.H. Pathology and molecular genetics of pancreatic neoplasms. Cancer J. 2012, 18, 492–501. [Google Scholar] [CrossRef]
- Hruban, R.H.; Canto, M.I.; Goggins, M.; Schulick, R.; Klein, A.P. Update on familial pancreatic cancer. Adv. Surg. 2010, 44, 293–311. [Google Scholar] [CrossRef]
- Yeo, T.P. Demographics, epidemiology, and inheritance of pancreatic ductal adenocarcinoma. Semin. Oncol. 2015, 42, 8–18. [Google Scholar] [CrossRef]
- Becker, A.E.; Hernandez, Y.G.; Frucht, H.; Lucas, A.L. Pancreatic ductal adenocarcinoma: Risk factors, screening, and early detection. World J. Gastroenterol. 2014, 20, 11182–11198. [Google Scholar] [CrossRef]
- Chen, F.; Roberts, N.J.; Klein, A.P. Inherited pancreatic cancer. Chin. Clin. Oncol. 2017, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Landi, S. Genetic predisposition and environmental risk factors to pancreatic cancer: A review of the literature. Mutat. Res. 2009, 681, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Del Chiaro, M.; Segersvard, R.; Lohr, M.; Verbeke, C. Early detection and prevention of pancreatic cancer: Is it really possible today? World J. Gastroenterol. 2014, 20, 12118–12131. [Google Scholar] [CrossRef]
- Raimondi, S.; Maisonneuve, P.; Lowenfels, A.B. Epidemiology of pancreatic cancer: An overview. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 699–708. [Google Scholar] [CrossRef]
- Sessa, F.; Solcia, E.; Capella, C.; Bonato, M.; Scarpa, A.; Zamboni, G.; Pellegata, N.S.; Ranzani, G.N.; Rickaert, F.; Klöppel, G. Intraductal papillary-mucinous tumours represent a distinct group of pancreatic neoplasms: An investigation of tumour cell differentiation and K-ras, p53 and c-erbB-2 abnormalities in 26 patients. Virchows Arch. 1994, 425, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Reshkin, S.J.; Cardone, R.A.; Koltai, T. Genetic Signature of Human Pancreatic Cancer and Personalized Targeting. Cells 2024, 13, 602. [Google Scholar] [CrossRef]
- Zhao, Y.; Tang, J.; Jiang, K.; Liu, S.Y.; Aicher, A.; Heeschen, C. Liquid biopsy in pancreatic cancer—Current perspective and future outlook. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188868. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Casolino, R.; Paiella, S.; Azzolina, D.; Beer, P.A.; Corbo, V.; Lorenzoni, G.; Gregori, D.; Golan, T.; Braconi, C.; Froeling, F.E.; et al. Homologous recombination deficiency in pancreatic cancer: A systematic review and prevalence meta-analysis. J. Clin. Oncol. 2021, 39, 2617–2631. [Google Scholar] [CrossRef]
- Golan, T.; Khvalevsky, E.Z.; Hubert, A.; Gabai, R.M.; Hen, N.; Segal, A.; Domb, A.; Harari, G.; Ben David, E.; Raskin, S.; et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 2015, 6, 24560–24570. [Google Scholar] [CrossRef]
- Blankenstein, T.; Coulie, P.G.; Gilboa, E.; Jaffee, E.M. The determinants of tumour immunogenicity. Nat. Rev. Cancer 2012, 12, 307–313. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Farhangnia, P.; Khorramdelazad, H.; Nickho, H.; Delbandi, A.A. Current and future immunotherapeutic approaches in pancreatic cancer treatment. J. Hematol. Oncol. 2024, 17, 40. [Google Scholar] [CrossRef] [PubMed]
- Falcomatà, C.; Bärthel, S.; Schneider, G.; Rad, R.; Schmidt-Supprian, M.; Saur, D. Context-specific determinants of the immunosuppressive tumor microenvironment in pancreatic cancer. Cancer Discov. 2023, 13, 278–297. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, R.; Debnath, D.; Hartley, M.L.; Noel, M.S. The role of immunotherapy in pancreatic cancer. Curr. Oncol. 2022, 29, 6864–6892. [Google Scholar] [CrossRef] [PubMed]
