Abstract
The aim of this study was to elucidate molecular profiling in HER2-low tumors based on a promising dataset. A total of 615 consecutive HER2-negative breast cancer samples were assayed. The genomic mutations in the two groups with different HER2 expression levels (HER2-0 vs. HER2-low) were compared. The mutation types obtained via next-generation targeted sequencing were correlated with the clinicopathological features of the patients with HER2-0 and HER2-low breast cancer. The results showed that there was a significantly higher percentage of receptor-positive (ER/PR) tumors and more low-level Ki-67 tumors, but a lower incidence of stage I/II tumors in the HER2-low group compared to the HER2-0 group. There was a significantly higher frequency of 17.62% (65/369) for PIK3CA_SNA in the HER2-low group than in the HER2-0 group, which had a frequency of only 9.35% (23/246) (p = 0.006). When the called gene alterations in the triple-negative breast cancer (TNBC) group were compared with those in the luminal-like breast cancer group, there was a significantly high frequency of 28.17% (140/497) for ERBB2_SNA in a luminal-like group than in the TNBC group(16.95% (20/118)).We conclude that the early detection of PIK3CA mutations is likely to be important and might help therapeutic decision making in patients with HER2-low tumors.
1. Introduction
Breast cancer is a complex disease that displays heterogeneity at the genomic, transcriptomic, and proteomic levels, as well as existing in a variety of different cellular microenvironments [1]. A simplified categorization, based on the expression level of the estrogen receptor (ER), the progesterone receptor (PR), and the human epidermal growth factor 2 (HER2), has been adopted in clinical practice, and this has allowed breast cancers to be grouped into three distinct subtypes: luminal-like tumors (ER-positive and/or PR-positive and HER2-negative), HER2-positive tumors (HER2-positive, any ER, and PR-positive), and triple-negative (ER-negative, PR-negative, and HER2-negative) tumors. The recommended strategies to treat these tumors vary distinctly on account of their different biology and responsiveness to treatment. Anti-HER2 therapies have been demonstrated to have promising efficacy against HER2-positive breast cancers; however, the benefits of such a treatment have not yet been translated to those tumors that do not have HER2 overexpression [2]. The addition of trastuzumab to chemotherapy treatment does not improve invasive disease-free survival among patients when the breast cancer does not overexpress HER2. Previous guidelines have therefore limited the application of HER2-directed therapies only toward tumors that show the overexpression and/or amplification of HER2; this is defined as a score of 3+ on immunohistochemical (IHC) analysis or HER2 gene amplification defined via in situ hybridization (ISH) [3].
HER2-low expression, currently defined as an IHC score of 1+ or 2+ with no amplification of the HER2 gene when analyzed via ISH assay [4,5], has currently resulted in tumors being classified as luminal-like or triple-negative breast cancers. Among patients with HER2-low tumors, progressing from the first line of therapy for metastatic breast cancers means that chemotherapy is often considered the next strategy. Breast cancer with a low expression of HER2 is a new area of interest in breast cancer research. Recent clinical trials have demonstrated significant clinical benefit when new-generation antibody–drug conjugate (ADC) treatments are implemented for metastatic breast cancer patients with HER2-low tumors [6]. These promising results have created a brand-new landscape in the era of ADC and have broken through the borders of previous subtype groupings. Here, we analyzed the clinicopathological features and explored the molecular profiling of HER2-low tumors using a previously established dataset and tried to determine if there are significant differences between HER2-0 tumors and HER2-low tumors in terms of their molecular profiles.
2. Results
2.1. A Comparison of the Clinical and Pathological Features between the HER2-0 and HER2-Low Groups
In our database, 671 patients were recorded as HER2-negative. When correlated with NGS target sequencing data, those tumor samples with a non-pass quality and a coverage of <250 were excluded. This gave a total of 615 patients with HER2-negative tumors, namely HER2-0 (IHC score 0, n = 246) and HER2-low (IHC score 1+/2+ and FISH negative, n = 369), and these were enrolled in the present study (Supplementary Information Figure S1). Those with HER2 IHC 3+ or IHC 2+/FISH+ were excluded. The clinicopathological features of this cohort study stratified by their HER2-0 and HER2-low status tumors were then analyzed. Clinical parameters, such as age, tumor size, and lymph node status, showed no statistically significant differences between HER2-0 and HER2-low tumors (Table 1).

