Characterization of Novel Trypanosoma cruzi-Specific Antigen with Potential Use in the Diagnosis of Chagas Disease
Abstract
:1. Introduction
2. Results
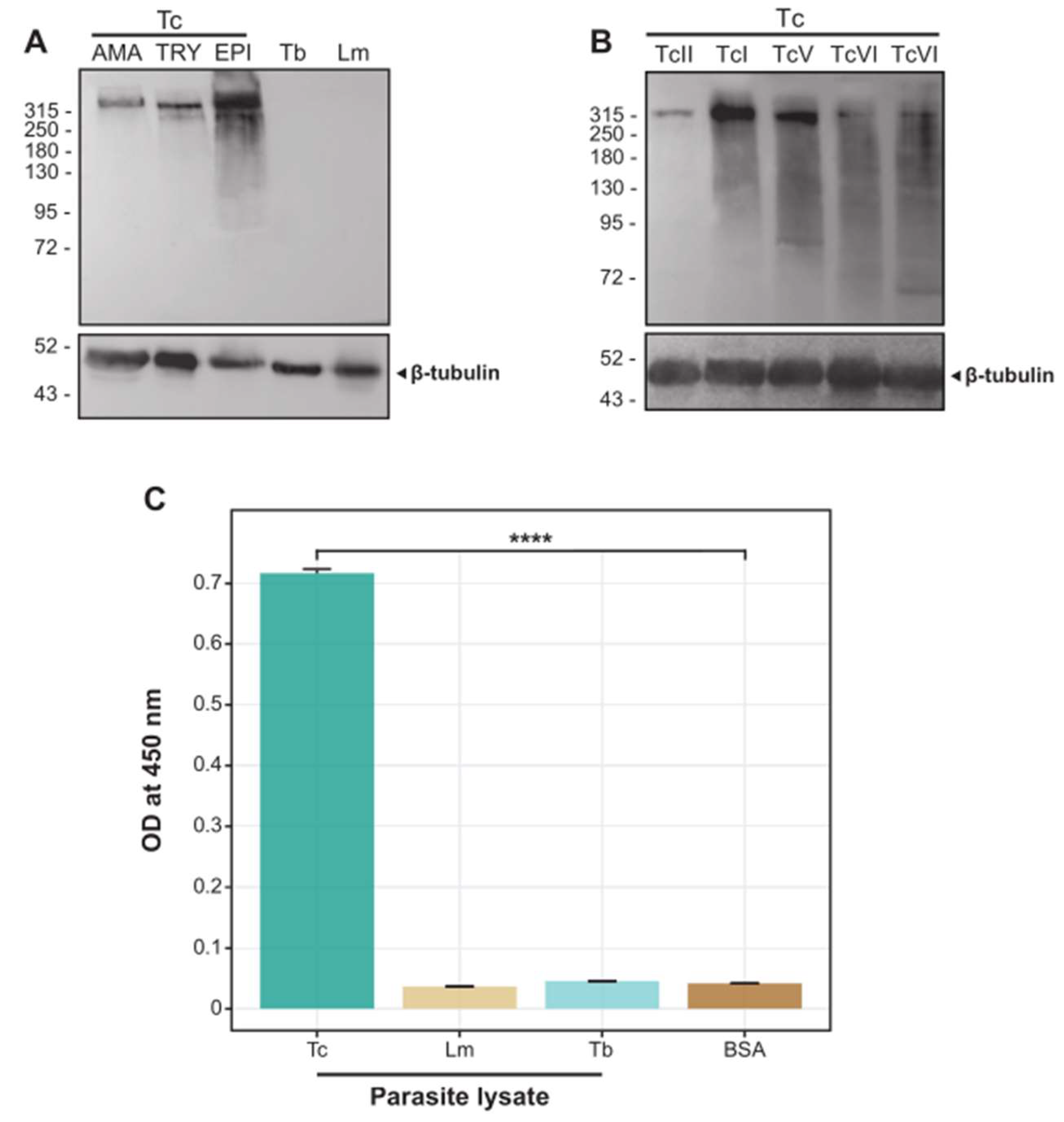
2.1. The scFv 6B6 Specifically Recognizes a Protein from T. cruzi but Not from Related Pathogens
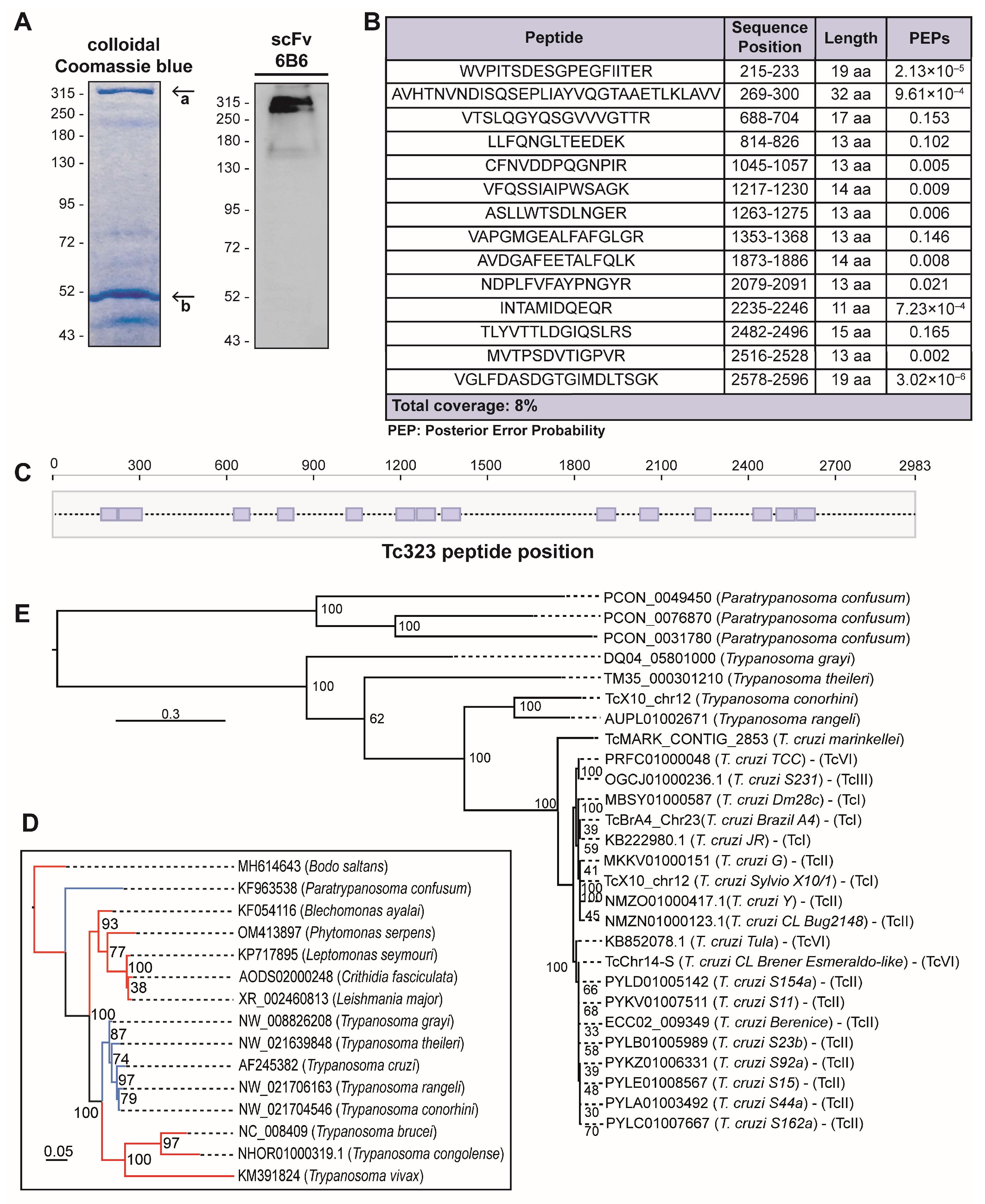
2.2. Tc323 Is the Target Antigen of scFv 6B6
2.3. Tc323 Localizes Intracellularly and Exhibits a Membrane-Associated Pattern
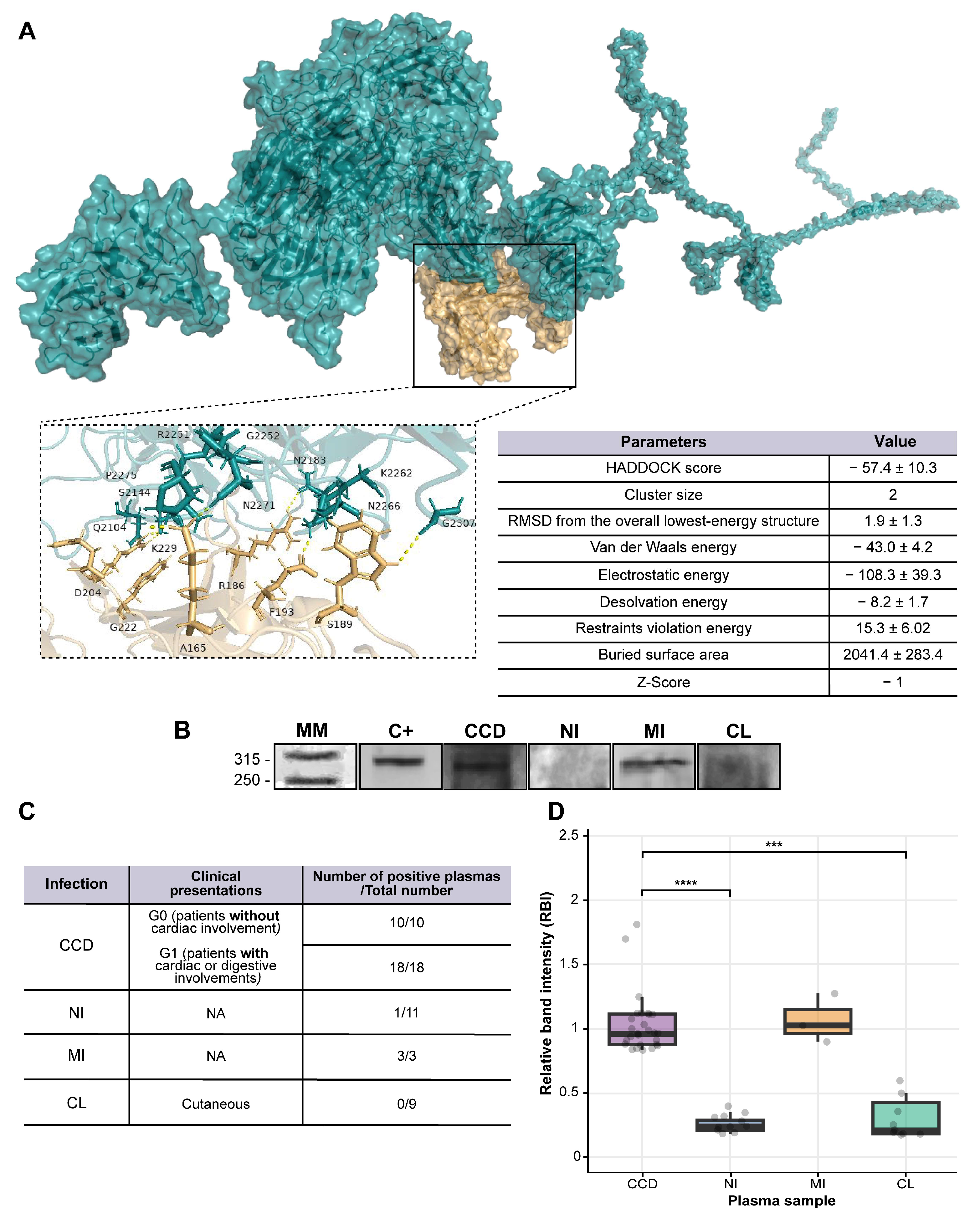
2.4. Tc323 Is Immunogenic
3. Discussion
4. Materials and Methods
4.1. Production of scFv 6B6 and Chim m6B6 Antibodies
4.2. Immunoprecipitation and Nano-LC Mass Spectrometry
4.3. In Silico Predictions and Phylogeny Analyses
4.4. Parasite Strains and Growth Conditions
4.5. Preparation of Cell Extracts
4.6. Enzyme-Linked Immunosorbent Assay (ELISA)
4.7. Western Blot
4.8. Immunofluorescence Microscopy
4.9. Cytometry
4.10. Ethics Statement
4.11. Study Population and Human Sample Collection
4.12. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hotez, P.J.; Bottazzi, M.E.; Franco-Paredes, C.; Ault, S.K.; Periago, M.R. The Neglected Tropical Diseases of Latin America and the Caribbean: A Review of Disease Burden and Distribution and a Roadmap for Control and Elimination. PLoS Negl. Trop. Dis. 2008, 2, e300. [Google Scholar] [CrossRef] [PubMed]
- Henrique Santana, K.; Gustavo Rodrigues Oliveira, L.; Barros de Castro, D.; Pereira, M. Epidemiology of Chagas Disease in Pregnant Women and Congenital Transmission of Trypanosoma cruzi in the Americas: Systematic Review and Meta-Analysis. Trop. Med. Int. Health 2020, 25, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Shikanai-Yasuda, M.A.; Carvalho, N.B. Oral Transmission of Chagas Disease. Clin. Infect. Dis. 2012, 54, 845–852. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Fourth WHO Report on Neglected Tropical Diseases: Integrating Neglected Tropical Diseases into Global Health and Development; WHO: Geneva, Switzerland, 2017; ISBN 9789241565448. [Google Scholar]
- Conners, E.E.; Vinetz, J.M.; Weeks, J.R.; Brouwer, K.C. A Global Systematic Review of Chagas Disease Prevalence among Migrants Graphical Abstract HHS Public Access. Acta Trop. 2016, 156, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Molina, J.A.; Molina, I. Chagas Disease. Lancet 2017, 391, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Prata, A. Clinical and Epidemiological Aspects of Chagas Disease.Lancet Infection Disease. Clin. Epidemiol. Asp. Chagas Dis. 2001, 1, 92–100. [Google Scholar] [CrossRef]
