Transgenerational Transmission of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) Effects in Human Granulosa Cells: The Role of MicroRNAs
Abstract
1. Introduction
2. Results
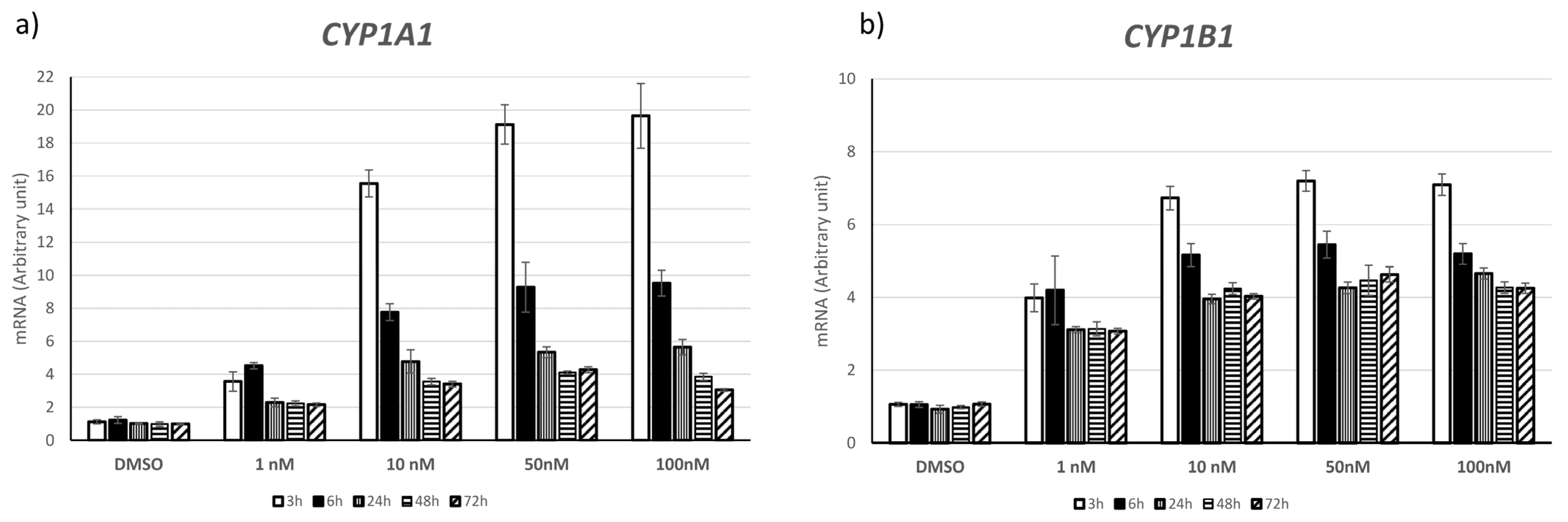
2.1. Choice of TCDD Concentration and Exposure Times
2.2. Characterization of the Morphology and Viability of Human Granulosa Cells
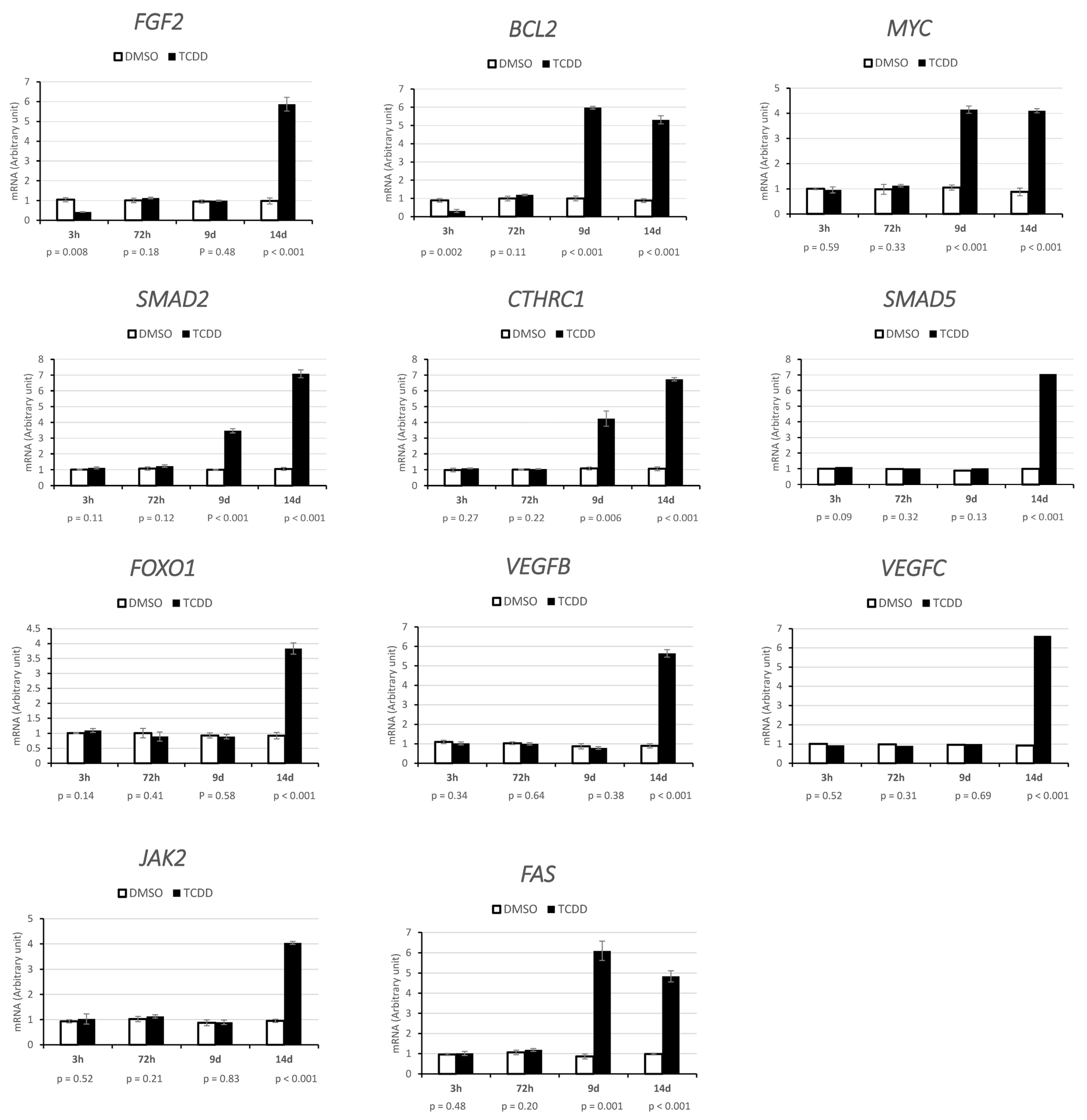
2.3. CYP1A1 and CYP1B1 Expression after Acute and Chronic TCDD Exposure and on Day 9 and 14 after Chronic Exposure
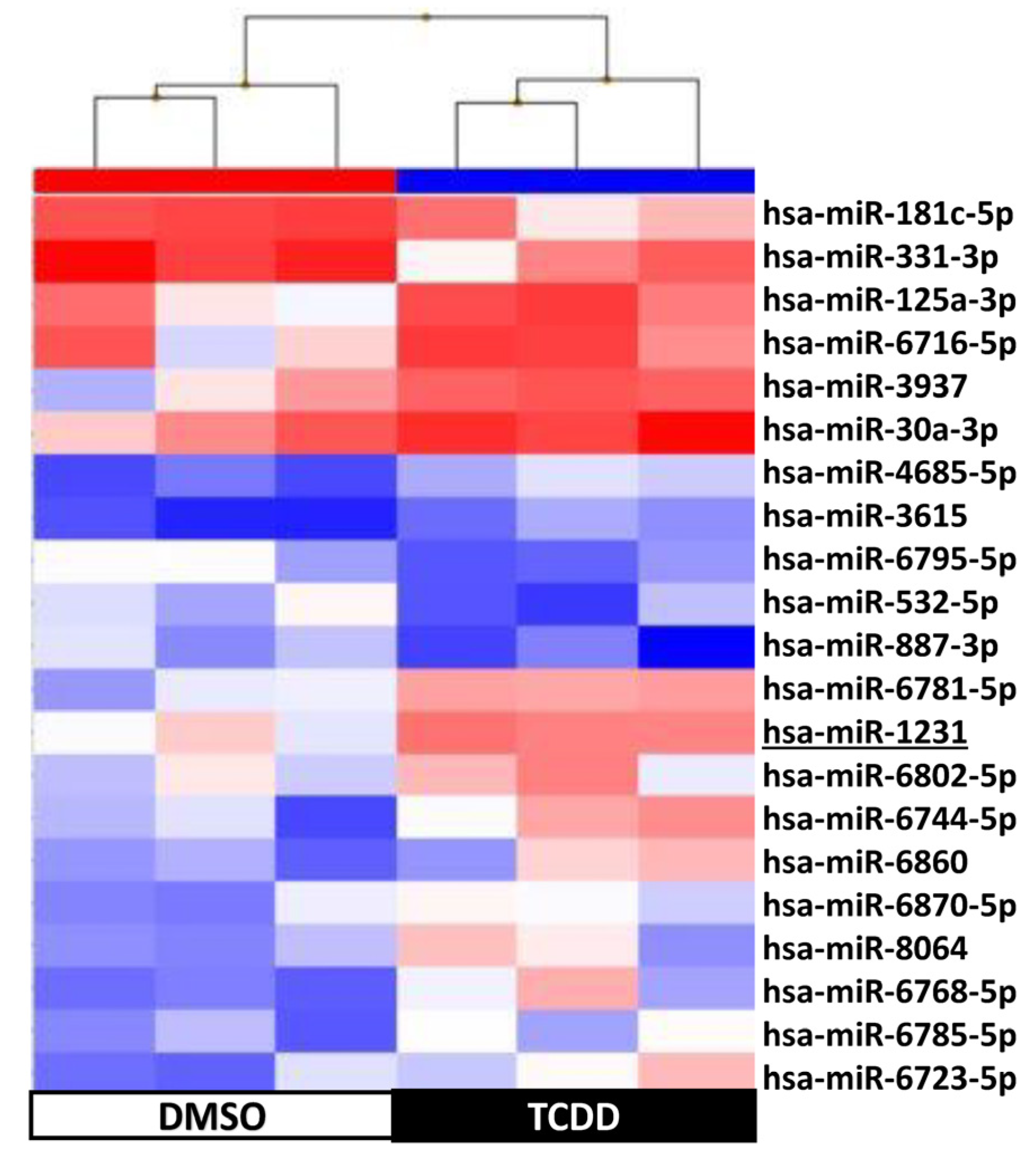
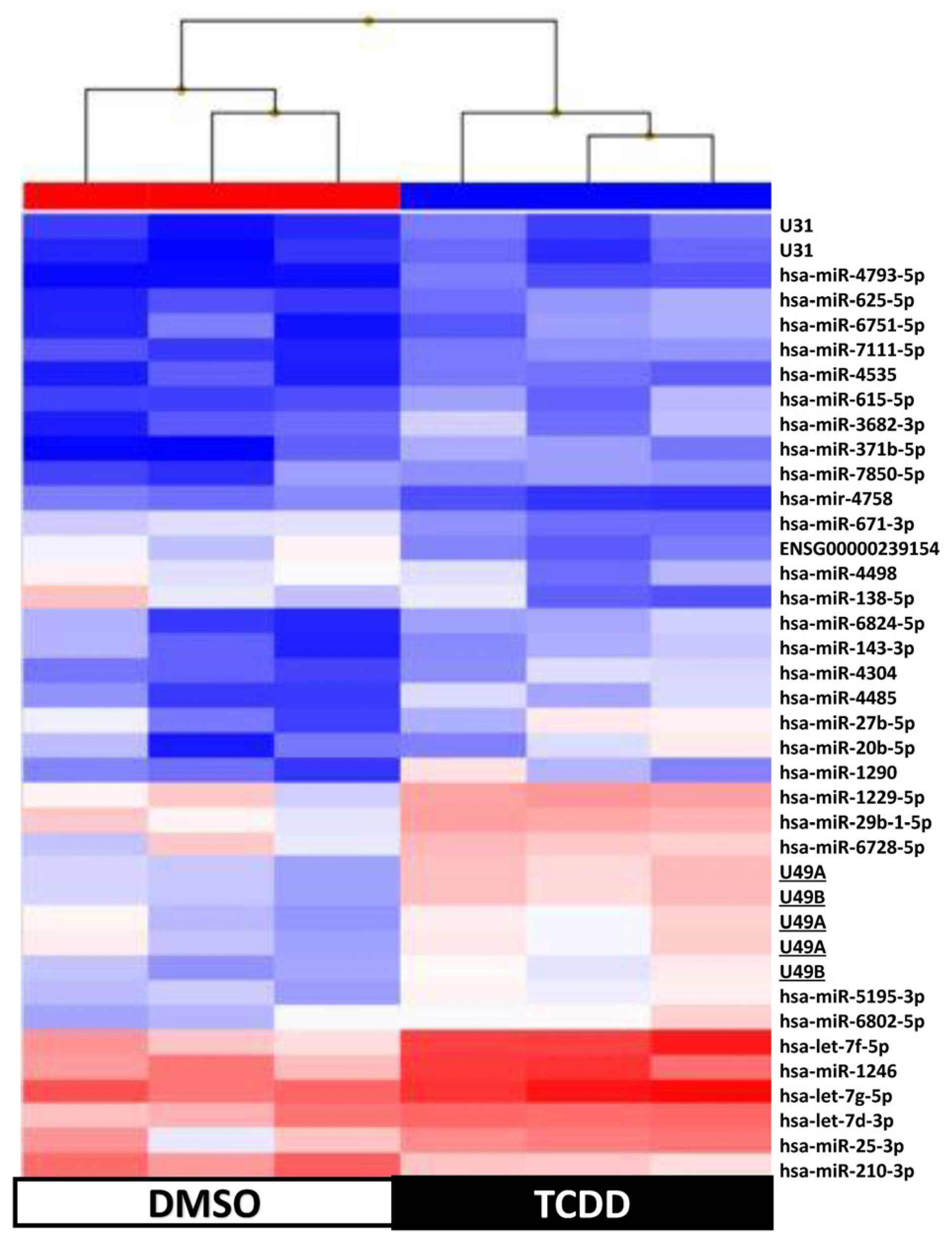
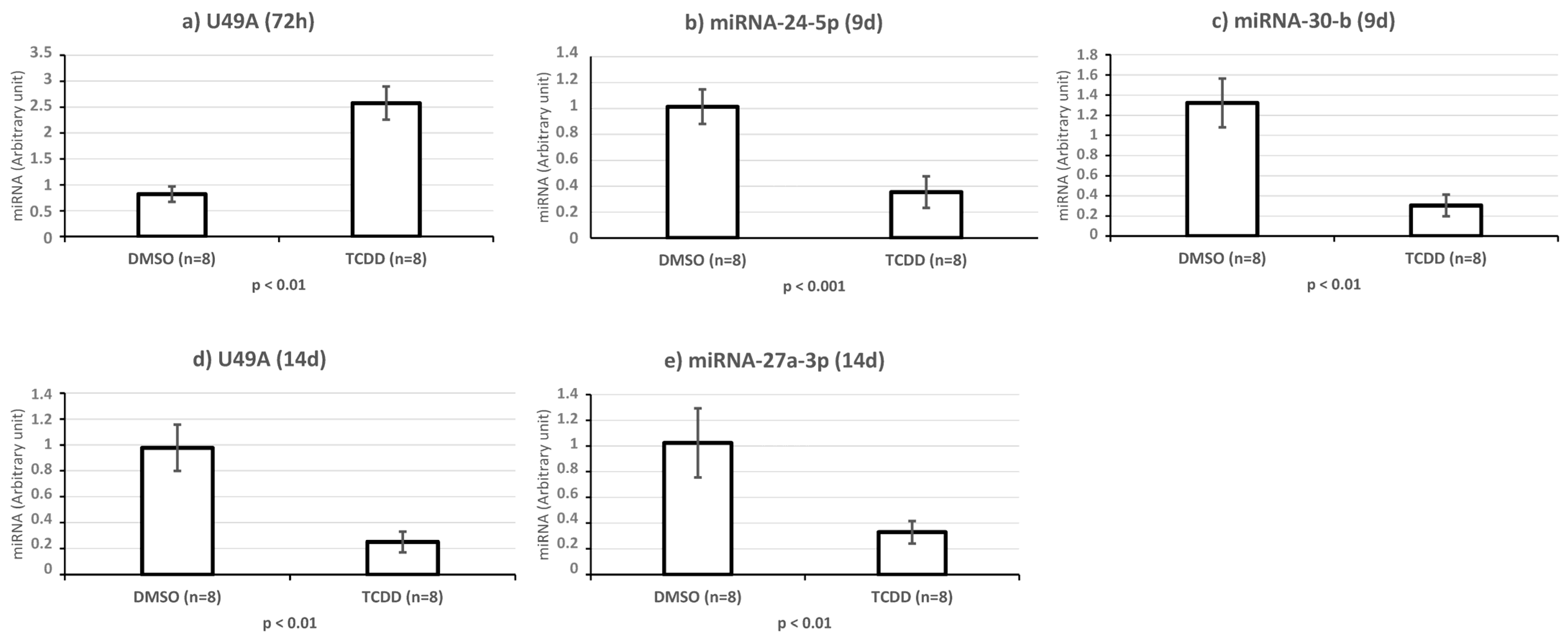
2.4. KGN Cell miRNome Profile in Function of the Exposure Status
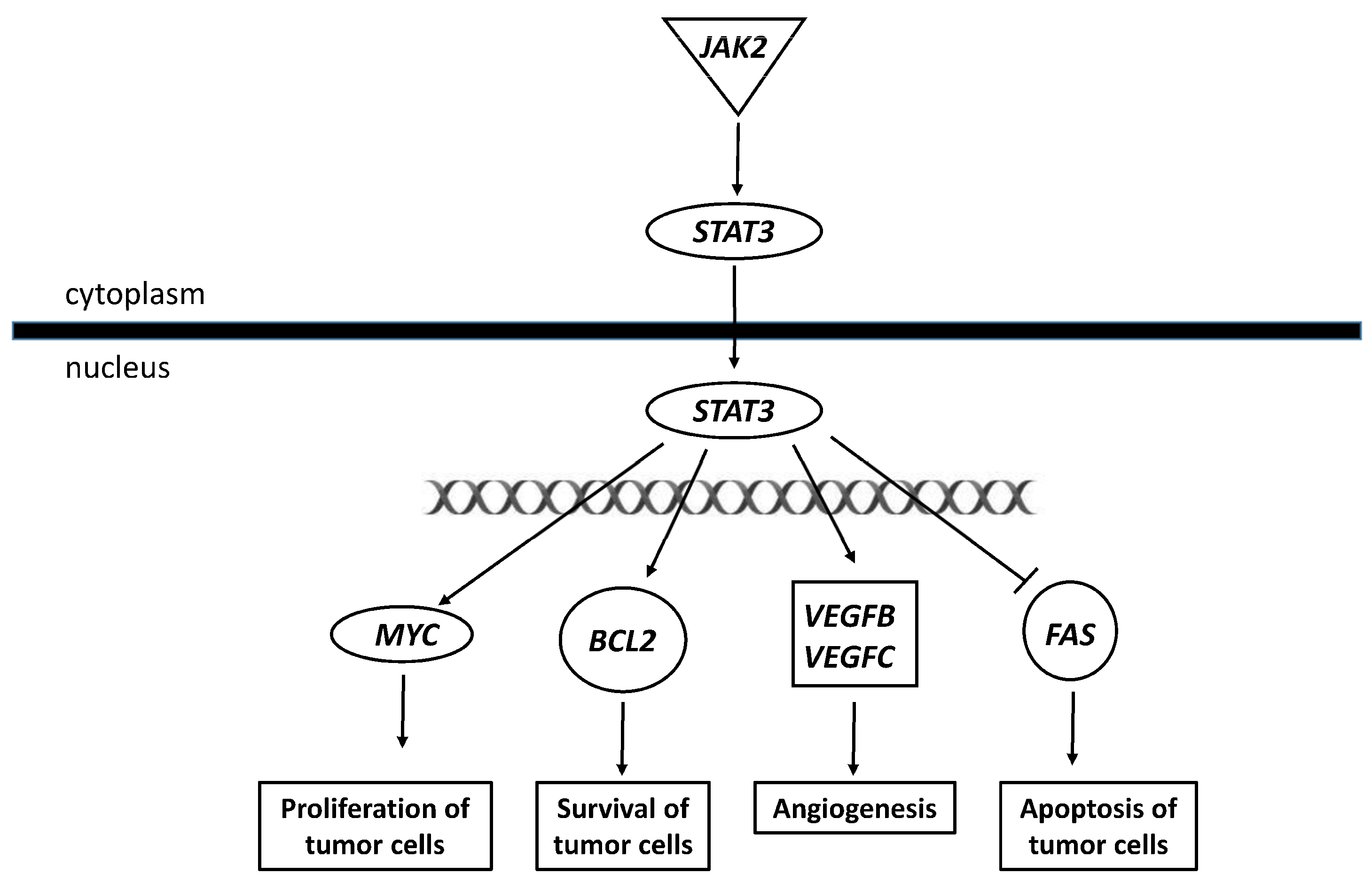
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Line
4.3. Exposure Schedules
4.4. Cell Counts
4.5. RNA Extraction
4.6. Quantitative RT-PCR
4.7. Microarray Hybridization and Data Analysis
4.8. TaqMan miRNA Assays
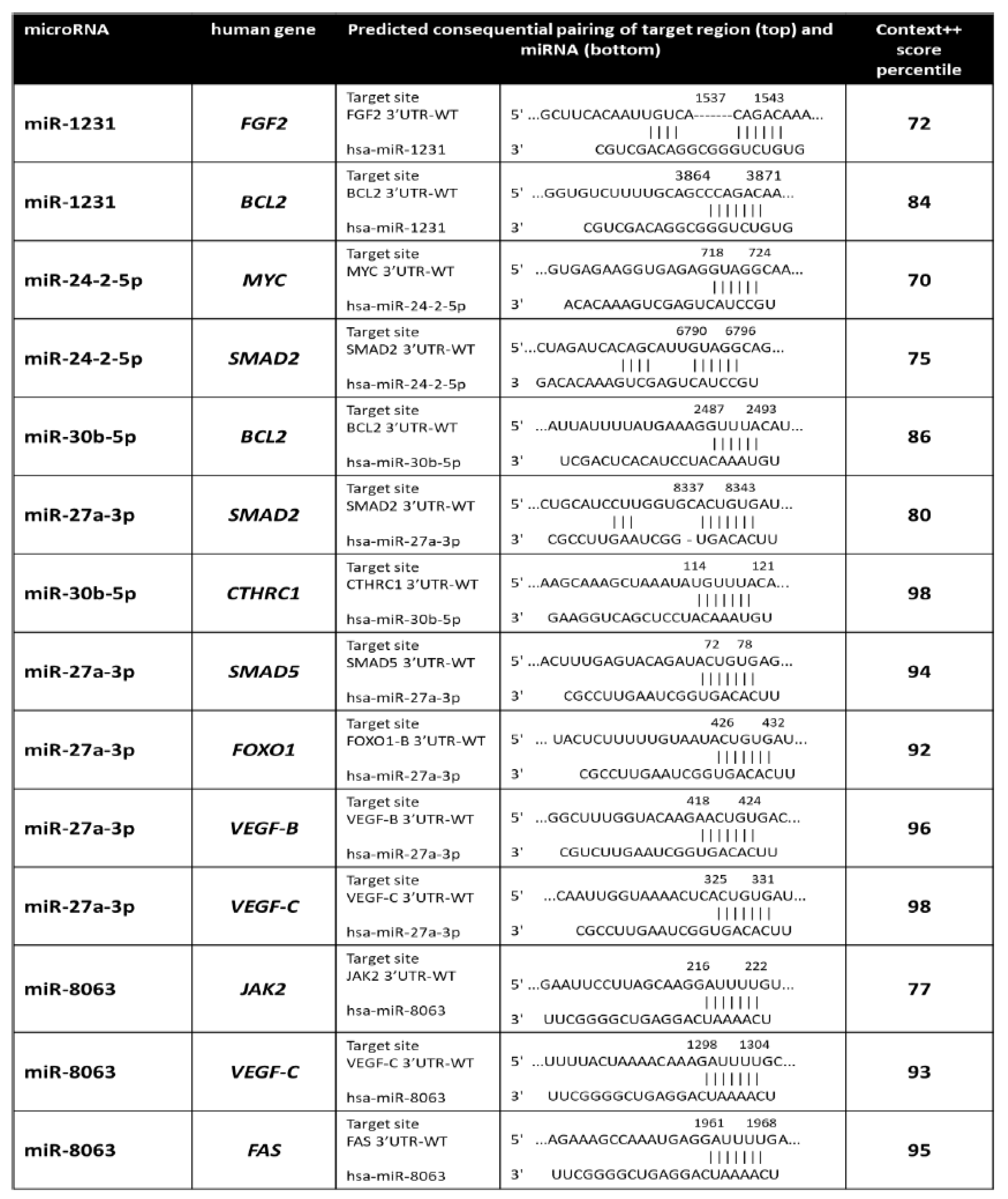
4.9. MiRNA Target Prediction
4.10. Expression of miRNA Target Genes
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scsukova, S.; Rollerova, E.; Bujnakova Mlynarcikova, A. Impact of endocrine disrupting chemicals on onset and development of female reproductive disorders and hormone-related cancer. Reprod. Biol. 2016, 16, 243–254. [Google Scholar] [CrossRef]
- Modica, R.; Benevento, E.; Colao, A. Endocrine-disrupting chemicals (EDCs) and cancer: New perspectives on an old relationship. J. Endocrinol. Investig. 2023, 46, 667–677. [Google Scholar] [CrossRef]
- Del Pup, L.; Mantovani, A.; Luce, A.; Cavaliere, C.; Facchini, G.; Di Francia, R.; Caraglia, M.; Berretta, M. Endocrine disruptors and female cancer: Informing the patients (Review). Oncol. Rep. 2015, 34, 3–11. [Google Scholar] [PubMed]
- Rogers, R.E.; Chai, S.; Pask, A.J.; Mattiske, D.M. Prenatal exposure to diethylstilbestrol has long-lasting, transgenerational impacts on fertility and reproductive development. Toxicol. Sci. 2023, 195, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Moran, F.M.; Conley, A.J.; Corbin, C.J.; Enan, E.; VandeVoort, C.; Overstreet, J.W.; Lasley, B.L. 2,3,7,8-tetrachlorodibenzo-p-dioxin decreases estradiol production without altering the enzyme activity of cytochrome P450 aromatase of human luteinized granulosa cells in vitro. Biol. Reprod. 2000, 62, 1102–1108. [Google Scholar] [CrossRef]
- Gaspari, L.; Paris, F.; Kalfa, N.; Soyer-Gobillard, M.O.; Sultan, C.; Hamamah, S. Experimental Evidence of 2,3,7,8-Tetrachlordibenzo-p-Dioxin (TCDD) Transgenerational Effects on Reproductive Health. Int. J. Mol. Sci. 2021, 22, 9091. [Google Scholar] [CrossRef]
- Orlowska, K.; Swigonska, S.; Sadowska, A.; Ruszkowska, M.; Nynca, A.; Molcan, T.; Zmijewska, A.; Ciereszko, R.E. Proteomic changes of aryl hydrocarbon receptor (AhR)-silenced porcine granulosa cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). PLoS ONE 2019, 14, e0223420. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.M.; Ohshima, K.; Watanabe, G.; Taya, K.; Strawn, E.Y.; Hutz, R.J. TCDD increases inhibin A production by human luteinized granulosa cells in vitro. J. Reprod. Dev. 2006, 52, 523–528. [Google Scholar] [CrossRef]
