Assessment of Genetic Stability in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Using Droplet Digital PCR
Abstract
1. Introduction
2. Results
2.1. Generation and Cardiac Gene Expression of hiPSC-Derived CMs (hiPSC-CMs)
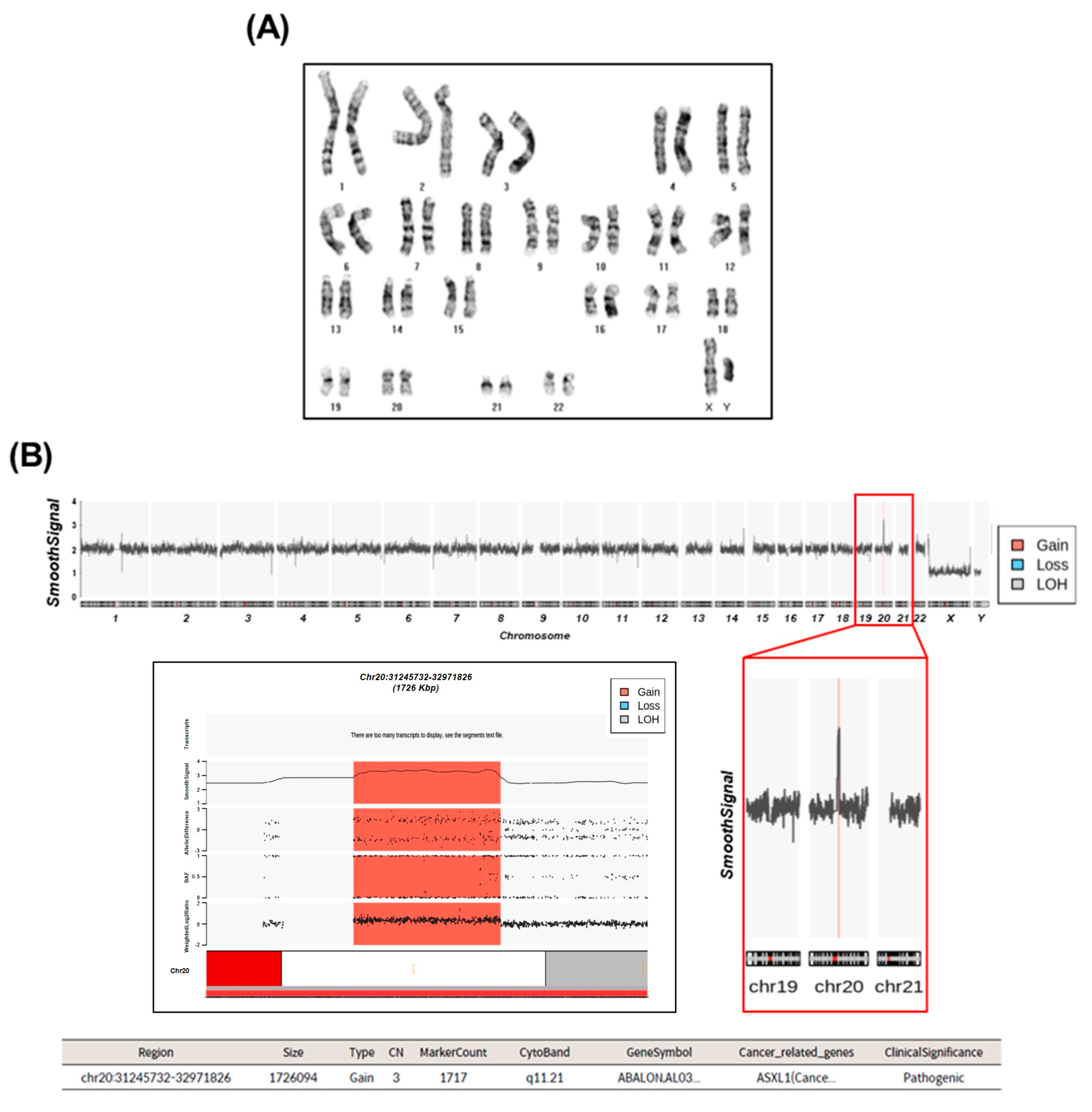
2.2. Cytogenetic Analysis
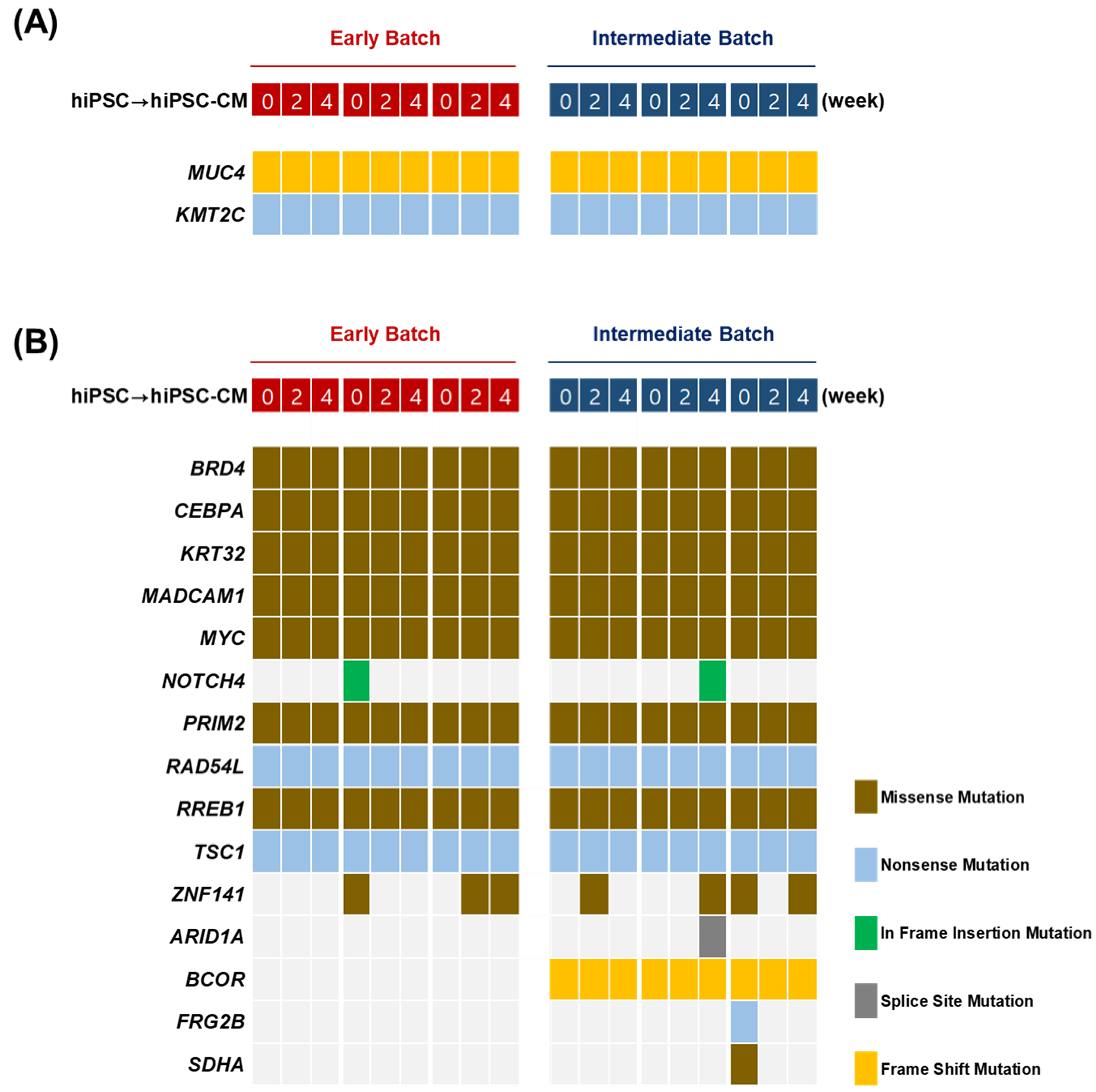
2.3. Whole-Exome Sequencing
2.4. Targeted Sequencing
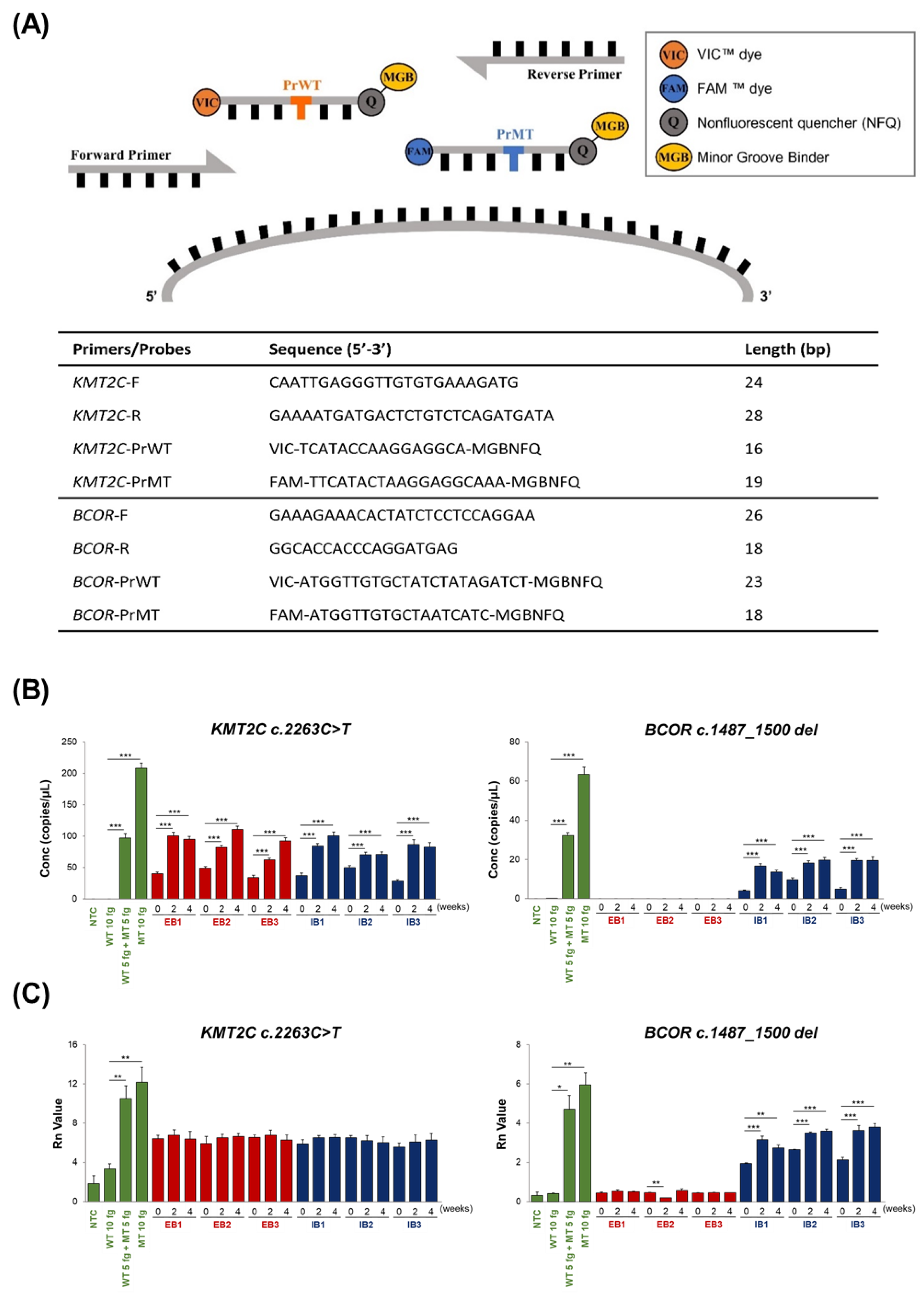
2.5. Real-Time PCR and ddPCR
2.6. Validation
2.6.1. Precision
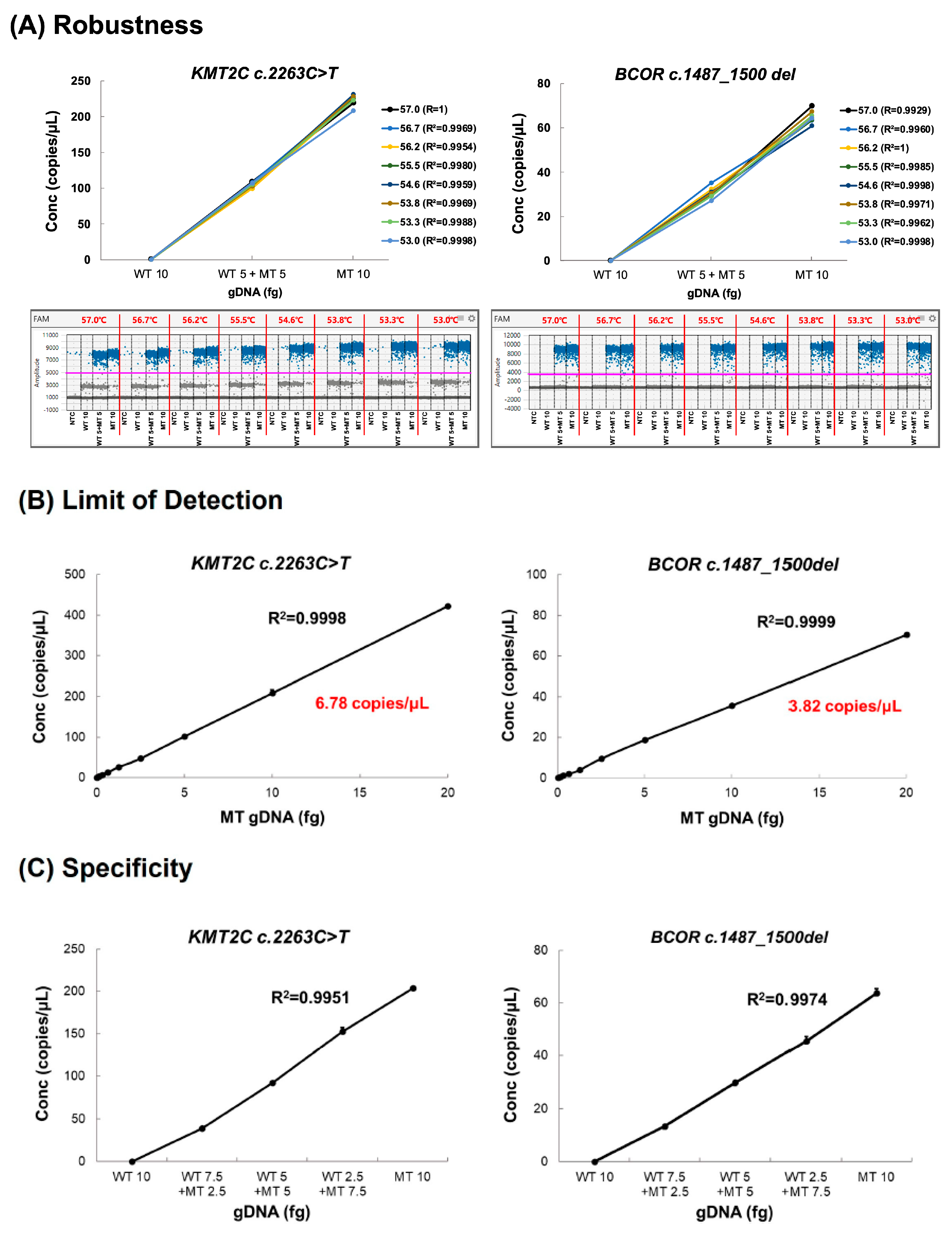
2.6.2. Robustness
2.6.3. Limit of Detection (LOD)
2.6.4. Specificity
3. Discussion
4. Materials and Methods
4.1. hiPSC Culture
4.2. Differentiation of hiPSCs into CMs
4.3. Karyotyping
4.4. CytoscanHD Chip Analysis
4.5. Whole-Exome Sequencing
4.6. Targeted Sequencing
4.7. Control Template Generation
4.8. ddPCR Analysis
4.9. Evaluation of Differentiation into Cardiomyocyte Using RT-qPCR
4.10. Detection of Tumorigenic Variants Using Real-Time PCR
4.11. Validation of ddPCR for hiPSC-CMs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hoang, D.M.; Pham, P.T.; Bach, T.Q.; Ngo, A.T.L.; Nguyen, Q.T.; Phan, T.T.K.; Nguyen, G.H.; Le, P.T.T.; Hoang, V.T.; Forsyth, N.R.; et al. Stem cell-based therapy for human diseases. Signal Transduct. Target. Ther. 2022, 7, 272. [Google Scholar] [CrossRef]
- Rivera-Ordaz, A.; Peli, V.; Manzini, P.; Barilani, M.; Lazzari, L. Critical Analysis of cGMP Large-Scale Expansion Process in Bioreactors of Human Induced Pluripotent Stem Cells in the Framework of Quality by Design. BioDrugs 2021, 35, 693–714. [Google Scholar] [CrossRef]
- Peterson, S.E.; Loring, J.F. Genomic instability in pluripotent stem cells: Implications for clinical applications. J. Biol. Chem. 2014, 289, 4578–4584. [Google Scholar] [CrossRef]