- Rassi, A.; Rassi, A.; Marin-Neto, J.A. Chagas Disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Gomes, Y.M.; Lorena, V.M.B.; Luquetti, A.O. Diagnosis of Chagas Disease: What Has Been Achieved? What Remains to Be Done with Regard to Diagnosis and Follow up Studies? Mem. Inst. Oswaldo Cruz 2009, 104, 115–121. [Google Scholar] [CrossRef]
- Santos, F.L.N.; De Souza, W.V.; Da Silva Barros, M.; Nakazawa, M.; Krieger, M.A.; De Miranda Gomes, Y. Chronic Chagas Disease Diagnosis: A Comparative Performance of Commercial Enzyme Immunoassay Tests. Am. J. Trop. Med. Hyg. 2016, 94, 1034–1039. [Google Scholar] [CrossRef]
- Journal, P.A. Síntesis de Evidencia: Guía Para El Diagnóstico y El Tratamiento de La Enfermedad de Chagas. Rev. Panam. Salud Pública 2020, 44, 1–7. [Google Scholar] [CrossRef]
- Rivera, H.N.; McAuliffe, I.; Aderohunmu, T.L.; Wiegand, R.E.; Montgomery, S.P.; Bradbury, R.S.; Handali, S. Evaluation of the Performance of Ortho T. Cruzi ELISA Test System for the Detection of Antibodies to Trypanosoma cruzi. J. Clin. Microbiol. 2022, 60, e0013422. [Google Scholar] [CrossRef] [PubMed]
- Caballero, Z.C.; Sousa, O.E.; Marques, W.P.; Saez-Alquezar, A.; Umezawa, E.S. Evaluation of Serological Tests to Identify Trypanosoma cruzi Infection in Humans and Determine Cross-Reactivity with Trypanosoma rangeli and leishmania Spp. Clin. Vaccine Immunol. 2007, 14, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Daltro, R.T.; Leony, L.M.; Maron Freitas, N.E.; Oliveira Silva, Â.A.; Santos, E.F.; Del-Rei, R.P.; Felinto Brito, M.E.; Brandão-Filho, S.P.; Gomes, Y.M.; Silva, M.S.; et al. Cross-Reactivity Using Chimeric Trypanosoma cruzi Antigens: Diagnostic Performance in Settings Where Chagas Disease and American Cutaneous or Visceral Leishmaniasis Are Coendemic. J. Clin. Microbiol. 2019, 57, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chiaramonte, M.G.; Zwirner, N.W.; Caropresi, S.L.; Taranto, N.J.; Malchiodi, E.L. Trypanosoma cruzi and Leishmania Spp. Human Mixed Infection. Am. J. Trop. Med. Hyg. 1996, 54, 271–273. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, N.M.; Myler, P.J.; Bartholomeu, D.C.; Nilsson, D.; Aggarwal, G.; Tran, A.N.; Ghedin, E.; Worthey, E.A.; Delcher, A.L.; Blandin, G.; et al. The Genome Sequence of Trypanosoma cruzi, Etiologic Agent of Chagas Disease. Science 2005, 309, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Frank, F.M.; Fernández, M.M.; Taranto, N.J.; Cajal, S.P.; Margni, R.A.; Castro, E.; Thomaz-Soccol, V.; Malchiodi, E.L.; Fernandez, M.M.; Taranto, N.J.; et al. Characterization of Human Infection by Leishmania Spp. in the Northwest of Argentina: Immune Response, Double Infection with Trypanosoma cruzi and Species of Leishmania Involved. Parasitology 2003, 126, S0031182002002585. [Google Scholar] [CrossRef] [PubMed]
- Vega Benedetti, A.F.; Cimino, R.O.; Cajal, P.S.; Juarez, M.D.V.; Villalpando, C.A.; Gil, J.F.; Marcipar, I.S.; Krolewiecki, A.J.; Nasser, J.R. Performance of Different Trypanosoma cruzi Antigens in the Diagnosis of Chagas Disease in Patients with American Cutaneous Leishmaniasis from a Co-Endemic Region in Argentina. Trop. Med. Int. Health 2013, 18, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Izeta-Alberdi, A.; Ibarra-Cerdeña, C.N.; Moo-Llanes, D.A.; Ramsey, J.M. Geographical, Landscape and Host Associations of Trypanosoma cruzi DTUs and Lineages. Parasit. Vectors 2016, 9, 631. [Google Scholar] [CrossRef]
- Stevens, J.R.; Noyes, H.A.; Schofield, C.J.; Gibson, W. The Molecular Evolution of Trypanosomatidae. Adv. Parasitol. 2001, 48, 1–56. [Google Scholar]
- Herreros-Cabello, A.; Callejas-Hernández, F.; Fresno, M.; Gironès, N. Comparative Proteomic Analysis of Trypomastigotes from Trypanosoma cruzi Strains with Different Pathogenicity. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 76, 104041. [Google Scholar] [CrossRef]
- Grippo, V.; Mahler, E.; Elias, F.E.; Cauerhff, A.; Gómez, K.A.; Tentori, M.C.; Ruiz, A.; Vigliano, C.A.; Laguens, R.P.; Berek, C.; et al. The Heavy Chain Variable Segment Gene Repertoire in Chronic Chagas′ Heart Disease. J. Immunol. 2009, 183, 8015–8025. [Google Scholar] [CrossRef] [PubMed]
- Grippo, V.; Niborski, L.L.; Gomez, K.A.; Levin, M.J. Human Recombinant Antibodies against Trypanosoma cruzi Ribosomal P2β Protein. Parasitology 2011, 138, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Niborski, L.L.; Potenza, M.; Chirivi, R.G.S.; Simonetti, L.; Ossowski, M.S.; Grippo, V.; May, M.; Staquicini, D.I.; Parodi-Talice, A.; Robello, C.; et al. Recombinant Antibody against Trypanosoma cruzi from Patients with Chronic Chagas Heart Disease Recognizes Mammalian Nervous System. EBioMedicine 2021, 63, 103206. [Google Scholar] [CrossRef] [PubMed]