- Tai, P.T.; Nishijo, M.; Kido, T.; Nakagawa, H.; Maruzeni, S.; Naganuma, R.; Anh, N.T.; Morikawa, Y.; Luong, H.V.; Anh, T.H.; et al. Dioxin concentrations in breast milk of Vietnamese nursing mothers: A survey four decades after the herbicide spraying. Environ. Sci. Technol. 2011, 45, 6625–6632. [Google Scholar] [CrossRef]
- Weiss, J.; Papke, O.; Bignert, A.; Jensen, S.; Greyerz, E.; Agostoni, C.; Besana, R.; Riva, E.; Giovannini, M.; Zetterstrom, R. Concentrations of dioxins and other organochlorines (PCBs, DDTs, HCHs) in human milk from Seveso, Milan and a Lombardian rural area in Italy: A study performed 25 years after the heavy dioxin exposure in Seveso. Acta Paediatr. 2003, 92, 467–472. [Google Scholar] [CrossRef]
- Prokopec, S.D.; Viluksela, M.; Miettinen, H.M.; Boutros, P.C.; Pohjanvirta, R. Transgenerational epigenetic and transcriptomic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure in rat. Arch. Toxicol. 2020, 94, 1613–1624. [Google Scholar] [CrossRef]
- Eskenazi, B.; Mocarelli, P.; Warner, M.; Samuels, S.; Vercellini, P.; Olive, D.; Needham, L.L.; Patterson, D.G., Jr.; Brambilla, P.; Gavoni, N.; et al. Serum dioxin concentrations and endometriosis: A cohort study in Seveso, Italy. Environ. Health Perspect. 2002, 110, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Eskenazi, B.; Warner, M.; Mocarelli, P.; Samuels, S.; Needham, L.L.; Patterson, D.G., Jr.; Lippman, S.; Vercellini, P.; Gerthoux, P.M.; Brambilla, P.; et al. Serum dioxin concentrations and menstrual cycle characteristics. Am. J. Epidemiol. 2002, 156, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Eskenazi, B.; Warner, M.; Samuels, S.; Young, J.; Gerthoux, P.M.; Needham, L.; Patterson, D.; Olive, D.; Gavoni, N.; Vercellini, P.; et al. Serum dioxin concentrations and risk of uterine leiomyoma in the Seveso Women’s Health Study. Am. J. Epidemiol. 2007, 166, 79–87. [Google Scholar] [CrossRef]
- Eskenazi, B.; Warner, M.; Marks, A.R.; Samuels, S.; Needham, L.; Brambilla, P.; Mocarelli, P. Serum dioxin concentrations and time to pregnancy. Epidemiology 2010, 21, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Eskenazi, B.; Warner, M.; Brambilla, P.; Signorini, S.; Ames, J.; Mocarelli, P. The Seveso accident: A look at 40years of health research and beyond. Environ. Int. 2018, 121 Pt 1, 71–84. [Google Scholar] [CrossRef]
- Eskenazi, B.; Ames, J.; Rauch, S.; Signorini, S.; Brambilla, P.; Mocarelli, P.; Siracusa, C.; Holland, N.; Warner, M. Dioxin exposure associated with fecundability and infertility in mothers and daughters of Seveso, Italy. Hum. Reprod. 2021, 36, 794–807. [Google Scholar] [CrossRef]
- Le, T.N.; Johansson, A. Impact of chemical warfare with agent orange on women’s reproductive lives in Vietnam: A pilot study. Reprod. Health Matters 2001, 9, 156–164. [Google Scholar]
- Chaffin, C.L.; Peterson, R.E.; Hutz, R.J. In utero and lactational exposure of female Holtzman rats to 2,3,7,8-tetrachlorodibenzo-p-dioxin: Modulation of the estrogen signal. Biol. Reprod. 1996, 55, 62–67. [Google Scholar] [CrossRef]
- Hirakawa, T.; Minegishi, T.; Abe, K.; Kishi, H.; Inoue, K.; Ibuki, Y.; Miyamoto, K. Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of follicle-stimulating hormone receptors during cell differentiation in cultured granulosa cells. Endocrinology 2000, 141, 1470–1476. [Google Scholar] [CrossRef]
