Prognostic Significance of Glycolysis-Related Genes in Lung Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
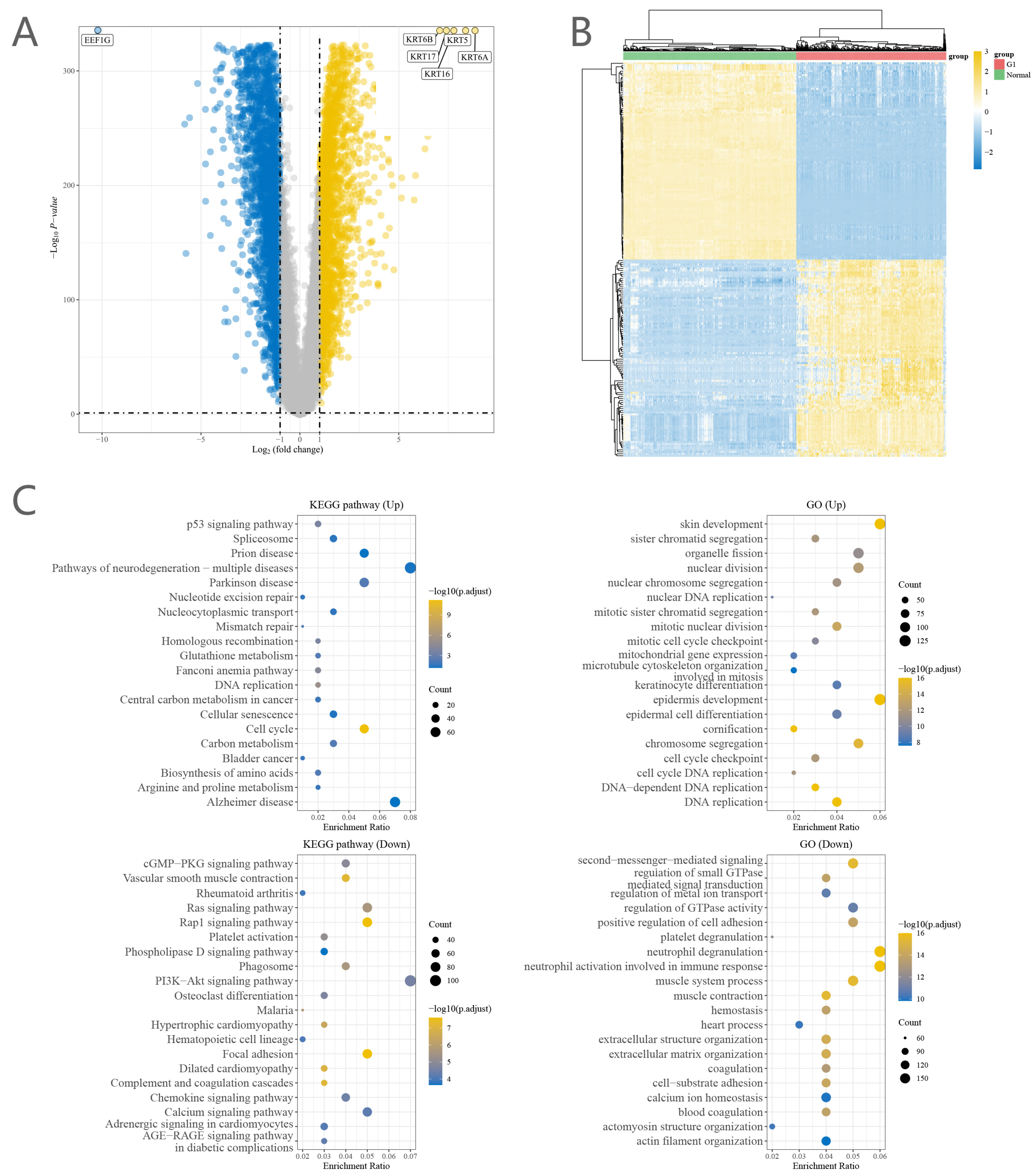
2.1. Identification of DEGs with Prognostic Effects
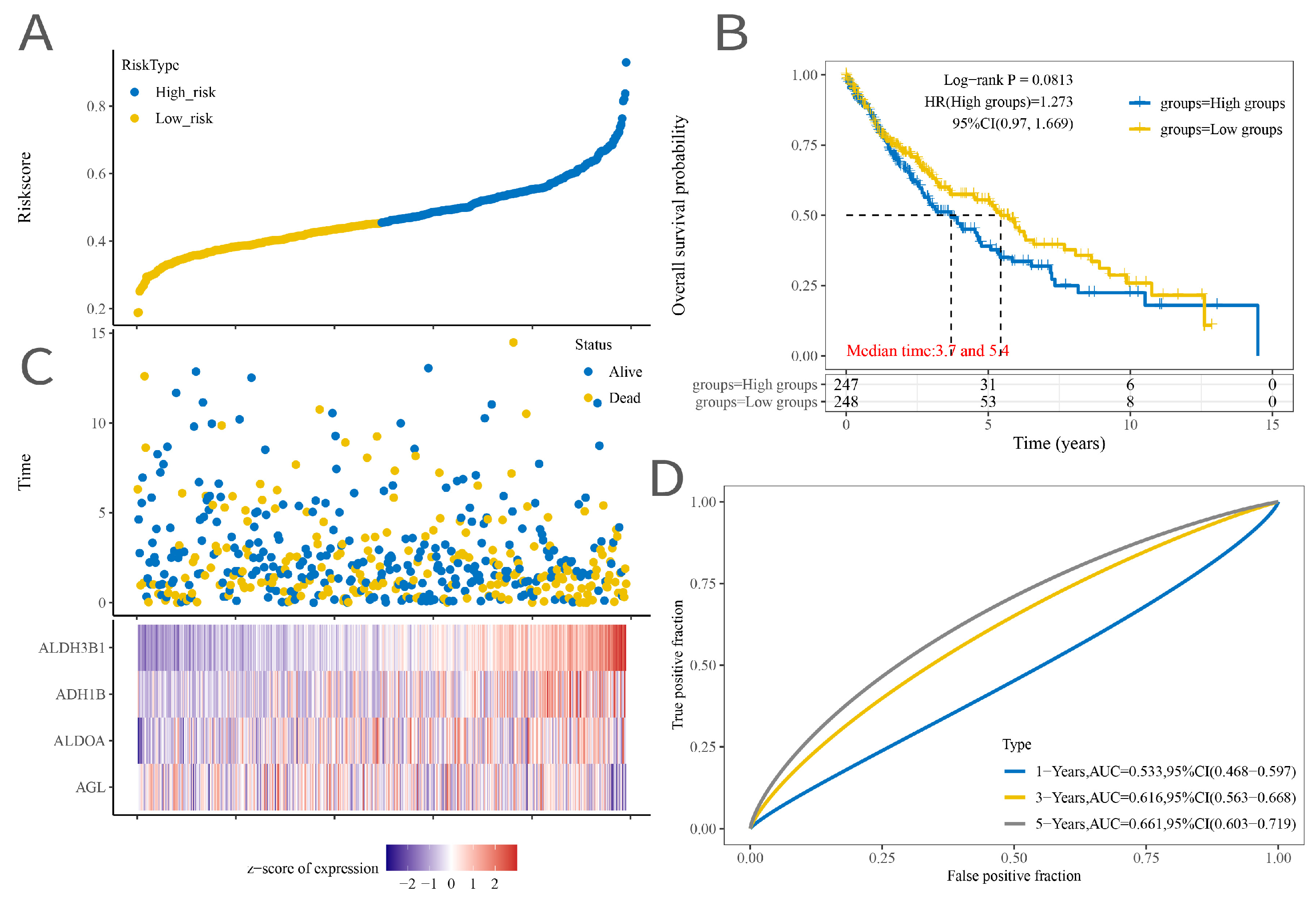
2.2. Signature Model Construction
2.3. Construction of a Nomogram Model
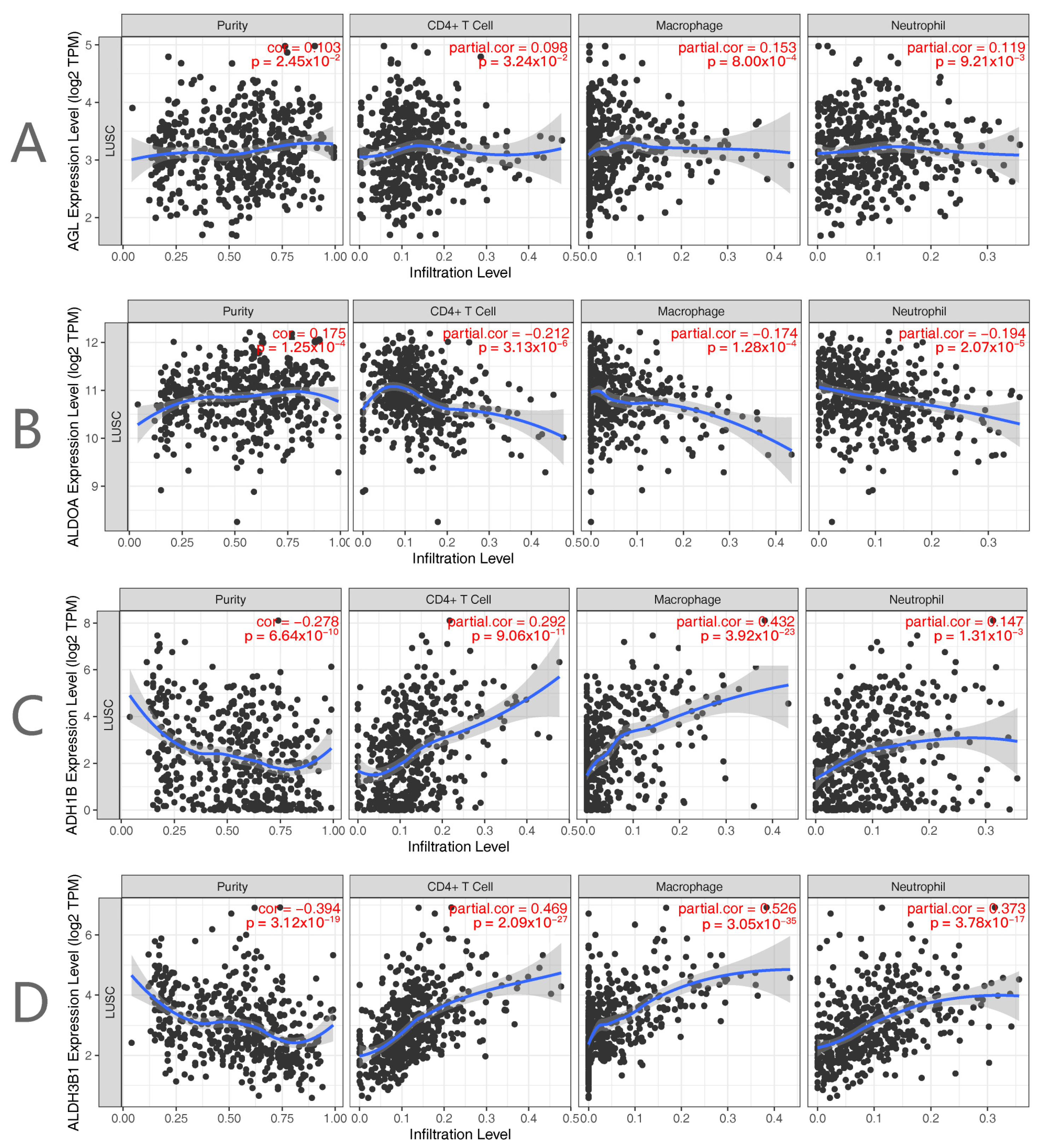
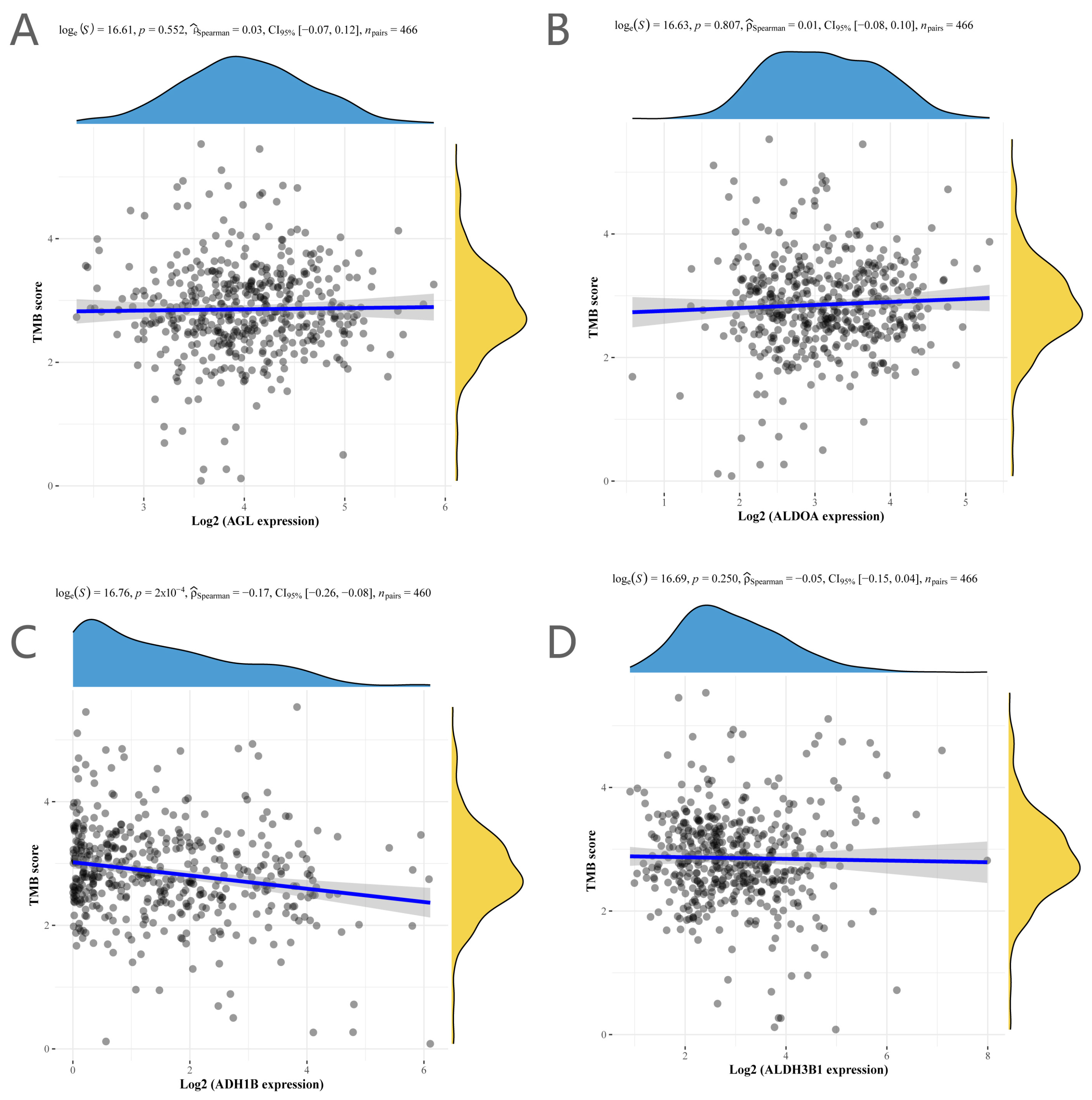
2.4. Immunological Profile and Tumor Mutational Load Analysis
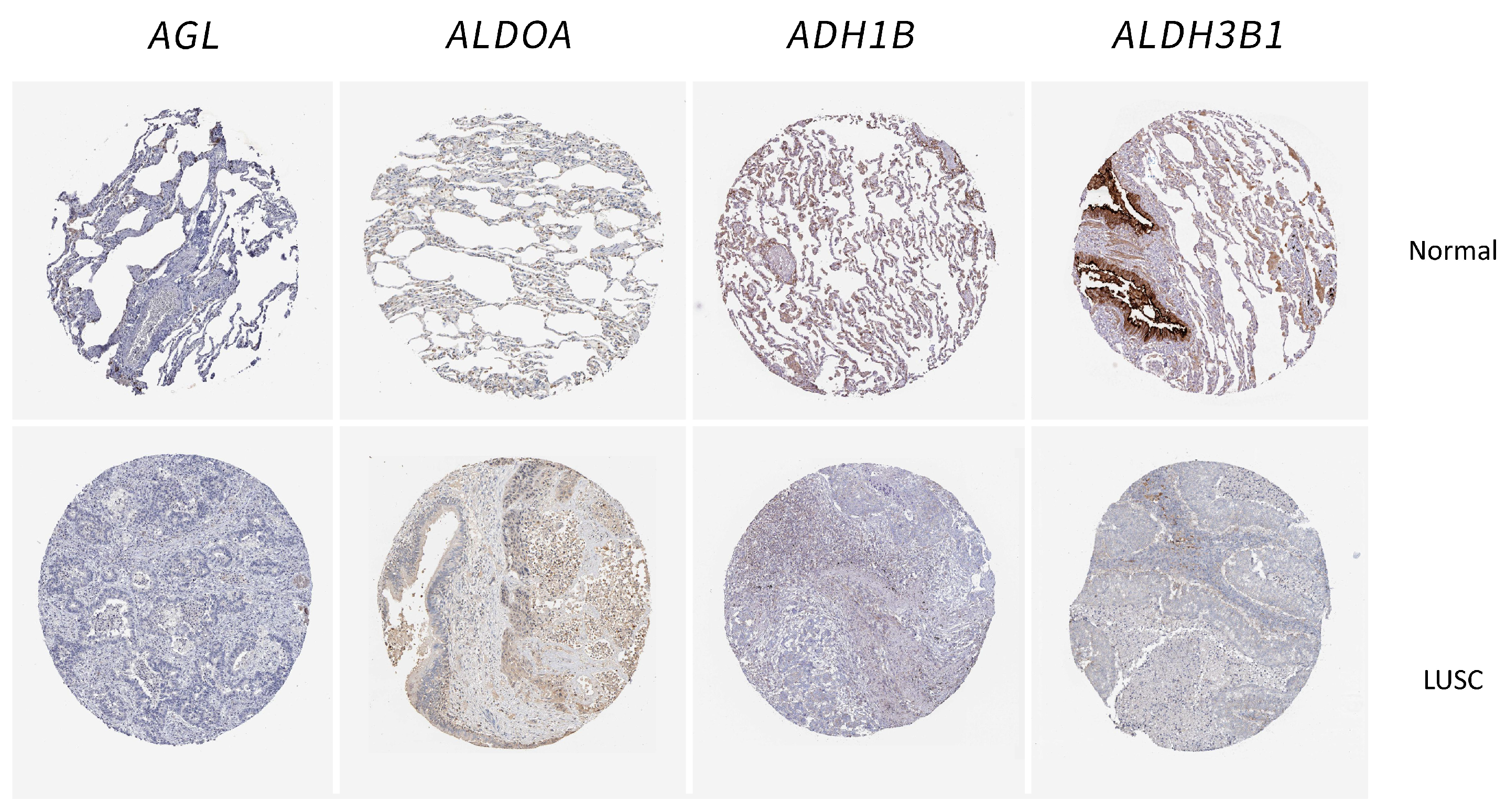
2.5. Validation of the Glycolysis-Related Genes in Tissue Samples
3. Materials and Methods
3.1. Data Acquisition
3.2. DEG Identification
3.3. Enrichment Analysis
3.4. Signature Model Construction
3.5. Nomogram Model Construction
3.6. Immune Infiltration and Tumor Mutational Load Analysis
3.7. Immunohistochemistry
4. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Travis, W.D.; Dacic, S.; Sholl, L.M.; Wistuba, I.I. Pathologic Assessment of Lung Squamous Cell Carcinoma After Neoadjuvant Immunotherapy. J. Thorac. Oncol. 2021, 16, e9–e10. [Google Scholar] [CrossRef]
- Conti, L.; Gatt, S. Squamous-Cell Carcinoma of the Lung. N. Engl. J. Med. 2018, 379, e17. [Google Scholar] [CrossRef] [PubMed]
- Kujtan, L.; Kancha, R.K.; Gustafson, B.; Douglass, L.; Ward, C.R.; Buzard, B.; Subramanian, J. Squamous cell carcinoma of the lung: Improving the detection and management of immune-related adverse events. Expert Rev. Anticancer Ther. 2022, 22, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Reinfeld, B.I.; Rathmell, W.K.; Kim, T.K.; Rathmell, J.C. The therapeutic implications of immunosuppressive tumor aerobic glycolysis. Cell. Mol. Immunol. 2022, 19, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Ghosh, S.; Kumar, S. Tumor glycolysis, an essential sweet tooth of tumor cells. Semin. Cancer Biol. 2022, 86 Pt 3, 1216–1230. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, M.; Xia, M.; Guo, Y.; Zu, X.; Zhong, J. Ubiquitination regulation of aerobic glycolysis in cancer. Life Sci. 2022, 292, 120322. [Google Scholar] [CrossRef]
- Li, Y.; Gu, J.; Xu, F.; Zhu, Q.; Ge, D.; Lu, C. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data. Sci. Rep. 2018, 8, 15834. [Google Scholar] [CrossRef]
- Yang, L.; Wei, S.; Zhang, J.; Hu, Q.; Hu, W.; Cao, M.; Zhang, L.; Wang, Y.; Wang, P.; Wang, K. Construction of a predictive model for immunotherapy efficacy in lung squamous cell carcinoma based on the degree of tumor-infiltrating immune cells and molecular typing. J. Transl. Med. 2022, 20, 364. [Google Scholar] [CrossRef] [PubMed]
- Kawai, H.; Saito, Y.; Demura, R.; Odaka, H.; Takahashi, S.; Takahashi, K.; Kurokawa, H.; Enomoto, K. Case of advanced pulmonary squamous