- Alfaleh, M.A.; Alsaab, H.O.; Mahmoud, A.B.; Alkayyal, A.A.; Jones, M.L.; Mahler, S.M.; Hashem, A.M. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front. Immunol. 2020, 11, 1986. [Google Scholar] [CrossRef] [PubMed]
- Belicky, S.; Damborsky, P.; Zapatero-Rodríguez, J.; O’Kennedy, R.; Tkac, J. Full-Length Antibodies versus Single-Chain Antibody Fragments for a Selective Impedimetric Lectin-Based Glycoprofiling of Prostate Specific Antigen. Electrochim. Acta 2017, 246, 399–405. [Google Scholar] [CrossRef]
- Dos Santos, G.P.; Abukawa, F.M.; Souza-Melo, N.; Alcântara, L.M.; Bittencourt-Cunha, P.; Moraes, C.B.; Jha, B.K.; McGwire, B.S.; Moretti, N.S.; Schenkman, S. Cyclophilin 19 Secreted in the Host Cell Cytosol by Trypanosoma cruzi Promotes ROS Production Required for Parasite Growth. Cell. Microbiol. 2021, 23, e13295. [Google Scholar] [CrossRef]
- Bangs, J.D.; Uyetake, L.; Brickman, M.J.; Balber, A.E.; Boothroyd, J.C. Molecular Cloning and Cellular Localization of a BiP Homologue in Trypanosoma Brucei. Divergent ER Retention Signals in a Lower Eukaryote. J. Cell Sci. 1993, 105, 1101–1113. [Google Scholar] [CrossRef] [PubMed]
- Moraes, M.C.S.; Jesus, T.C.L.; Hashimoto, N.N.; Dey, M.; Schwartz, K.J.; Alves, V.S.; Avila, C.C.; Bangs, J.D.; Dever, T.E.; Schenkman, S.; et al. Novel Membrane-Bound EIF2alpha Kinase in the Flagellar Pocket of Trypanosoma Brucei. Eukaryot. Cell 2007, 6, 1979–1991. [Google Scholar] [CrossRef]
- Lo, Y.T.; Shih, T.C.; Pai, T.W.; Ho, L.P.; Wu, J.L.; Chou, H.Y. Conformational Epitope Matching and Prediction Based on Protein Surface Spiral Features. BMC Genom. 2021, 22, 116. [Google Scholar] [CrossRef]
- Riera, C.; Verges, M.; Iniesta, L.; Fisa, R.; Gállego, M.; Tebar, S.; Portús, M. Short Report: Identification of a Western Blot Pattern for the Specific Diagnosis of Trypanosoma cruzi Infection in Human Sera. Am. J. Trop. Med. Hyg. 2012, 86, 412–416. [Google Scholar] [CrossRef]
- Ferreira, A.W.; Belem, Z.R.; Lemos, E.A.; Reed, S.G.; Campos-Neto, A. Enzyme-Linked Immunosorbent Assay for Serological Diagnosis of Chagas’ Disease Employing a Trypanosoma cruzi Recombinant Antigen That Consists of Four Different Peptides. J. Clin. Microbiol. 2001, 39, 4390–4395. [Google Scholar] [CrossRef] [PubMed]