- Chaffin, C.L.; Brogan, R.S.; Peterson, R.E.; Hutz, R.J.; Wehrenberg, W.B. Modulation of growth axis gene expression by in utero and lactational exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in the weaning Holtzman rat. Endocrine 1996, 5, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Chaffin, C.L.; Heimler, I.; Rawlins, R.G.; Wimpee, B.A.; Sommer, C.; Hutz, R.J. Estrogen receptor and aromatic hydrocarbon receptor in the primate ovary. Endocrine 1996, 5, 315–321. [Google Scholar] [CrossRef]
- Ruszkowska, M.; Sadowska, A.; Nynca, A.; Orlowska, K.; Swigonska, S.; Molcan, T.; Paukszto, L.; Jastrzebski, J.P.; Ciereszko, R.E. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on the transcriptome of aryl hydrocarbon receptor (AhR) knock-down porcine granulosa cells. PeerJ 2020, 8, e8371. [Google Scholar] [CrossRef]
- Pieklo, R.; Grochowalski, A.; Gregoraszczuk, E.L. 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters follicular steroidogenesis in time- and cell-specific manner. Exp. Clin. Endocrinol. Diabetes 2000, 108, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Grochowalski, A.; Chrzaszcz, R.; Pieklo, R.; Gregoraszczuk, E.L. Estrogenic and antiestrogenic effect of in vitro treatment of follicular cells with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Chemosphere 2001, 43, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Jablonska, O.; Piasecka, J.; Ostrowska, M.; Sobocinska, N.; Wasowska, B.; Ciereszko, R.E. The expression of the aryl hydrocarbon receptor in reproductive and neuroendocrine tissues during the estrous cycle in the pig. Anim. Reprod. Sci. 2011, 126, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Sadowska, A.; Nynca, A.; Ruszkowska, M.; Paukszto, L.; Myszczynski, K.; Orlowska, K.; Swigonska, S.; Molcan, T.; Jastrzebski, J.P.; Ciereszko, R.E. Transcriptional profiling of porcine granulosa cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Chemosphere 2017, 178, 368–377. [Google Scholar] [CrossRef]
- Ruszkowska, M.; Nynca, A.; Paukszto, L.; Sadowska, A.; Swigonska, S.; Orlowska, K.; Molcan, T.; Jastrzebski, J.P.; Ciereszko, R.E. Identification and characterization of long non-coding RNAs in porcine granulosa cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Anim. Sci. Biotechnol. 2018, 9, 72. [Google Scholar] [CrossRef]
- Orlowska, K.; Swigonska, S.; Sadowska, A.; Ruszkowska, M.; Nynca, A.; Molcan, T.; Ciereszko, R.E. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the proteome of porcine granulosa cells. Chemosphere 2018, 212, 170–181. [Google Scholar] [CrossRef]
- Katic, J.; Cemeli, E.; Baumgartner, A.; Laubenthal, J.; Bassano, I.; Stolevik, S.B.; Granum, B.; Namork, E.; Nygaard, U.C.; Lovik, M.; et al. Evaluation of the genotoxicity of 10 selected dietary/environmental compounds with the in vitro micronucleus cytokinesis-block assay in an interlaboratory comparison. Food Chem. Toxicol. 2010, 48, 2612–2623. [Google Scholar] [CrossRef]
- Korkalainen, M.; Huumonen, K.; Naarala, J.; Viluksela, M.; Juutilainen, J. Dioxin induces genomic instability in mouse embryonic fibroblasts. PLoS ONE 2012, 7, e37895. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huumonen, K.; Korkalainen, M.; Viluksela, M.; Lahtinen, T.; Naarala, J.; Juutilainen, J. Role of microRNAs and DNA Methyltransferases in Transmitting Induced Genomic Instability between Cell Generations. Front. Public. Health 2014, 2, 139. [Google Scholar] [CrossRef]