Table 1.
Clinical presentations by different immunohistochemical scores in HER2-negativebreast cancer.
However, notably, there was a significantly lower incidence of stage I/II samples and a significantly higher percentage of receptor (ER/PR) positive samples in the HER2-low group compared to the HER2-0 group. In contrast, there was a significantly higher percentage of tumors grade III and Ki-67 ≥ 30% in the HER2-0 group compared to the HER2-low group (Table 2) (p < 0.05, Chi-Square test or Fisher’s exact test).

Table 2.
Pathological features by different immunohistochemical scores in HER-2 negative breast cancer.
2.2. A Comparison of Mutation Types between the HER2-0 and HER2-Low Groups
The Venn diagram comparison between HER2-0 and HER2-low groups is shown in Supplementary Information Figure S1. The results of the TMO assay revealed that the average number of called variants in the HER2-0 group was 4.57 (SD: 5.06, range: 1~68), while those in the HER2-low group were5.60 (SD: 13.37, range: 1~190). The mutation types of called variants were SNA, such as synonymous, missense, insertion/deletion (Indel), or frameshift, and SA, such as fusion, truncation, and CNA. The average number of each mutation type among HER2-0 and HER2-low breast cancers with at least one variant was demonstrated in Table 3.

Table 3.
Comparisons of mutation types between HER2-0 and HER2-low tumors.
2.3. Actionable Genes between HER2-0 and HER2-Low Groups
Based on ESCAT criteria, the actionable gene variants were AKT2, BCRA1, BRCA2, ERBB2, ERBB3, PI3KCA, PTEN, and MDM2. The average actionable gene variants among HER2-0 and HER2-low breast cancers are shown in Table 4.

Table 4.
Comparisons of actionable genes between HER2-0 and HER2-low tumors.
2.4. Comparison of Mutational Alterations Present between the HER2-0 and HER2-Low Groups
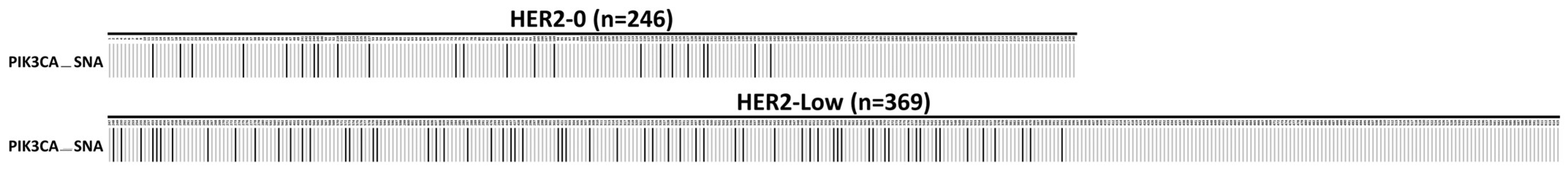
When the called gene alterations in the HER2-0 group were compared with those in the HER2-low group, there was a significantly higher frequency of 17.62% (65/369) for PIK3CA_SNA in HER2-low group than in the HER2-0 group, which had sample frequency of 9.35% (23/246) (p = 0.006, Chi-Square test). The oncoplot is presented in Figure 1.

Figure 1.
An oncoplot comparison of single nucleotide alteration (SNA) between HER2-0 and HER2-low groups. The PIK3CA_SNA is at exon 20 (the H1047R mutation). The HER2-0 is defined as IHC score 0 group (n = 246) and the HER2-low as IHC score 1+/2+ and FISH negative group (n = 369). The x-axis indicates subject number, while y-axis indicates PIK3CA_SNA mutation. The gray bars indicate no alteration compared to the reference genome, and the back bars indicate gene alteration noticed.