- FDA. Guidance for Industry: Preclinical Assessment of Investigational Cellular and Gene Therapy Product. 2013. Available online: https://www.fda.gov/media/87564/download (accessed on 19 December 2023).
- EMA. Guidelines on Quality, Non-Clinical and Clinical Aspects of Medicinal Products Containing Genetically Modified Cells. 2021. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-quality-non-clinical-and-clinical-aspects-medicinal-products-containing-genetically-modified-cells-revision-1_en.pdf (accessed on 19 December 2023).
- MFDS. Guideline on Quality, Non-Clinical and Clinical Assessment of Stem Cell Therapy Product. 2018. Available online: https://www.mfds.go.kr/docviewer/skin/doc.html?fn=20230725032527588.pdf&rs=/docviewer/result/data0011/15265/1/202312 (accessed on 19 December 2023).
- ICH. Preclinical Safety Evaluation of Biotechnology-Derived Pharmaceuticals S6 (R1). 2011. Available online: https://database.ich.org/sites/default/files/S6_R1_Guideline_0.pdf (accessed on 19 December 2023).
- Lee, T.H.; Kang, T.H. DNA Oxidation and Excision Repair Pathways. Int. J. Mol. Sci. 2019, 20, 6092. [Google Scholar] [CrossRef] [PubMed]
- Novo, N.; Romero-Tamayo, S.; Marcuello, C.; Boneta, S.; Blasco-Machin, I.; Velazquez-Campoy, A.; Villanueva, R.; Moreno-Loshuertos, R.; Lostao, A.; Medina, M.; et al. Beyond a platform protein for the degradosome assembly: The Apoptosis-Inducing Factor as an efficient nuclease involved in chromatinolysis. PNAS Nexus 2023, 2, pgac312. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.V.; Harrison, C.J.; Buck, G.A.; Richards, S.M.; Secker-Walker, L.M.; Martineau, M.; Vance, G.H.; Cherry, A.M.; Higgins, R.R.; Fielding, A.K.; et al. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): Analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood 2007, 109, 3189–3197. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, F.C.; Kim, A.S.; Mosse, C.A.; Shaver, A.C.; Yenamandra, A.; Seegmiller, A.C. Limited Utility of Fluorescence In Situ Hybridization for Recurrent Abnormalities in Acute Myeloid Leukemia at Diagnosis and Follow-up. Am. J. Clin. Pathol. 2018, 149, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Akkari, Y.M.N.; Baughn, L.B.; Dubuc, A.M.; Smith, A.C.; Mallo, M.; Dal Cin, P.; Diez Campelo, M.; Gallego, M.S.; Granada Font, I.; Haase, D.T.; et al. Guiding the global evolution of cytogenetic testing for hematologic malignancies. Blood 2022, 139, 2273–2284. [Google Scholar] [CrossRef]
- Jo, H.Y.; Han, H.W.; Jung, I.; Ju, J.H.; Park, S.J.; Moon, S.; Geum, D.; Kim, H.; Park, H.J.; Kim, S.; et al. Development of genetic quality tests for good manufacturing practice-compliant induced pluripotent stem cells and their derivatives. Sci. Rep. 2020, 10, 3939. [Google Scholar] [CrossRef]
- Yuan, Y.; Chung, C.Y.; Chan, T.F. Advances in optical mapping for genomic research. Comput. Struct. Biotechnol. J. 2020, 18, 2051–2062. [Google Scholar] [CrossRef]
- Hartman, P.; Beckman, K.; Silverstein, K.; Yohe, S.; Schomaker, M.; Henzler, C.; Onsongo, G.; Lam, H.C.; Munro, S.; Daniel, J.; et al. Next generation sequencing for clinical diagnostics: Five year experience of an academic laboratory. Mol. Genet. Metab. Rep. 2019, 19, 100464. [Google Scholar] [CrossRef] [PubMed]
- Doostparast Torshizi, A.; Wang, K. Next-generation sequencing in drug development: Target identification and genetically stratified clinical trials. Drug Discov. Today 2018, 23, 1776–1783. [Google Scholar] [CrossRef]
- Morganti, S.; Tarantino, P.; Ferraro, E.; D’Amico, P.; Duso, B.A.; Curigliano, G. Next Generation Sequencing (NGS): A Revolutionary Technology in Pharmacogenomics and Personalized Medicine in Cancer. Adv. Exp. Med. Biol. 2019, 1168, 9–30. [Google Scholar] [PubMed]