- Bunkar, N.; Pathak, N.; Lohiya, N.K.; Mishra, P.K. Epigenetics: A key paradigm in reproductive health. Clin. Exp. Reprod. Med. 2016, 43, 59–81. [Google Scholar] [CrossRef] [PubMed]
- Szymanowska, A.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Amero, P. Non-Coding RNAs: Foes or Friends for Targeting Tumor Microenvironment. Non-Coding RNA 2023, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Enan, E.; Moran, F.; VandeVoort, C.A.; Stewart, D.R.; Overstreet, J.W.; Lasley, B.L. Mechanism of toxic action of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in cultured human luteinized granulosa cells. Reprod. Toxicol. 1996, 10, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Enan, E.; Lasley, B.; Stewart, D.; Overstreet, J.; Vandevoort, C.A. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) modulates function of human luteinizing granulosa cells via cAMP signaling and early reduction of glucose transporting activity. Reprod. Toxicol. 1996, 10, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Heimler, I.; Rawlins, R.G.; Owen, H.; Hutz, R.J. Dioxin perturbs, in a dose- and time-dependent fashion, steroid secretion, and induces apoptosis of human luteinized granulosa cells. Endocrinology 1998, 139, 4373–4379. [Google Scholar] [CrossRef]
- Son, D.S.; Ushinohama, K.; Gao, X.; Taylor, C.C.; Roby, K.F.; Rozman, K.K.; Terranova, P.F. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) blocks ovulation by a direct action on the ovary without alteration of ovarian steroidogenesis: Lack of a direct effect on ovarian granulosa and thecal-interstitial cell steroidogenesis in vitro. Reprod. Toxicol. 1999, 13, 521–530. [Google Scholar] [CrossRef]
- Dasmahapatra, A.K.; Wimpee, B.A.; Trewin, A.L.; Wimpee, C.F.; Ghorai, J.K.; Hutz, R.J. Demonstration of 2,3,7,8-tetrachlorodibenzo-p-dioxin attenuation of P450 steroidogenic enzyme mRNAs in rat granulosa cell in vitro by competitive reverse transcriptase-polymerase chain reaction assay. Mol. Cell Endocrinol. 2000, 164, 5–18. [Google Scholar] [CrossRef]
- Hirakawa, T.; Minegishi, T.; Abe, K.; Kishi, H.; Ibuki, Y.; Miyamoto, K. Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of luteinizing hormone receptors during cell differentiation in cultured granulosa cells. Arch. Biochem. Biophys. 2000, 375, 371–376. [Google Scholar] [CrossRef]
- Gregoraszczuk, E.L. Dioxin exposure and porcine reproductive hormonal activity. Cad. Saude Publica 2002, 18, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Moran, F.M.; Lohstroh, P.; VandeVoort, C.A.; Chen, J.; Overstreet, J.W.; Conley, A.J.; Lasley, B.L. Exogenous steroid substrate modifies the effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on estradiol production of human luteinized granulosa cells in vitro. Biol. Reprod. 2003, 68, 244–251. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wojtowicz, A.; Tomanek, M.; Augustowska, K.; Gregoraszczuk, E.L. Aromatic hydrocarbon receptor (AhR) in the porcine theca and granulosa cells: Effect of TCDD, PCB 126 and PCB 153 on the expression of AhR. Endocr. Regul. 2005, 39, 109–118. [Google Scholar] [PubMed]
- Vidal, J.D.; Vandevoort, C.A.; Marcus, C.B.; Lazarewicz, N.R.; Conley, A.J. 2,3,7,8-tetrachlorodibenzo-p-dioxin induces CYP1B1 expression in human luteinized granulosa cells. Arch. Biochem. Biophys. 2005, 439, 53–60. [Google Scholar] [CrossRef]