cell carcinoma cured by resection through preoperative induction of immune checkpoint inhibitor. Thorac. Cancer 2018, 9, 495–497. [Google Scholar] [CrossRef]
- Qiu, Y.; Li, H.; Xie, J.; Qiao, X.; Wu, J. Identification of ABCC5 among ATP-Binding Cassette Transporter Family as a New Biomarker for Hepatocellular Carcinoma Based on Bioinformatics Analysis. Int. J. Gen. Med. 2021, 14, 7235–7246. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Chen, L.; Sun, Q.; Li, H.; Wei, W.; Wu, D.; Hu, Y.; Zhu, Z.; Shi, J.; Wang, M. An immune subtype-related prognostic signature of hepatocellular carcinoma based on single-cell sequencing analysis. Aging 2022, 14, 3276–3292. [Google Scholar] [CrossRef]
- Xie, J.; Li, H.; Chen, L.; Cao, Y.; Hu, Y.; Zhu, Z.; Wang, M.; Shi, J. A Novel Pyroptosis-Related lncRNA Signature for Predicting the Prognosis of Skin Cutaneous Melanoma. Int. J. Gen. Med. 2021, 14, 6517–6527. [Google Scholar] [CrossRef]
- Chang, Y.C.; Kim, C.H. Molecular Research of Glycolysis. Int. J. Mol. Sci. 2022, 23, 5052. [Google Scholar] [CrossRef]
- Chelakkot, C.; Chelakkot, V.S.; Shin, Y.; Song, K. Modulating Glycolysis to Improve Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 2606. [Google Scholar] [CrossRef]
- Guin, S.; Pollard, C.; Ru, Y.; Ritterson Lew, C.; Duex, J.E.; Dancik, G.; Owens, C.; Spencer, A.; Knight, S.; Holemon, H.; et al. Role in tumor growth of a glycogen debranching enzyme lost in glycogen storage disease. J. Natl. Cancer Inst. 2014, 106, dju062. [Google Scholar] [CrossRef] [PubMed]
- Guin, S.; Ru, Y.; Agarwal, N.; Lew, C.R.; Owens, C.; Comi, G.P.; Theodorescu, D. Loss of Glycogen Debranching Enzyme AGL Drives Bladder Tumor Growth via Induction of Hyaluronic Acid Synthesis. Clin. Cancer Res. 2016, 22, 1274–1283. [Google Scholar] [CrossRef] [PubMed]