- Real, F.X. A catastrophic hypothesis for pancreas cancer progression. Gastroenterology 2003, 124, 1958–1964. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Liu, X.; Zhang, Y.; Liu, Y.; Wang, Y.; Guo, S.; Jin, X.; Zhang, J.; Guan, Y.; Liu, Y. Frontiers and future of immunotherapy for pancreatic cancer: From molecular mechanisms to clinical application. Front. Immunol. 2024, 15, 1383978. [Google Scholar] [CrossRef]
- Sanchez, K.; Page, D.B.; Urba, W. Immunotherapy toxicities. Surg. Oncol. Clin. 2019, 28, 387–401. [Google Scholar] [CrossRef]
- Eso, Y.; Seno, H. Current status of treatment with immune checkpoint inhibitors for gastrointestinal, hepatobiliary, and pancreatic cancers. Ther. Adv. Gastroenterol. 2020, 13, 1756284820948773. [Google Scholar] [CrossRef]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Zhao, Y.; Samal, E.; Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005, 436, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Naguibneva, I.; Ameyar-Zazoua, M.; Polesskaya, A.; Ait-Si-Ali, S.; Groisman, R.; Souidi, M.; Cuvellier, S.; Harel-Bellan, A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 2006, 8, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Pichiorri, F.; Palumbo, T.; Iuliano, R.; Cimmino, A.; Aqeilan, R.; Volinia, S.; Bhatt, D.; Alder, H.; Marcucci, G.; et al. MicroRNA fingerprints during human mega-karyocytopoiesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5078–5083. [Google Scholar] [CrossRef] [PubMed]
- Dalmay, T.; Edwards, D.R. MicroRNAs and the hallmarks cancer. Oncogene 2006, 25, 6170–6175. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Hwang, H.W.; Mendell, J.T. MicroRNAs in cell proliferation, cell death and tumorigenesis. Br. J. Cancer 2006, 94, 776–780. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Mitra, R.; Maulik, U.; Zhang, M.Q. Development of the human cancer mikroRNA network. Silence 2010, 1, 6. [Google Scholar] [CrossRef]
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.; Alitheen, N.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef]
- Sandesc, M.; Dinu, A.; Rogobete, A.F.; Bedreag, O.H.; Sandesc, D.; Papurica, M.; Bratu, L.M.; Negoita, S.; Vernic, C.; Popovici, S.E.; et al. Circulating microRNAs Expressions as Genetic Biomarkers in Pancreatic Cancer Patients Continuous Non-Invasive Monitoring. Clin. Lab. 2017, 63, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Itani, M.M.; Nassar, F.J.; Tfayli, A.H.; Talhouk, R.S.; Chamandi, G.K.; Itani, A.R.S.; Makoukji, J.; Boustany, R.M.N.; Hou, L.; Zgheib, N.K.; et al. Signature of Four Circulating microRNAs as Potential Biomarkers for Diagnosing Early-Stage Breast Cancer. Int. J. Mol. Sci. 2021, 22, 6121. [Google Scholar] [CrossRef]
- Szabo, A.; Gurlich, R.; Liberko, M.; Soumarova, R.; Vernerova, Z.; Mandys, V.; Popov, A. Expression of selected microRNAs in pancreatic ductal adenocarcinoma: Is there a relation to tumor morphology, progression and patient’s outcome? Neoplasma 2020, 67, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, H.; Wu, W.; Jiang, T.; Qiu, Z. Regulation of miR-155 affects pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1. Oncol. Rep. 2013, 30, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Wang, M.; Liu, Z. A panel of 8 miRNAs as a novel diagnostic biomarker in pancreatic cancer. Medicine 2020, 99, e22261. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, R.L.; Liu, S.; Tai, M.C.; Rajapakshe, K.; Huang, Y.; Longton, G.; DeCapite, C.; Hurd, M.W.; Paris, P.L.; Kirkwood, K.S.; et al. Plasma miRNA Biomarkers in Limited Volume Samples for Detection of Early-stage Pancreatic Cancer. Cancer Prev. Res. 2021, 14, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Yabushita, S.; Fukamachi, K.; Tanaka, H.; Sumida, K.; Deguchi, Y.; Sukata, T.; Kawamura, S.; Uwagawa, S.; Suzui, M.; Tsuda, H. Circulating microRNAs in serum of human K-ras oncogene transgenic rats with pancreatic ductal adenocarcinomas. Pancreas 2012, 41, 1013–1018. [Google Scholar] [CrossRef]