- Araujo, F.G. Analysis of Trypanosoma cruzi Antigens Bound by Specific Antibodies and by Antibodies to Related Trypanosomatids. Infect. Immun. 1986, 53, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Malchiodi, E.L.; Chiaramonte, M.G.; Taranto, N.J.; Zwirner, N.W.; Margni, R.A. Cross-Reactivity Studies and Differential Serodiagnosis of Human Infections Caused by Trypanosoma cruzi and Leishmania Spp; Use of Immunoblotting and ELISA with a Purified Antigen. Clin. Exp. Immunol. 1994, 97, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Herreros-Cabello, A.; Callejas-Hernández, F.; Gironès, N.; Fresno, M. Trypanosoma cruzi Genome: Organization, Multi-Gene Families, Transcription, and Biological Implications. Genes 2020, 11, 1196. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, M.T.; Taciana, K.; Silva, S.; Neves, L.X.; Jean, M.; Toledo, D.O.; Castro-borges, W.; De Lana, M. Differential Expression of Proteins in Genetically Distinct Trypanosoma cruzi Samples (TcI and TcII DTUs ) Isolated from Chronic Chagas Disease Cardiac Patients. Parasites Vectors 2018, 11, 1–11. [Google Scholar] [CrossRef]
- De Oliveira, M.T.; Branquinho, R.T.; Alessio, G.D.; Mello, C.G.C.; Nogueira-de-Paiva, N.C.; Carneiro, C.M.; de Ornelas Toledo, M.J.; Reis, A.B.; Martins-Filho, O.A.M.; de Lana, M. TcI, TcII and TcVI Trypanosoma cruzi Samples from Chagas Disease Patients with Distinct Clinical Forms and Critical Analysis of in Vitro and in Vivo Behavior, Response to Treatment and Infection Evolution in Murine Model. Acta Trop. 2017, 167, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna, C.; Nakayasu, E.S.; Pereira, M.G.; Lourenço, D.; de Souza, W.; Almeida, I.C.; Cunha-E-Silva, N.L. Subcellular Proteomics of Trypanosoma cruzi Reservosomes. Proteomics 2009, 9, 1782–1794. [Google Scholar] [CrossRef]
- Ulrich, P.N.; Jimenez, V.; Park, M.; Martins, V.P.; Atwood, J., 3rd; Moles, K.; Collins, D.; Rohloff, P.; Tarleton, R.; Moreno, S.N.J.; et al. Identification of Contractile Vacuole Proteins in Trypanosoma cruzi. PLoS ONE 2011, 6, e18013. [Google Scholar] [CrossRef]
- Queiroz, R.M.L.; Charneau, S.; Motta, F.N.; Santana, J.M.; Roepstorff, P.; Ricart, C.A.O. Comprehensive Proteomic Analysis of Trypanosoma cruzi Epimastigote Cell Surface Proteins by Two Complementary Methods. J. Proteome Res. 2013, 12, 3255–3263. [Google Scholar] [CrossRef]
- Brossas, J.-Y.; Gulin, J.E.N.; Bisio, M.M.C.; Chapelle, M.; Marinach-Patrice, C.; Bordessoules, M.; Palazon Ruiz, G.; Vion, J.; Paris, L.; Altcheh, J.; et al. Secretome Analysis of Trypanosoma cruzi by Proteomics Studies. PLoS ONE 2017, 12, e0185504. [Google Scholar] [CrossRef]