- Horling, K.; Santos, A.N.; Fischer, B. The AhR is constitutively activated and affects granulosa cell features in the human cell line KGN. Mol. Hum. Reprod. 2011, 17, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Swedenborg, E.; Kotka, M.; Seifert, M.; Kanno, J.; Pongratz, I.; Ruegg, J. The aryl hydrocarbon receptor ligands 2,3,7,8-tetrachlorodibenzo-p-dioxin and 3-methylcholanthrene regulate distinct genetic networks. Mol. Cell Endocrinol. 2012, 362, 39–47. [Google Scholar] [CrossRef]
- Ernst, J.; Jann, J.C.; Biemann, R.; Koch, H.M.; Fischer, B. Effects of the environmental contaminants DEHP and TCDD on estradiol synthesis and aryl hydrocarbon receptor and peroxisome proliferator-activated receptor signalling in the human granulosa cell line KGN. Mol. Hum. Reprod. 2014, 20, 919–928. [Google Scholar] [CrossRef]
- Jablonska, O.; Piasecka-Srader, J.; Nynca, A.; Kolomycka, A.; Robak, A.; Wasowska, B.; Ciereszko, R.E. 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters steroid secretion but does not affect cell viability and the incidence of apoptosis in porcine luteinised granulosa cells. Acta Vet. Hung. 2014, 62, 408–421. [Google Scholar] [CrossRef]
- Piasecka-Srader, J.; Kolomycka, A.; Nynca, A.; Ciereszko, R.E. Effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin and phytoestrogen genistein on the activity and the presence of steroidogenic enzyme proteins in cultured granulosa cells of pigs. Anim. Reprod. Sci. 2014, 148, 171–181. [Google Scholar] [CrossRef]
- Piasecka-Srader, J.; Sadowska, A.; Nynca, A.; Orlowska, K.; Jablonska, M.; Jablonska, O.; Petroff, B.K.; Ciereszko, R.E. The combined effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin and the phytoestrogen genistein on steroid hormone secretion, AhR and ERbeta expression and the incidence of apoptosis in granulosa cells of medium porcine follicles. J. Reprod. Dev. 2016, 62, 103–113. [Google Scholar] [CrossRef][Green Version]
- Das, D.N.; Panda, P.K.; Sinha, N.; Mukhopadhyay, S.; Naik, P.P.; Bhutia, S.K. DNA damage by 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced p53-mediated apoptosis through activation of cytochrome P450/aryl hydrocarbon receptor. Environ. Toxicol. Pharmacol. 2017, 55, 175–185. [Google Scholar] [CrossRef]
- Nynca, A.; Sadowska, A.; Paukszto, L.; Molcan, T.; Ruszkowska, M.; Swigonska, S.; Orlowska, K.; Myszczynski, K.; Jastrzebski, J.P.; Ciereszko, R.E. Temporal changes in the transcriptomic profile of granulosa cells of pigs treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Anim. Reprod. Sci. 2019, 207, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Molcan, T.; Swigonska, S.; Nynca, A.; Sadowska, A.; Ruszkowska, M.; Orlowska, K.; Ciereszko, R.E. Is CYP1B1 involved in the metabolism of dioxins in the pig? Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Rattan, S.; Flaws, J.A. The epigenetic impacts of endocrine disruptors on female reproduction across generationsdagger. Biol. Reprod. 2019, 101, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Montjean, D.; Neyroud, A.S.; Yefimova, M.G.; Benkhalifa, M.; Cabry, R.; Ravel, C. Impact of Endocrine Disruptors upon Non-Genetic Inheritance. Int. J. Mol. Sci. 2022, 23, 3350. [Google Scholar] [CrossRef]
- Patino-Garcia, D.; Cruz-Fernandes, L.; Bunay, J.; Orellana, R.; Moreno, R.D. Daily exposure to phthalates and alkylphenols alters miR biogenesis and expression in mice ovaries. J. Mol. Endocrinol. 2020, 65, 175–186. [Google Scholar] [CrossRef]