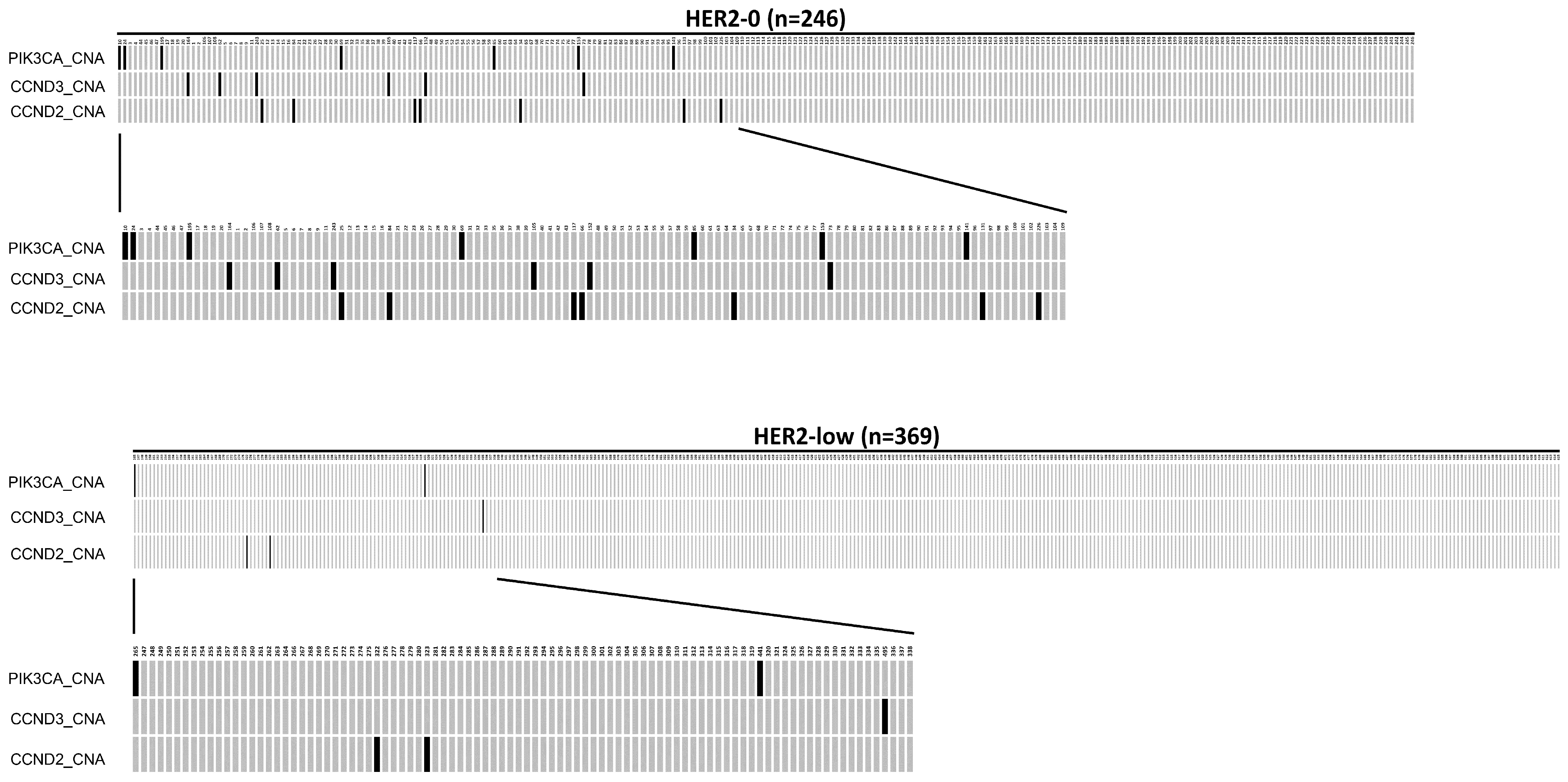
Among the 246 HER2-0 patients, 112 (45.5%) tumors showed copy number alteration (CNA), while 185 tumors (50.1%) in the 369 HER2-low patients had CNA mutations. In addition, there was a significantly higher frequency of PIK3CA_CNA (2.85%), CCND3_CNA (2.44%), and CCND2_CNA (2.85%) mutations in the HER2-0 group than in the HER2-low group, which had frequencies of 0.54%, 0.27%, and 0.54%, respectively (p < 0.05, Chi-Square test). The oncoplot result is presented in Figure 2. The mutation details of Figure 1 and Figure 2 are shown in Table 5.