- Di Resta, C.; Galbiati, S.; Carrera, P.; Ferrari, M. Next-generation sequencing approach for the diagnosis of human diseases: Open challenges and new opportunities. Ejifcc 2018, 29, 4–14. [Google Scholar]
- Zhong, Y.; Xu, F.; Wu, J.; Schubert, J.; Li, M.M. Application of Next Generation Sequencing in Laboratory Medicine. Ann. Lab. Med. 2021, 41, 25–43. [Google Scholar] [CrossRef]
- Sullivan, S.; Stacey, G.N.; Akazawa, C.; Aoyama, N.; Baptista, R.; Bedford, P.; Bennaceur Griscelli, A.; Chandra, A.; Elwood, N.; Girard, M.; et al. Quality control guidelines for clinical-grade human induced pluripotent stem cell lines. Regen. Med. 2018, 13, 859–866. [Google Scholar] [CrossRef]
- Koehler, K.R.; Tropel, P.; Theile, J.W.; Kondo, T.; Cummins, T.R.; Viville, S.; Hashino, E. Extended passaging increases the efficiency of neural differentiation from induced pluripotent stem cells. BMC Neurosci. 2011, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Cantor, E.L.; Shen, F.; Jiang, G.; Tan, Z.; Cunningham, G.M.; Wu, X.; Philips, S.; Schneider, B.P. Passage number affects differentiation of sensory neurons from human induced pluripotent stem cells. Sci. Rep. 2022, 12, 15869. [Google Scholar] [CrossRef]
- Hu, S.; Zhao, M.T.; Jahanbani, F.; Shao, N.Y.; Lee, W.H.; Chen, H.; Snyder, M.P.; Wu, J.C. Effects of cellular origin on differentiation of human induced pluripotent stem cell-derived endothelial cells. JCI Insight 2016, 1, e85558. [Google Scholar] [CrossRef]
- Hussein, S.M.; Batada, N.N.; Vuoristo, S.; Ching, R.W.; Autio, R.; Narva, E.; Ng, S.; Sourour, M.; Hamalainen, R.; Olsson, C.; et al. Copy number variation and selection during reprogramming to pluripotency. Nature 2011, 471, 58–62. [Google Scholar] [CrossRef]