- Richmond, C.S.; Oldenburg, D.; Dancik, G.; Meier, D.R.; Weinhaus, B.; Theodorescu, D.; Guin, S. Glycogen debranching enzyme (AGL) is a novel regulator of non-small cell lung cancer growth. Oncotarget 2018, 9, 16718–16730. [Google Scholar] [CrossRef]
- Chang, Y.C.; Chiou, J.; Yang, Y.F.; Su, C.Y.; Lin, Y.F.; Yang, C.N.; Lu, P.-J.; Huang, M.-S.; Yang, C.-J.; Hsiao, M. Therapeutic Targeting of Aldolase A Interactions Inhibits Lung Cancer Metastasis and Prolongs Survival. Cancer Res. 2019, 79, 4754–4766. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lin, J.D.; Zuo, X.Y.; Zhuang, Y.X.; Hong, C.Q.; Zhang, G.J.; Cui, X.-J.; Cui, Y.-K. Elevated transcriptional levels of aldolase A (ALDOA) associates with cell cycle-related genes in patients with NSCLC and several solid tumors. BioData Min. 2017, 10, 6. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, M.; Li, L.; Huang, Z. ALDH3B1 Is an Independent Prognostic Biomarker of Lung Adenocarcinoma. Technol. Cancer Res. Treat. 2020, 19, 1–9. [Google Scholar] [CrossRef]
- Lindahl, R. Aldehyde dehydrogenases and their role in carcinogenesis. Crit. Rev. Biochem. Mol. Biol. 1992, 27, 283–335. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadasah, S.F. Prognostic Significance of Glycolysis-Related Genes in Lung Squamous Cell Carcinoma. Int. J. Mol. Sci. 2024, 25, 1143. https://doi.org/10.3390/ijms25021143
Kadasah SF. Prognostic Significance of Glycolysis-Related Genes in Lung Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2024; 25(2):1143. https://doi.org/10.3390/ijms25021143
Chicago/Turabian StyleKadasah, Sultan F. 2024. "Prognostic Significance of Glycolysis-Related Genes in Lung Squamous Cell Carcinoma" International Journal of Molecular Sciences 25, no. 2: 1143. https://doi.org/10.3390/ijms25021143