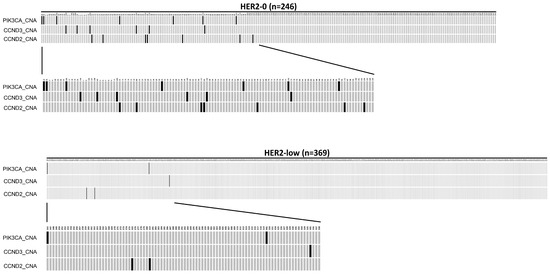
Figure 2.
An oncoplot comparison of copy number alteration (CAN) between HER2-0 and HER2-low groups. The HER2-0 is defined as IHC score 0 group (n = 246) and the HER2-low as IHC score 1+/2+ and FISH negative group (n = 369). The x-axis indicates subject number, while y-axis indicates gene alterations such as PIK3CA_CNA, CCND2_CNA, and CCND3_CNA mutations. The gray bars indicate no alteration compared to the reference genome, and the back bars indicate gene alteration noticed.

Table 5.
Comparison of mutation alterations between HER2-0 and HER 2-low expression tumors.
2.5. Comparison of the Mutational Alterations between TNBC Breast Cancer and Luminal-Like Breast Cancer in HER2-Low Tumors
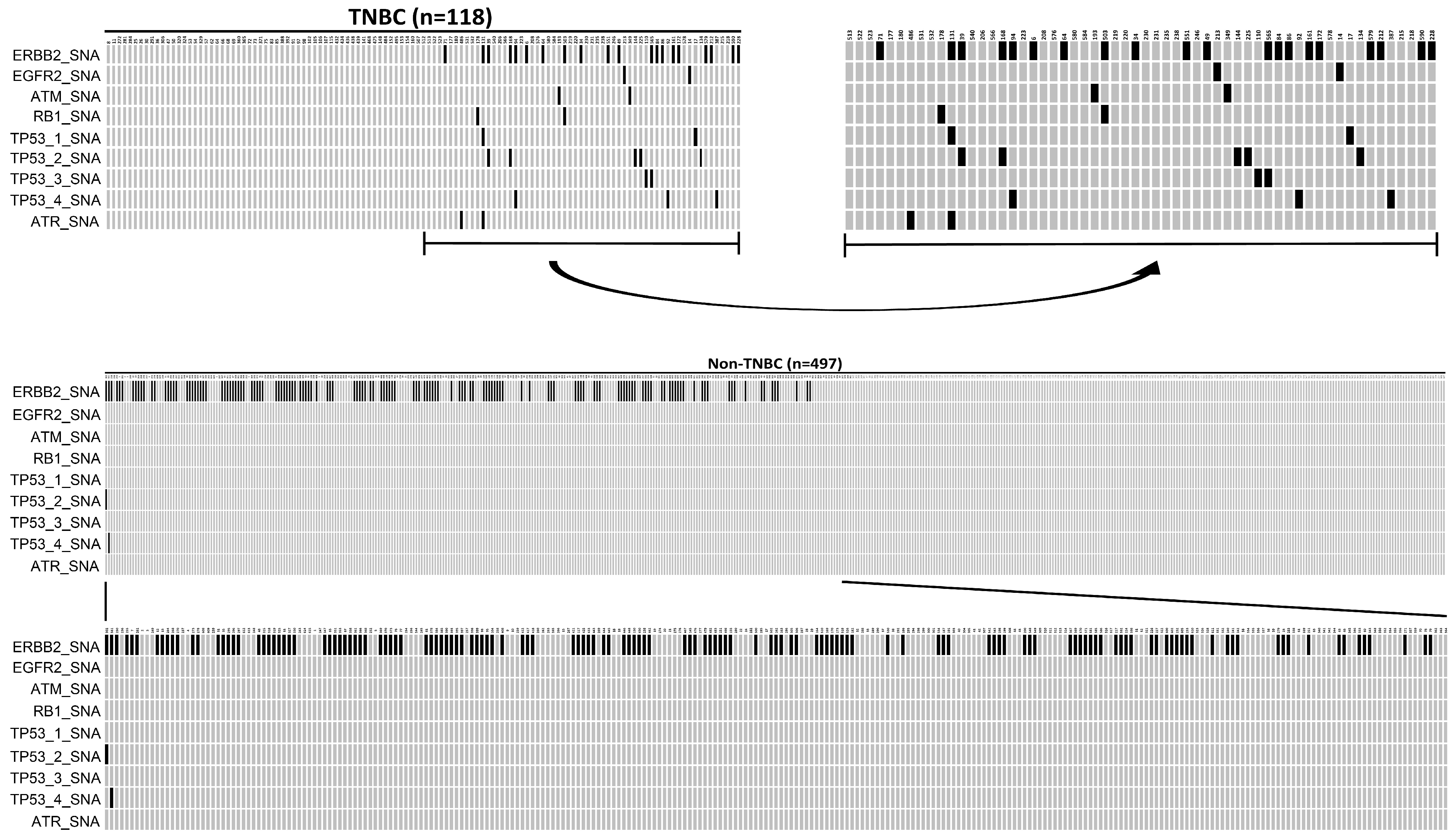
In this 615-patient cohort study, 118 (19.19%) were TNBC, while 497 (80.81%) were luminal-like tumors. When the called gene alterations in the TNBC group were compared with those identified in the luminal-like group, there was a significantly high sample frequency of 28.17% (140/497) for ERBB2_SNA in a luminal-like group compared