- Suva, M.L.; Riggi, N.; Bernstein, B.E. Epigenetic reprogramming in cancer. Science 2013, 339, 1567–1570. [Google Scholar] [CrossRef]
- Ji, J.; Ng, S.H.; Sharma, V.; Neculai, D.; Hussein, S.; Sam, M.; Trinh, Q.; Church, G.M.; McPherson, J.D.; Nagy, A.; et al. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells 2012, 30, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Kitamura, T. ASXL1 mutation in clonal hematopoiesis. Exp. Hematol. 2020, 83, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Balasubramani, A.; Larjo, A.; Bassein, J.A.; Chang, X.; Hastie, R.B.; Togher, S.M.; Lahdesmaki, H.; Rao, A. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat. Commun. 2015, 6, 7307. [Google Scholar] [CrossRef] [PubMed]
- Astolfi, A.; Fiore, M.; Melchionda, F.; Indio, V.; Bertuccio, S.N.; Pession, A. BCOR involvement in cancer. Epigenomics 2019, 11, 835–855. [Google Scholar] [CrossRef] [PubMed]
- Chai, X.; Tao, Q.; Li, L. The role of RING finger proteins in chromatin remodeling and biological functions. Epigenomics 2023, 15, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Benzaquen, J.; Morris, L.G.T.; Ilie, M.; Hofman, P. Mutations in KMT2C, BCOR and KDM5C Predict Response to Immune Checkpoint Blockade Therapy in Non-Small Cell Lung Cancer. Cancers 2022, 14, 2816. [Google Scholar] [CrossRef]
- Garcia-Sanz, P.; Trivino, J.C.; Mota, A.; Perez Lopez, M.; Colas, E.; Rojo-Sebastian, A.; Garcia, A.; Gatius, S.; Ruiz, M.; Prat, J.; et al. Chromatin remodelling and DNA repair genes are frequently mutated in endometrioid endometrial carcinoma. Int. J. Cancer 2017, 140, 1551–1563. [Google Scholar] [CrossRef]
- Salk, J.J.; Schmitt, M.W.; Loeb, L.A. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 2018, 19, 269–285. [Google Scholar] [CrossRef]