- Toyama, H.; Mathews, F.S.; Adachi, O.; Matsushita, K. Quinohemoprotein Alcohol Dehydrogenases: Structure, Function, and Physiology. Arch. Biochem. Biophys. 2004, 428, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Rucker, R.; Chowanadisai, W.; Nakano, M. Potential Physiological Importance of Pyrroloquinoline Quinone. Altern. Med. Rev. 2009, 14, 268–277. [Google Scholar] [PubMed]
- Anthony, C. Pyrroloquinoline Quinone (PQQ) and Quinoprotein Enzymes. Antioxid. Redox Signal. 2001, 3, 757–774. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Ishida, T.; Yoshida, M.; Samejima, M.; Ohno, H.; Igarashi, K.; Nakamura, N. Crystal Structure of the Catalytic and Cytochrome b Domains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase. Appl. Environ. Microbiol. 2019, 85, e01692-19. [Google Scholar] [CrossRef] [PubMed]
- Campos, F.M.F.; Liarte, D.B.; Mortara, R.A.; Romanha, A.J.; Murta, S.M.F. Characterization of a Gene Encoding Alcohol Dehydrogenase in Benznidazole-Susceptible and -Resistant Populations of Trypanosoma cruzi. Acta Trop. 2009, 111, 56–63. [Google Scholar] [CrossRef] [PubMed]
- González, L.; García-Huertas, P.; Triana-Chávez, O.; García, G.A.; Murta, S.M.F.; Mejía-Jaramillo, A.M. Aldo-Keto Reductase and Alcohol Dehydrogenase Contribute to Benznidazole Natural Resistance in Trypanosoma cruzi. Mol. Microbiol. 2017, 106, 704–718. [Google Scholar] [CrossRef]
- Skalický, T.; Dobáková, E.; Wheeler, R.J.; Tesařová, M.; Flegontov, P.; Jirsová, D.; Votýpka, J.; Yurchenko, V.; Ayala, F.J.; Lukeš, J. Extensive Flagellar Remodeling during the Complex Life Cycle of Paratrypanosoma, an Early-Branching Trypanosomatid. Proc. Natl. Acad. Sci. USA 2017, 114, 11757–11762. [Google Scholar] [CrossRef]
- Haag, J.; O’hUigin, C.; Overath, P. The Molecular Phylogeny of Trypanosomes: Evidence for an Early Divergence of the Salivaria. Mol. Biochem. Parasitol. 1998, 91, 37–49. [Google Scholar] [CrossRef]
- Cervantes-Landín, A.Y.; Martínez, I.; Schabib, M.; Espinoza, B. High Molecular Weight Proteins of Trypanosoma cruzi Reduce Cross-Reaction with Leishmania Spp. in Serological Diagnosis Tests. BioMed Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- Passos, V.M.; Volpini, A.C.; Braga, E.M.; Lacerda, P.A.; Ouaissi, A.; Lima-Martins, M.V.; Krettli, A.U. Differential Serodiagnosis of Human Infections Caused by Trypanosoma cruzi and Leishmania Spp. Using ELISA with a Recombinant Antigen (RTc24). Mem. Inst. Oswaldo Cruz 1997, 92, 791–793. [Google Scholar] [CrossRef]