- Xie, J.R.; Jiang, Y.Y.; Xu, W.; Tao, J.Z. MiR-1231 correlates tumor prognosis and inhibits cell growth in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8308–8313. [Google Scholar]
- Wang, R.; Wang, L.; Wang, L.; Cui, Z.; Cheng, F.; Wang, W.; Yang, X. FGF2 Is Protective Towards Cisplatin-Induced KGN Cell Toxicity by Promoting FTO Expression and Autophagy. Front Endocrinol. 2022, 13, 890623. [Google Scholar] [CrossRef]
- Liu, P.; Zhao, L.; Zitvogel, L.; Kepp, O.; Kroemer, G. The BCL2 inhibitor venetoclax mediates anticancer effects through dendritic cell activation. Cell Death Differ. 2023, 30, 2447–2451. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, Q.; Guo, S.; Zhang, Q.; Tang, J.; Liu, G.; Kong, D.; Li, J.; Yan, S.; Wang, R.; et al. 2, 3, 7, 8-Tetrachlorodibenzo-p-dioxin promotes endothelial cell apoptosis through activation of EP3/p38MAPK/Bcl-2 pathway. J. Cell Mol. Med. 2017, 21, 3540–3551. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Kong, W.; Zheng, S.; Yu, C.; Yu, Y.; Xu, Y.; Ye, L.; Shao, Y. Prognostic significance of microRNA miR-24 in cancers: A meta-analysis. Bioengineered 2021, 12, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, N.; Tang, Q.; Sheng, H.; Long, S.; Wu, W. MicroRNA-24 in Cancer: A Double Side Medal With Opposite Properties. Front. Oncol. 2020, 10, 553714. [Google Scholar] [CrossRef]
- Zeng, M.; Kwiatkowski, N.P.; Zhang, T.; Nabet, B.; Xu, M.; Liang, Y.; Quan, C.; Wang, J.; Hao, M.; Palakurthi, S.; et al. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. eLife 2018, 7, e39030. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, C.; Ju, Z.; Jiao, D.; Hu, D.; Qi, L.; Liu, S.; Wu, X.; Zhao, C. Kruppel-like factors in tumors: Key regulators and therapeutic avenues. Front. Oncol. 2023, 13, 1080720. [Google Scholar] [CrossRef]
- Cherukunnath, A.; Davargaon, R.S.; Ashraf, R.; Kamdar, U.; Srivastava, A.K.; Tripathi, P.P.; Chatterjee, N.; Kumar, S. KLF8 is activated by TGF-beta1 via Smad2 and contributes to ovarian cancer progression. J. Cell Biochem. 2022, 123, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, J.; Wang, J.; Chen, X.; Zhu, Y.; Chen, Y. Mir-30b-3p affects the migration and invasion function of ovarian cancer cells by targeting the CTHRC1 gene. Biol. Res. 2020, 53, 10. [Google Scholar] [CrossRef]
- Hou, M.; Cheng, Z.; Shen, H.; He, S.; Li, Y.; Pan, Y.; Feng, C.; Chen, X.; Zhang, Y.; Lin, M.; et al. High expression of CTHRC1 promotes EMT of epithelial ovarian cancer (EOC) and is associated with poor prognosis. Oncotarget 2015, 6, 35813–35829. [Google Scholar] [CrossRef]
- Mei, D.; Zhu, Y.; Zhang, L.; Wei, W. The Role of CTHRC1 in Regulation of Multiple Signaling and Tumor Progression and Metastasis. Mediators Inflamm. 2020, 2020, 9578701. [Google Scholar] [CrossRef]
- Li, X.; Xu, M.; Ding, L.; Tang, J. MiR-27a: A Novel Biomarker and Potential Therapeutic Target in Tumors. J. Cancer 2019, 10, 2836–2848. [Google Scholar] [CrossRef]
- Wang, Z.; Ji, G.; Wu, Q.; Feng, S.; Zhao, Y.; Cao, Z.; Tao, C. Integrated microarray meta-analysis identifies miRNA-27a as an oncogene in ovarian cancer by inhibiting FOXO1. Life Sci. 2018, 210, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xiang, J.; Shen, J.; Zou, X.; Zhai, S.; Yin, Y.; Li, P.; Wang, X.; Sun, Q. Oncogenic MicroRNA-27a is a target for genistein in ovarian cancer cells. Anticancer Agents Med. Chem. 2013, 13, 1126–1132. [Google Scholar] [CrossRef]