- Shin, S.; Park, J. Characterization of sequence-specific errors in various next-generation sequencing systems. Mol. Biosyst. 2016, 12, 914–922. [Google Scholar] [CrossRef]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef]
- Bacher, U.; Shumilov, E.; Flach, J.; Porret, N.; Joncourt, R.; Wiedemann, G.; Fiedler, M.; Novak, U.; Amstutz, U.; Pabst, T. Challenges in the introduction of next-generation sequencing (NGS) for diagnostics of myeloid malignancies into clinical routine use. Blood Cancer J. 2018, 8, 113. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Q.; Wang, Q.; Jia, P.; Zhao, Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: Features and perspectives. BMC Bioinform. 2013, 14 (Suppl. S11), S1. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Lu, H.M.; Chen, J.; Li, S.; Elliott, A.M. Sanger Confirmation Is Required to Achieve Optimal Sensitivity and Specificity in Next-Generation Sequencing Panel Testing. J. Mol. Diagn. 2016, 18, 923–932. [Google Scholar] [CrossRef]
- Della Starza, I.; De Novi, L.A.; Santoro, A.; Salemi, D.; Spinelli, O.; Tosi, M.; Soscia, R.; Paoloni, F.; Cappelli, L.V.; Cavalli, M.; et al. Digital Droplet PCR Is a Reliable Tool to Improve Minimal Residual Disease Stratification in Adult Philadelphia-Negative Acute Lymphoblastic Leukemia. J. Mol. Diagn. 2022, 24, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Eric Capo, G.S. Shuntaro Koizumi, Isolde Puts, Fredrik Olajos, Helena Königsson, Jan Karlsson, Pär Byström., Droplet digital PCR applied to environmental DNA, a promising method to estimate fish population abundance from humic-rich aquatic ecosystems. Environ. DNA 2021, 3, 343–352. [Google Scholar] [CrossRef]
- Baker, C.S.; Steel, D.; Nieukirk, S.; Klinck, H. Environmental DNA (eDNA) from the Wake of the Whales: Droplet Digital PCR for Detection and Species Identification. Front. Mar. Sci. 2018, 5, 133. [Google Scholar] [CrossRef]