to the TNBC group, where the frequency was 16.95% (20/118) (p = 0.014, Fisher’s exact test). In contrast, there was a significantly higher sample frequency of SA (ATR/ATM) and SNA (FGFR2, RB1, TP53) in the TNBC group compared to the luminal-like group (p < 0.05, Fisher’s exact test). The oncoplot result is presented in Figure 3.The mutation details of Figure 3 are shown in Table 6.
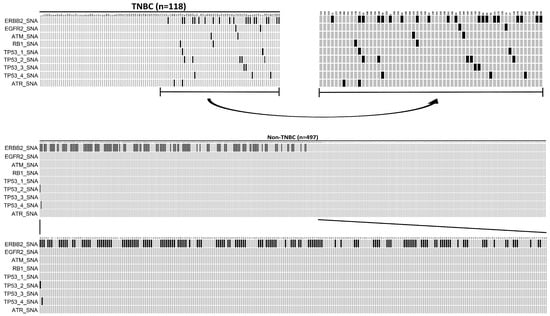
Figure 3.
An oncoplot comparison of mutations between triple-negative breast cancer (TNBC) and Luminal-like groups. TNBC is defined as ER (-), PR (-), and HER2 (-), while luminal-like is defined as ER (+) or PR (+) and HER2 (-).The x-axis indicates subject number, while y-axis indicates gene alterations such as ERBB2_SNA, EGFR2_SNA, TAM_SNA, RB1_SNA, TP53-1_SNA, TP53-2_SNA, TP53-3_SNA, TP53-4_SNA, and ATR_SNA mutations. The gray bars indicate no alteration compared to the reference genome, and the back bars indicate gene alteration noticed.

Table 6.
Comparison of mutation alterations between triple-negative tumors and luminal-like tumors in HER 2-negative breast cancer.