- Nie, M.; Yu, S.; Peng, S.; Fang, Y.; Wang, H.; Yang, X. miR-23a and miR-27a promote human granulosa cell apoptosis by targeting SMAD5. Biol. Reprod. 2015, 93, 98. [Google Scholar] [CrossRef]
- Weis-Banke, S.E.; Lerdrup, M.; Kleine-Kohlbrecher, D.; Mohammad, F.; Sidoli, S.; Jensen, O.N.; Yanase, T.; Nakamura, T.; Iwase, A.; Stylianou, A.; et al. Mutant FOXL2(C134W) Hijacks SMAD4 and SMAD2/3 to Drive Adult Granulosa Cell Tumors. Cancer Res. 2020, 80, 3466–3479. [Google Scholar] [CrossRef] [PubMed]
- Sopo, M.; Anttila, M.; Hamalainen, K.; Kivela, A.; Yla-Herttuala, S.; Kosma, V.M.; Keski-Nisula, L.; Sallinen, H. Expression profiles of VEGF-A, VEGF-D and VEGFR1 are higher in distant metastases than in matched primary high grade epithelial ovarian cancer. BMC Cancer 2019, 19, 584. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, L.; Geng, J.; Liu, L.; Xu, L. β-elemene inhibits oxygen-induced retinal neovascularization via promoting miR-27a and reducing VEGF expression. Mol. Med. Rep. 2019, 19, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Q.; Yuan, T.; Jiang, H.; Yang, Y.Y.; Wang, L.; Fu, R.M.; Luo, S.Q.; Zhang, T.; Wu, Z.Y.; Wen, K.M. MicroRNA-8063 targets heterogeneous nuclear ribonucleoprotein AB to inhibit the self-renewal of colorectal cancer stem cells via the Wnt/beta-catenin pathway. Oncol. Rep. 2021, 46, 219. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Miyamoto, M.; Aoyama, T.; Soyama, H.; Goto, T.; Hirata, J.; Suzuki, A.; Nagaoka, I.; Tsuda, H.; Furuya, K.; et al. JAK2/STAT3 pathway as a therapeutic target in ovarian cancers. Oncol. Lett. 2018, 15, 5772–5780. [Google Scholar] [CrossRef]
- Trisdale, S.K.; Schwab, N.M.; Hou, X.; Davis, J.S.; Townson, D.H. Molecular manipulation of keratin 8/18 intermediate filaments: Modulators of FAS-mediated death signaling in human ovarian granulosa tumor cells. J. Ovarian Res. 2016, 9, 8. [Google Scholar] [CrossRef]
- Valleron, W.; Laprevotte, E.; Gautier, E.F.; Quelen, C.; Demur, C.; Delabesse, E.; Agirre, X.; Prosper, F.; Kiss, T.; Brousset, P. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia 2012, 26, 2052–2060. [Google Scholar] [CrossRef]
- Zhou, F.; Liu, Y.; Rohde, C.; Pauli, C.; Gerloff, D.; Kohn, M.; Misiak, D.; Baumer, N.; Cui, C.; Gollner, S.; et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 2017, 19, 844–855. [Google Scholar] [CrossRef]
- Lin, L.M.; Pan, Q.; Sun, Y.M.; Wang, W.T. Small nucleolar RNA is potential as a novel player in leukemogenesis and clinical application. Blood Sci. 2021, 3, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Sun, Y.M.; Pan, Q.; Fang, K.; Chen, X.T.; Zeng, Z.C.; Chen, T.Q.; Zhu, S.X.; Huang, L.B.; Luo, X.Q.; et al. The snoRNA-like lncRNA LNC-SNO49AB drives leukemia by activating the RNA-editing enzyme ADAR1. Cell Discov. 2022, 8, 117. [Google Scholar] [CrossRef]
- Devlin, R.B.; Frampton, M.L.; Ghio, A.J. In vitro studies: What is their role in toxicology? Exp. Toxicol. Pathol. 2005, 57 (Suppl. S1), 183–188. [Google Scholar] [CrossRef] [PubMed]