- Sayour, N.V.; Toth, V.E.; Nagy, R.N.; Voros, I.; Gergely, T.G.; Onodi, Z.; Nagy, N.; Bodor, C.; Varadi, B.; Ruppert, M.; et al. Droplet Digital PCR Is a Novel Screening Method Identifying Potential Cardiac G-Protein-Coupled Receptors as Candidate Pharmacological Targets in a Rat Model of Pressure-Overload-Induced Cardiac Dysfunction. Int. J. Mol. Sci. 2023, 24, 13826. [Google Scholar] [CrossRef]
- He, Y.; Yan, W.; Dong, L.; Ma, Y.; Li, C.; Xie, Y.; Liu, N.; Xing, Z.; Xia, W.; Long, L.; et al. An effective droplet digital PCR method for identifying and quantifying meat adulteration in raw and processed food of beef (Bos taurus) and lamb (Ovis aries). Front. Sustain. Food Syst. 2023, 7, 1180301. [Google Scholar] [CrossRef]
- Maheshwari, Y.; Selvaraj, V.; Hajeri, S.; Yokomi, R. Application of droplet digital PCR for quantitative detection of Spiroplasma citri in comparison with real time PCR. PLoS ONE 2017, 12, e0184751. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Yun, J.H.; Park, J.W.; Seong, D.; Lee, S.H.; Park, K.D.; Lee, H.A.; Park, M. Effect of Xenogeneic Substances on the Glycan Profiles and Electrophysiological Properties of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Int. J. Stem Cells 2023, 16, 281–292. [Google Scholar] [CrossRef] [PubMed]

| GENE | Consequence | Protein Change | Variant Type | Tier (COSMIC) | Genomic Change | Chrom |
|---|---|---|---|---|---|---|
| KMT2C | Nonsense_Mutation | p.Gln755Ter | SNV | - | c.2263C > T | chr7 |
| BCOR | Frame_Shift_Del | p.Ile496AsnfsTer17 | Del | T3 | c.1487_1500del | chrX |
| Sample Type | KMT2C c.2263C > T Concentration (Copies/µL) | |||||||
|---|---|---|---|---|---|---|---|---|
| Analyst | A-1 | A-2 | A-3 | B-1 | B-2 | B-3 | Mean ± SD | %RSD |
| NTC | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 ± 0.00 | - |