- Smolarz, B.; Durczy’nski, A.; Romanowicz, H.; Hogendorf, P. The Role of microRNA in Pancreatic Cancer. Biomedicines 2021, 9, 1322. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Bhardwaj, A.; Singh, S.; Arora, S.; Wang, B.; Grizzle, W.E.; Singh, A.P. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis 2011, 32, 183–1839. [Google Scholar] [CrossRef]
- Bhatti, I.; Lee, A.; James, V.; Hall, R.I.; Lund, J.N.; Tufarelli, C.; Lobo, D.N.; Larvin, M. Knockdown of microRNA-21 inhibits proliferation and increases cell death by targeting programmed cell death 4 [PDCD4] in pancreatic ductal adenocarcinoma. J. Gastrointest. Surg. 2011, 15, 199–208. [Google Scholar] [CrossRef]
- Zhen, D.B.; Coveler, A.; Zanon, S.; Reni, M.; Chiorean, E.G. Biomarker-driven and molecularly targeted therapies for pancreatic adenocarcinoma. Semin. Oncol. 2018, 45, 107–115. [Google Scholar] [CrossRef]
- Liu, J.; Gao, J.; Du, Y.; Li, Z.; Ren, Y.; Gu, J.; Wang, X.; Gong, Y.; Wang, W.; Kong, X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. Cancer 2012, 131, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Nagao, Y.; Hisaoka, M.; Matsuyama, A.; Kanemitsu, S.; Hamada, T.; Fukuyama, T.; Nakano, R.; Uchiyama, A.; Kawamoto, M.; Yamaguchi, K.; et al. Association of microRNA-21 expression with its targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod. Pathol. 2012, 25, 112–121. [Google Scholar] [CrossRef]
- Poonpanichakul, T.; Shiao, M.S.; Jiravejchakul, N.; Matangkasombut, P.; Sirachainan, E.; Charoensawan, V.; Jinawath, N. Capturing tumour heterogeneity in pre- and post-chemotherapy colorectal cancer ascites-derived cells using single-cell RNA-sequencing. Biosci. Rep. 2021, 41, BSR20212093. [Google Scholar] [CrossRef] [PubMed]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Genes. 2022. Available online: https://www.aacr.org/professionals/research/aacr-project-genie/aacr-project-genie-data/11-most-common-cancer-typesin-the-aacr-project-genie-registry/pancreatic-cancer/ (accessed on 30 July 2024).
- Strickler, J.H.; Satake, H.; Hollebecque, A.; Sunakawa, Y.; Tomasini, P.; Bajor, D.L.; Schuler, M.H.; Yaeger, R.; George, T.J.; Garrido-Laguna, I.; et al. First data for sotorasib in patients with pancreatic cancer with KRAS p.G12C mutation: A phase I/II study evaluating efficacy and safety. JCO 2022, 40 (Suppl. S36), 360490. [Google Scholar] [CrossRef]
- Tesfaye, A.A.; Wang, H.; Hartley, M.L.; He, A.R.; Weiner, L.; Gabelia, N.; Kapanadze, L.; Shezad, M.; Brody, J.R.; Marshall, J.L.; et al. A Pilot trial of molecularly tailored therapy for patients with metastatic pancreatic ductal adenocarcinoma. J. Pancreat. Cancer 2019, 5, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Sohal, D.P.; Kennedy, E.B.; Cinar, P.; Conroy, T.; Copur, M.S.; Crane, C.H.; Garrido-Laguna, I.; Lau, M.W.; Johnson, T.; Krishnamurthi, S.; et al. Metastatic pancreatic cancer: ASCO guideline update. JCO 2020, 38, 3217–3230. [Google Scholar] [CrossRef]
- Kimura, H.; Yamamoto, H.; Harada, T.; Fumoto, K.; Osugi, Y.; Sada, R.; Maehara, N.; Hikita, H.; Mori, S.; Eguchi, H.; et al. CKAP4, a dkk1 receptor, is a biomarker in exosomes derived from pancreatic cancer and a molecular target for therapy. Clin. Cancer Res. 2019, 25, 1936–1947. [Google Scholar] [CrossRef]
- Kolbeinsson, H.M.; Chandana, S.; Wright, G.P.; Chung, M. Pancreatic Cancer: A Review of Current Treatment and Novel Therapies. J. Investig. Surg. 2023, 36, 2129884. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Y.; Zheng, Y.; Bao, Y.; Wang, P. Postoperative hepatic arterial infusion chemotherapy improved survival of pancreatic cancer after radical pancreatectomy: A retrospective study. Onco Targets Ther. 2018, 11, 903–907. [Google Scholar] [CrossRef]