- Truyens, C.; Dumonteil, E.; Alger, J.; Cafferata, M.L.; Ciganda, A.; Gibbons, L.; Herrera, C.; Sosa-Estani, S.; Buekens, P. Geographic Variations in Test Reactivity for the Serological Diagnosis of Trypanosoma cruzi Infection. J. Clin. Microbiol. 2021, 59, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Verani, J.R.; Seitz, A.; Gilman, R.H.; LaFuente, C.; Galdos-Cardenas, G.; Kawai, V.; de LaFuente, E.; Ferrufino, L.; Bowman, N.M.; Pinedo-Cancino, V.; et al. Geographic Variation in the Sensitivity of Recombinant Antigen-Based Rapid Tests for Chronic Trypanosoma cruzi Infection. Am. J. Trop. Med. Hyg. 2009, 80, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Zingales, B. Trypanosoma cruzi Genetic Diversity: Something New for Something Known about Chagas Disease Manifestations, Serodiagnosis and Drug Sensitivity. Acta Trop. 2017, 184, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Chirivi, R.G.S. Anti-Citrullinated Protein Antibodies as Novel Therapeutic Drugs in Rheumatoid Arthritis. J. Clin. Cell. Immunol. 2013, 01, 1–13. [Google Scholar] [CrossRef]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A Functional Genomic Resource for the Trypanosomatidae. Nucleic Acids Res. 2009, 38, 457–462. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: A Worldwide Hub of Protein Knowledge the UniProt Consortium. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Källberg, M.; Margaryan, G.; Wang, S.; Ma, J.; Xu, J. Raptorx Server: A Resource for Template-Based Protein Structure Modeling. Methods Mol. Biol. 2014, 1137, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Longhi, S.A.; Brandariz, S.B.; Lafon, S.O.; Niborski, L.L.; Luquetti, A.O.; Schijman, A.G.; Levin, M.J.; Gómez, K.A. Evaluation of In-House ELISA Using Trypanosoma cruzi Lysate and Recombinant Antigens for Diagnosis of Chagas Disease and Discrimination of Its Clinical Forms. Am. J. Trop. Med. Hyg. 2012, 87, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Kuschnir, E.; Sgammini, H.; Castro, R.; Evequoz, C.; Ledesma, R.; Brunetto, J. Evaluation of Cardiac Function by Radioisotopic Angiography, in Patients with Chronic Chagas Cardiopathy. Arq. Bras. Cardiol. 1985, 45, 249–256. [Google Scholar] [PubMed]