| WT 10 fg | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 ± 0.00 | - |
| WT 5 fg + MT 5 fg | 93.92 | 91.73 | 94.38 | 103.07 | 99.30 | 107.51 | 98.32 ± 5.57 | 5.67 |
| MT 10fg | 210.82 | 205.19 | 199.40 | 212.75 | 217.00 | 218.40 | 210.59 ± 6.60 | 3.13 |
| Sample Type | BCORc.1487_1500del Concentration (copies/µL) | |||||||
| Analyst | A-1 | A-2 | A-3 | B-1 | B-2 | B-3 | Mean ± SD | %RSD |
| NTC | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 ± 0.00 | - |
| WT 10 fg | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 ± 0.00 | - |
| WT 5 fg + MT 5 fg | 25.61 | 29.26 | 30.03 | 31.57 | 33.30 | 31.50 | 30.21 ± 2.42 | 8.01 |
| MT 10fg | 55.82 | 61.72 | 53.46 | 64.83 | 69.50 | 62.20 | 61.25 ± 5.36 | 8.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.W.; Bae, S.J.; Yun, J.H.; Kim, S.; Park, M. Assessment of Genetic Stability in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Using Droplet Digital PCR. Int. J. Mol. Sci. 2024, 25, 1101. https://doi.org/10.3390/ijms25021101
Park JW, Bae SJ, Yun JH, Kim S, Park M. Assessment of Genetic Stability in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Using Droplet Digital PCR. International Journal of Molecular Sciences. 2024; 25(2):1101. https://doi.org/10.3390/ijms25021101
Chicago/Turabian StylePark, Ji Won, Su Ji Bae, Jun Ho Yun, Sunhee Kim, and Misun Park. 2024. "Assessment of Genetic Stability in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Using Droplet Digital PCR" International Journal of Molecular Sciences 25, no. 2: 1101. https://doi.org/10.3390/ijms25021101
APA StylePark, J. W., Bae, S. J., Yun, J. H., Kim, S., & Park, M. (2024). Assessment of Genetic Stability in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Using Droplet Digital PCR. International Journal of Molecular Sciences, 25(2), 1101. https://doi.org/10.3390/ijms25021101