- Fitz-James, M.H.; Cavalli, G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022, 23, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Burgio, E.; Piscitelli, P.; Colao, A. Environmental Carcinogenesis and Transgenerational Transmission of Carcinogenic Risk: From Genetics to Epigenetics. Int. J. Environ. Res. Public Health 2018, 15, 1791. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Nishi, Y.; Yanase, T.; Mu, Y.; Oba, K.; Ichino, I.; Saito, M.; Nomura, M.; Mukasa, C.; Okabe, T.; Goto, K.; et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 2001, 142, 437–445. [Google Scholar] [CrossRef]


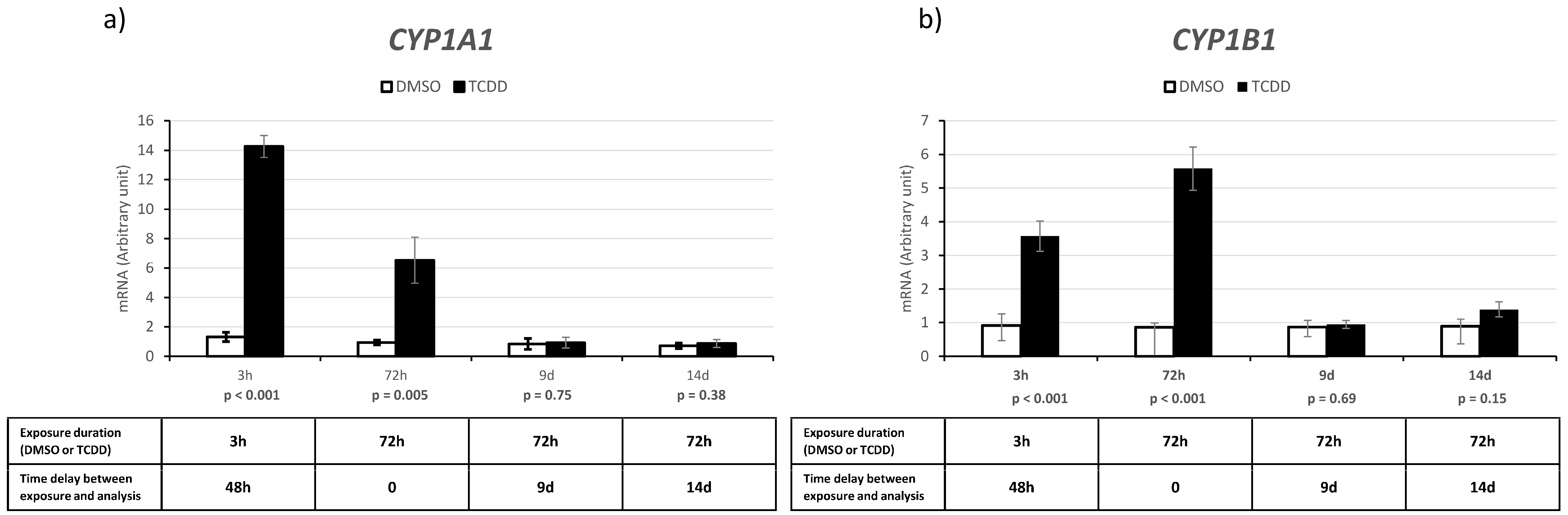
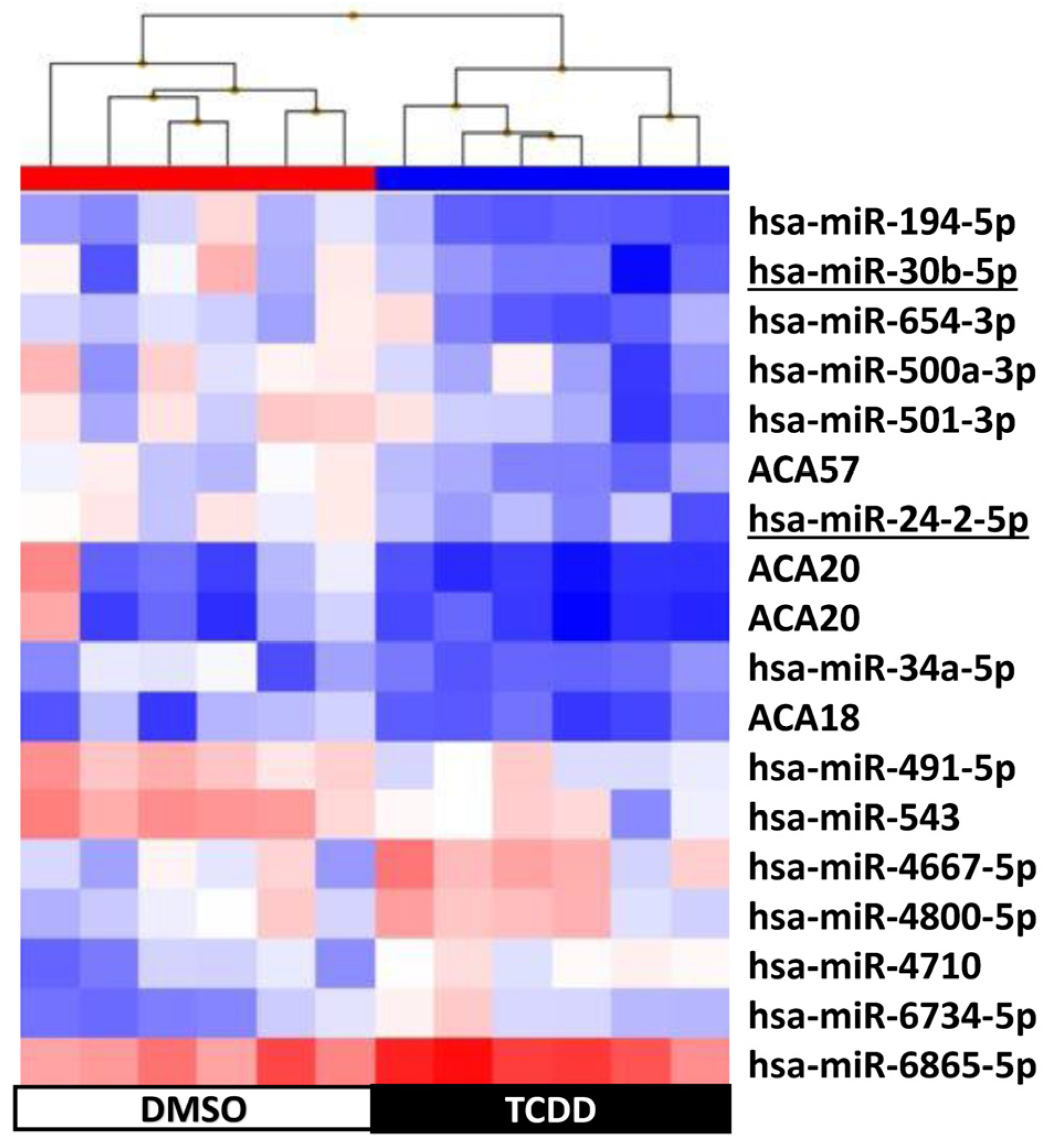
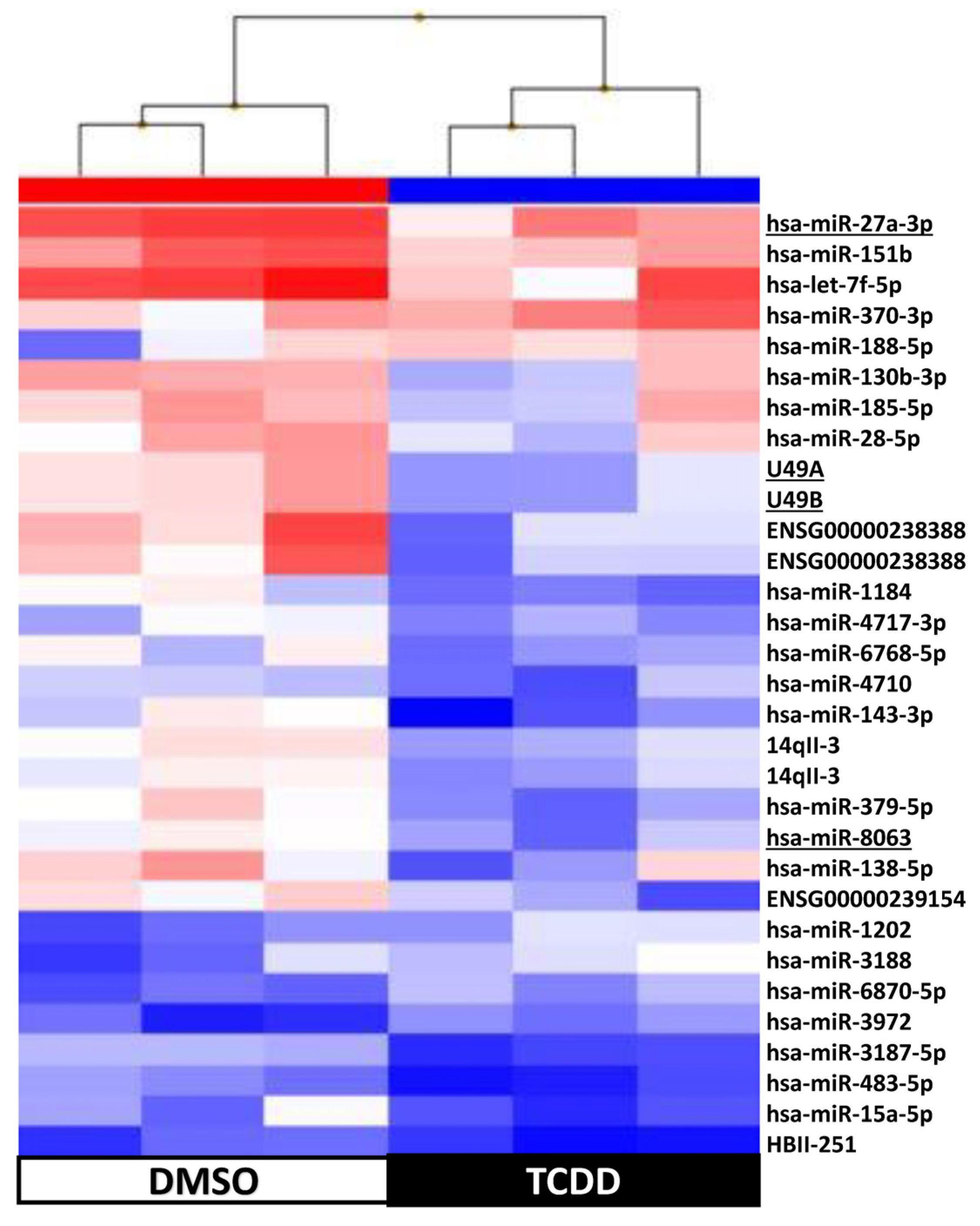
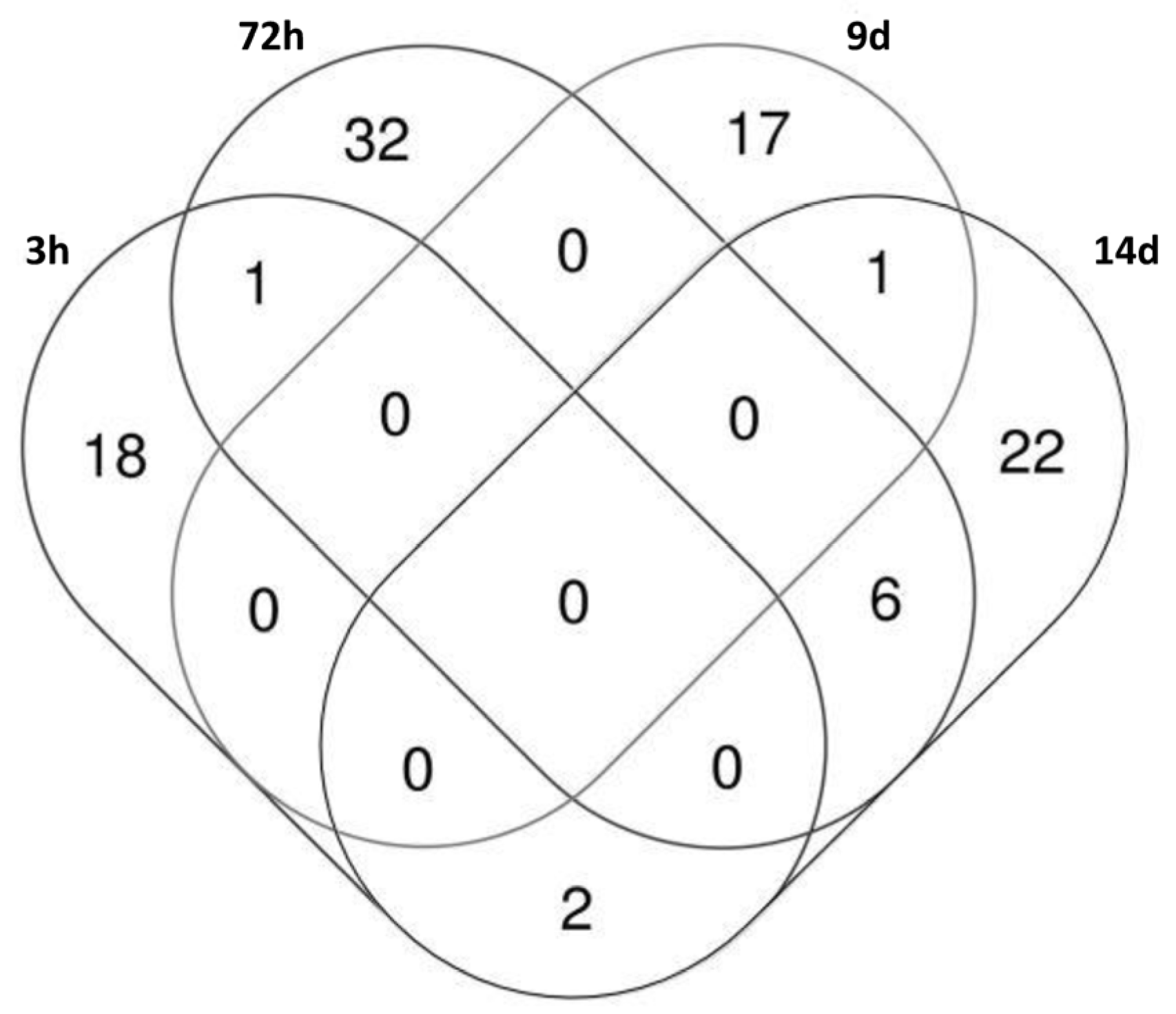
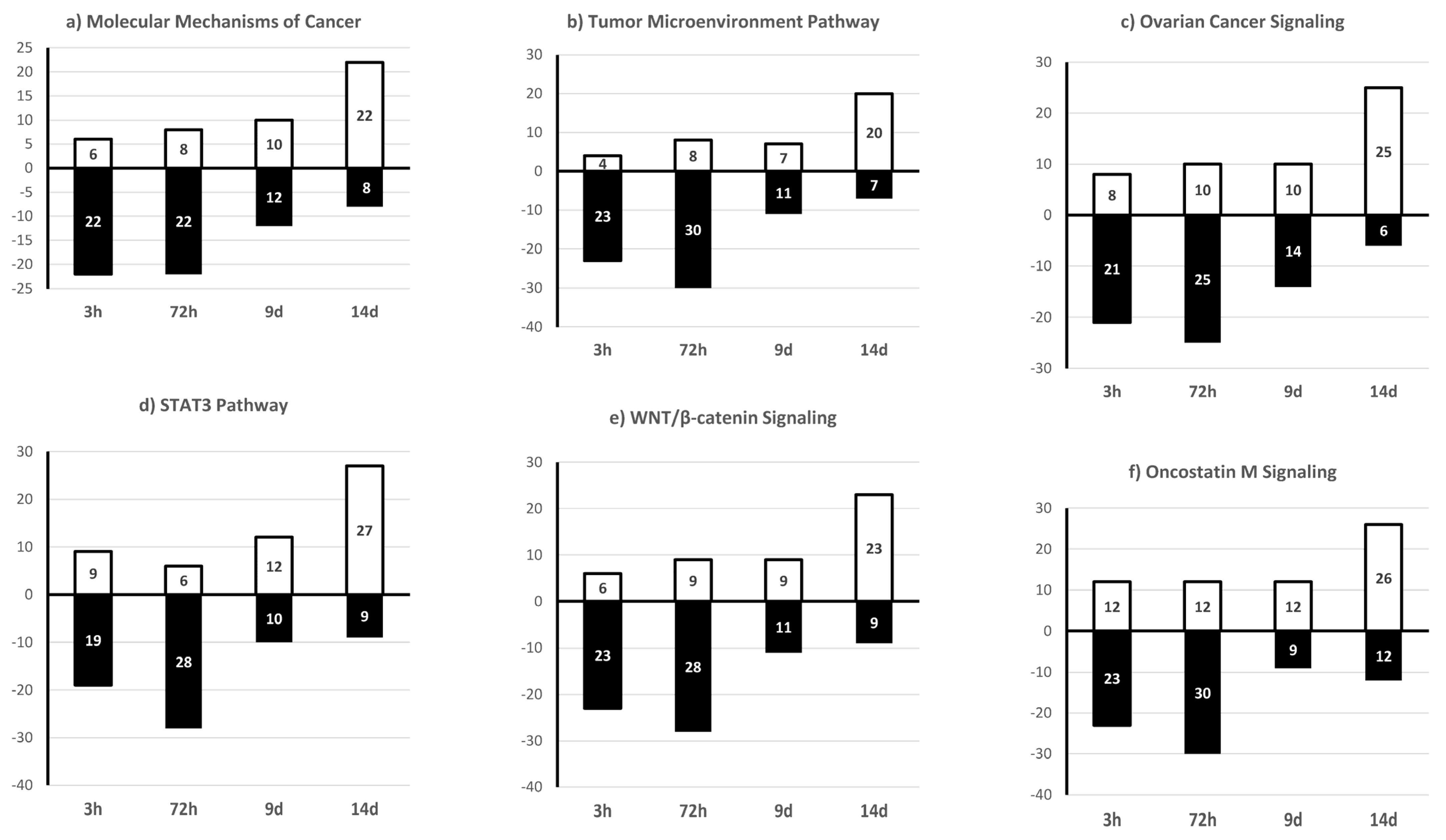
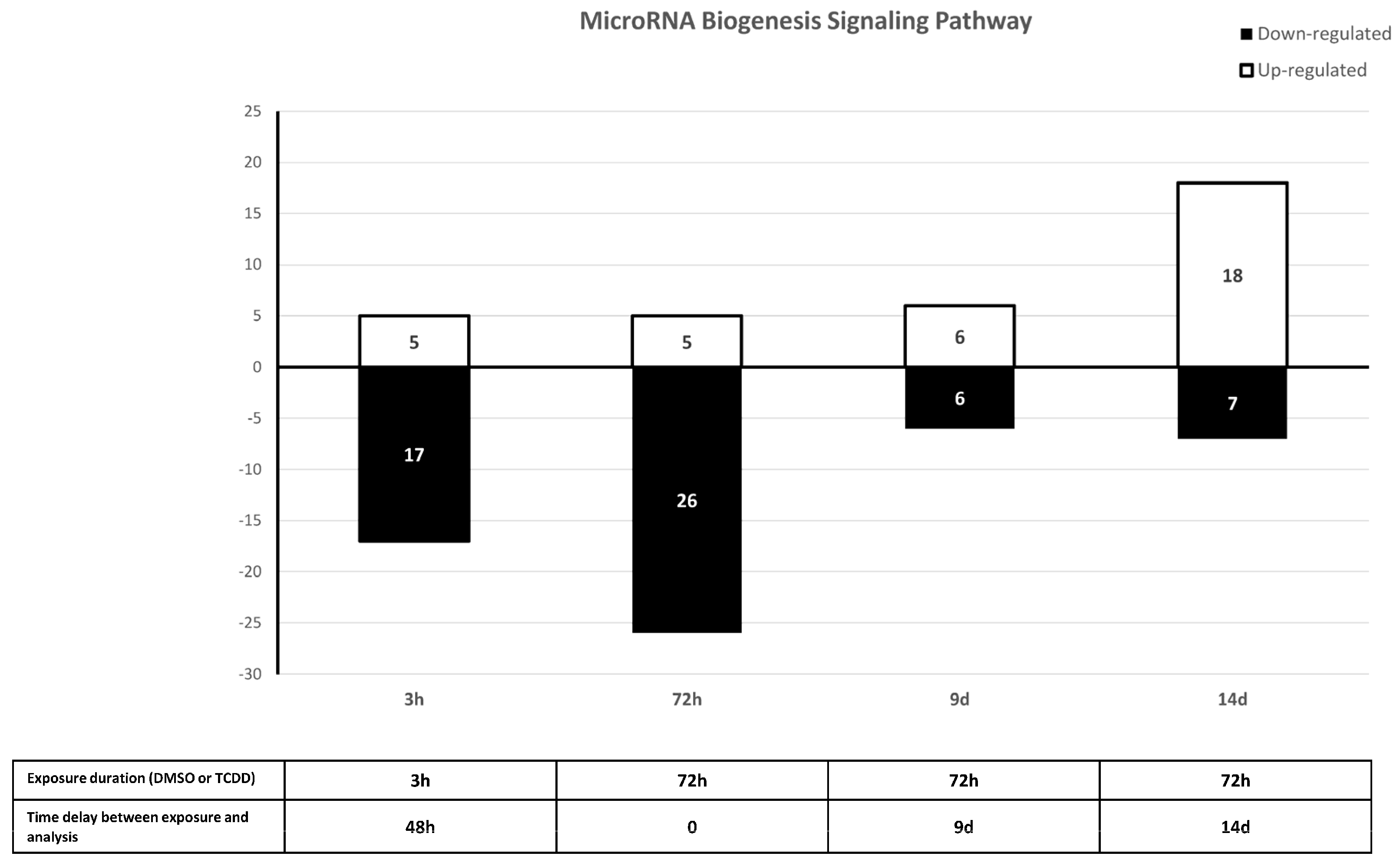
| 3 h | 72 h | 9 d | 14 d | |
|---|---|---|---|---|
| Exposure duration (DMSO or TCDD) | 3 h | 72 h | 72 h | 72 h |
| Time delay between exposure and analysis | 48 h | 0 | 9 d | 14 d |
| Number of differentially expressed snc-RNAs | 21 | 39 | 18 | 31 |
| Top Molecular and Cellular Function | 3 h | 72 h | 9 d | 14 d | |
|---|---|---|---|---|---|
| exposure duration (DMSO or TCDD) | 3 h | 72 h | 72 h | 72 h | |
| time delay between exposure and analysis | 48 h | 0 | 9 d | 14 d | |
| number of differentially expressed snc-RNAs | 21 | 39 | 18 | 31 | |
| Cellular development | ↓ | ↓ | = | ↑ | |
| number of genes | 1566 | 1774 | 848 | 1584 | |
| p-value range | 6.31 × 10−8–2.72 × 10−25 | 3.42E × 10−10–1.52 × 10−31 | 3.90 × 10−6–1.78 × 10−16 | 2.90 × 10−10–4.68 × 10−22 | |
| Cellular growth and proliferation | ↓ | ↓ | = | ↑ | |
| number of genes | 1472 | 1700 | 772 | 1454 | |
| p-value range | 6.31 × 10−8–2.72 × 10−25 | 3.42 × 10−10–1.52 × 10−31 | 3.90 × 10−6–3.74 × 10−18 | 1.42 × 10−10–2.37 × 10−25 | |
| Cell death and Survival | cell death | ↑ | ↑ | ↓ | |
| survival | ↓ | ↓ | ↑ | ||
| number of genes | 1644 | 942 | 1433 | ||
| p-value range | 4.27 × 10−10–3.85 × 10−33 | 3.14 × 10−6–1.82 × 10−18 | 1.92 × 10−10–5.24 × 10−24 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaspari, L.; Haouzi, D.; Gennetier, A.; Granes, G.; Soler, A.; Sultan, C.; Paris, F.; Hamamah, S. Transgenerational Transmission of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) Effects in Human Granulosa Cells: The Role of MicroRNAs. Int. J. Mol. Sci. 2024, 25, 1144. https://doi.org/10.3390/ijms25021144
Gaspari L, Haouzi D, Gennetier A, Granes G, Soler A, Sultan C, Paris F, Hamamah S. Transgenerational Transmission of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) Effects in Human Granulosa Cells: The Role of MicroRNAs. International Journal of Molecular Sciences. 2024; 25(2):1144. https://doi.org/10.3390/ijms25021144
Chicago/Turabian StyleGaspari, Laura, Delphine Haouzi, Aurélie Gennetier, Gaby Granes, Alexandra Soler, Charles Sultan, Françoise Paris, and Samir Hamamah. 2024. "Transgenerational Transmission of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) Effects in Human Granulosa Cells: The Role of MicroRNAs" International Journal of Molecular Sciences 25, no. 2: 1144. https://doi.org/10.3390/ijms25021144
APA StyleGaspari, L., Haouzi, D., Gennetier, A., Granes, G., Soler, A., Sultan, C., Paris, F., & Hamamah, S. (2024). Transgenerational Transmission of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) Effects in Human Granulosa Cells: The Role of MicroRNAs. International Journal of Molecular Sciences, 25(2), 1144. https://doi.org/10.3390/ijms25021144