- Dean, S.; Sunter, J.D.; Wheeler, R.J. TrypTag.Org: A Trypanosome Genome-Wide Protein Localisation Resource. Trends Parasitol. 2017, 33, 80–82. [Google Scholar] [CrossRef]
- Mudogo, C.N.; Werner, S.F.; Mogk, S.; Betzel, C.; Duszenko, M. The Conserved Hypothetical Protein Tb427.10.13790 Is Required for Cytokinesis in Trypanosoma Brucei. Acta Trop. 2018, 188, 34–40. [Google Scholar] [CrossRef]
- Durante, I.M.; Cámara, M.D.L.M.; Buscaglia, C.A. A Novel Trypanosoma cruzi Protein Associated to the Flagellar Pocket of Replicative Stages and Involved in Parasite Growth. PLoS ONE 2015, 10, e0130099. [Google Scholar] [CrossRef]
- Ruiz-Márvez, E.; Ramírez, C.A.; Rodríguez, E.R.; Flórez, M.M.; Delgado, G.; Guzmán, F.; Gómez-Puertas, P.; Requena, J.M.; Puerta, C.J. Molecular Characterization of TC964, a Novel Antigenic Protein from Trypanosoma cruzi. Int. J. Mol. Sci. 2020, 21, 2432. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ossowski, M.S.; Gallardo, J.P.; Niborski, L.L.; Rodríguez-Durán, J.; Lapadula, W.J.; Juri Ayub, M.; Chadi, R.; Hernandez, Y.; Fernandez, M.L.; Potenza, M.; et al. Characterization of Novel Trypanosoma cruzi-Specific Antigen with Potential Use in the Diagnosis of Chagas Disease. Int. J. Mol. Sci. 2024, 25, 1202. https://doi.org/10.3390/ijms25021202
Ossowski MS, Gallardo JP, Niborski LL, Rodríguez-Durán J, Lapadula WJ, Juri Ayub M, Chadi R, Hernandez Y, Fernandez ML, Potenza M, et al. Characterization of Novel Trypanosoma cruzi-Specific Antigen with Potential Use in the Diagnosis of Chagas Disease. International Journal of Molecular Sciences. 2024; 25(2):1202. https://doi.org/10.3390/ijms25021202
Chicago/Turabian StyleOssowski, Micaela S., Juan Pablo Gallardo, Leticia L. Niborski, Jessica Rodríguez-Durán, Walter J. Lapadula, Maximiliano Juri Ayub, Raúl Chadi, Yolanda Hernandez, Marisa L. Fernandez, Mariana Potenza, and et al. 2024. "Characterization of Novel Trypanosoma cruzi-Specific Antigen with Potential Use in the Diagnosis of Chagas Disease" International Journal of Molecular Sciences 25, no. 2: 1202. https://doi.org/10.3390/ijms25021202
APA StyleOssowski, M. S., Gallardo, J. P., Niborski, L. L., Rodríguez-Durán, J., Lapadula, W. J., Juri Ayub, M., Chadi, R., Hernandez, Y., Fernandez, M. L., Potenza, M., & Gómez, K. A. (2024). Characterization of Novel Trypanosoma cruzi-Specific Antigen with Potential Use in the Diagnosis of Chagas Disease. International Journal of Molecular Sciences, 25(2), 1202. https://doi.org/10.3390/ijms25021202