| Gene | Effects |
|---|---|
| CLDN4 | Impedes drug penetration into cells. Increases chemosensitivity. |
| LCN2 | Secretes glycoprotein that induces proliferation, angiogenesis, and invasion. |
| MSLN | Interacts with MUC18-favoring peritoneal metastasis and plays a role in drug resistance. |
| PSCA | A surface antigen correlated with advanced stages and tumor progression. |
| Over-expressed in metastasis. | |
| S100A4 | Increases tumor progression and promotes metastasis. |
| Inhibits apoptosis and promotes chemoresistance. | |
| SFN | Involved in tumor progression, considered a marker of poor prognosis. |
| TFF2 | Induces PC cell migration. Also shows anti-tumoral effects. |
| Circulating miRNAs | Expression Level |
|---|---|
| miR-21 | Increased |
| miR-155 | Increased |
| miR-196a | Increased |
| miR-210 | Increased |
| miR-16 | Increased |
| miR-21 | Increased |
| miR-155 | Increased |
| miR181a | Increased |
| miR-181b | Increased |
| miR-196a | Increased |
| miR-210 | Increased |
| miR-26b | Increased |
| miR-34a | Increased |
| miR-122 | Increased |
| miR-126 | Increased |
| miR-145 | Increased |
| miR-150 | Increased |
| miR-196a | Increased |
| miR-223 | Increased |
| miR-505 | Decreased |
| miR-636 | Decreased |
| miR-885.5p | Decreased |
| miR-18a | Decreased |
| miR-21 | Decreased |
| miR-221 | Decreased |
| miR-483-3p | Decreased |
| miR-20a | Decreased |
| miR-21 | Decreased |
| miR-24 | Decreased |
| miR-25 | Decreased |
| miR-99a | Decreased |
| miR-185 | Decreased |
| miR-191 | Decreased |
| miR-1246 | Decreased |
| miR-4644 | Decreased |
| miR-3976 | Decreased |
| miR-4306 | Decreased |
| let-7d | Decreased |
| Target | Drug | Trial Phase | NCT Number |
|---|---|---|---|
| KRAS | TCR-T (G12V) | I | NCT04146298 |
| Exosomes (G12D) | I | NCT03608631 | |
| LY 3537982 (G12C) * | I | NCT04956640 | |
| Anti-KRAS G12V mTCR PBL | I | NCT03190941 | |
| Anti-KRAS G12D mTCR PBL | I | NCT03745326 | |
| MRT X849 ** | II | NCT03785249 | |
| ELI-002 *** | I | NCT04853017 | |
| CDKN2A | SY 5609 (CDK7) | I | NCT04247126 |
| Palbociclib (CDK 4/6) *** | I | NCT03065062 | |
| BRCA 1 or 2 | Olaparib + Pembrolizumab | ||
| vs. Olaparib alone | II | NCT04548752 | |
| BRCA1, 2, PALB2 | Adjuvant Olaparib | ||
| vs. placebo (APOLLO trial) | II | NCT04858334 | |
| ATM | ATR inhibitors | ||
| PIK3CA | Taselisib (MAT CH) | II | NCT02465060 |
| ROS1 | Crizotinib (MAT CH) | II | NCT02465060 |
| BRAF | Dabrafenib/Trametinib | II | NCT02465060 |
| ERBB2 | Trastuzumab/Deruxtecan | II | NCT04482309 |
| CTNNB1 | Tegavivint | II | NCT04851119 |
| NF1 | Trametinib (MAT CH) | II | NCT02465060 |
| FGFR1 | Trametinib (MAT CH) | II | NCT02465060 |
| TP53 | SGT53 + GEM + NabP | II | NCT02340117 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pergolizzi, R.G.; Brower, S.T. Molecular Targets for the Diagnosis and Treatment of Pancreatic Cancer. Int. J. Mol. Sci. 2024, 25, 10843. https://doi.org/10.3390/ijms251910843
Pergolizzi RG, Brower ST. Molecular Targets for the Diagnosis and Treatment of Pancreatic Cancer. International Journal of Molecular Sciences. 2024; 25(19):10843. https://doi.org/10.3390/ijms251910843
Chicago/Turabian StylePergolizzi, Robert G., and Steven T. Brower. 2024. "Molecular Targets for the Diagnosis and Treatment of Pancreatic Cancer" International Journal of Molecular Sciences 25, no. 19: 10843. https://doi.org/10.3390/ijms251910843
APA StylePergolizzi, R. G., & Brower, S. T. (2024). Molecular Targets for the Diagnosis and Treatment of Pancreatic Cancer. International Journal of Molecular Sciences, 25(19), 10843. https://doi.org/10.3390/ijms251910843