3. Discussion
With the rapid advances in novel ADC regimens, it has become crucial to understand the biology and corresponding genomes of HER2-low tumors in order to define the population that can truly benefit from ADC. Here, we explored the genomic alterations in HER2-low tumors compared with HER2-0 tumors. The impressive results from the phase III DESTINY-Breast04 trial demonstrated the survival benefit of Trastuzumab deruxtecan (T-DXd) in patients with pretreated HER2-low metastatic breast cancer compared with conventional chemotherapy [6]. The revolutionary nature of ADC treatment has changed the destiny of advanced HER2-low breast cancer patients and upturned conventional treatment algorithms. Moreover, this development reshapes the way we think of the HER2-low expression phenotype, breaking through the borders between subtypes grouping and opening up discussion on how to recognize HER2-low tumors. Our work provides evidence to understand the difference in genomic mutations between the two entities, which were both defined as HER2-negative tumors before. To date, most studies of HER2-low tumors focused on the analysis of clinicopathological features and prognostic information, lacking the viewpoint from molecular profiling. Based on a previously comprehensive study, the VGH-TAYLOR trial, we elucidate the genomic alterations of HER2-low tumors from the updated 648 samples of Taiwanese breast cancer patients undergoing targeted sequencing [7,8]. Our study demonstrated that the HER2-low tumors presented more ER/PR-positive and higher percentage of low proliferative index (Ki-67 < 30%), and a significantly higher frequency for PIK3CA_SNA compared to HER2-0 tumors. In a previous study using large retrospective cohorts, the authors failed to find the major differences in clinicopathologic features or prognostic value between the HER2-low expression and others [9]. Another study collected retrospective PAM50 analyses to elucidate whether HER2-low and HER2-0 tumors significantly differ in gene expression, but ultimately, they found only minor differences between the two groups [4]. The study team of the DESTINY-Breast04 trial explored the potential biomarkers in collected baseline circulating tumor DNA (ctDNA) samples from 414 patients [10]. In the T-DXd and control arms, 51.3% and 54.0% of patients were found with ESR1 mutations, and 36.1% vs. 41.6% of patients presented PIK3CA mutations. In patients with prior CDK4/6 inhibitors, at least one CDK4/6 inhibitor resistance marker was observed in 71.5% and 70.2% of patients. However, the efficacy of T-DXd superior to the treatment of physician choices was consistently observed independent of ESR1, PIK3CA mutation, or known resistant markers of CDK4/6 inhibitors. Whether the amazing benefit from T-DXd can be extended to the HER2-0 tumors is unknown. The minimum threshold of HER2 expression to ensure the activity of T-DXd also still needs to be determined. Although the phase II DAISY trial showed inconclusive results on their primary endpoint, objective response rate (ORR), in the HER2 non-expressing cohort [11], the ongoing larger-scale study (ClinicalTrials.gov numbers, NCT04494425) may further answer the question on HER2-0 tumors. Here, our results may help to establish the fundamental knowledge on differences between the HER2-low and HER2-0 tumors.
A previous study indicated that ER located near the cell membrane is able to activate many receptor tyrosine kinases, including epidermal growth factor receptor (EGFR) and HER2/neu (HER2) [12], and it has been suggested that this might be a possible mechanism for the presence of tamoxifen resistance in ER(+)/HER2(+) breast cancers [13]. Recently, accumulating evidence has demonstrated the cross-talk between ER and HER2 signaling is able to help identify new therapeutic strategies, including the use of aromatase inhibitors, dual blockade (trastuzumab/perstuzumab), and CDK4/6 inhibitors, to treat various different breast cancer subtypes [14,15,16,17]. The fact that there is a significantly higher percentage of receptor(ER/PR) positivity in the HER2-low group compared to the HER2-0 group might be explained by the presence of ER-HER2 signaling cross-talk in this setting.
There is accumulating evidence suggesting that the phosphatidylinositol 3-kinase (PIK3) catalytic subunit PIK3CA plays an important role in human carcinogenesis. The PIK3CA (H1047R) mutation has been correlated with poor clinical prognosis not only in gastric carcinoma, glioblastoma, and colorectal carcinoma [18] but also in breast cancer [19,20]. Previously, mutations affecting the PIK3CA gene, which results in the hyperactivation of the alpha isoform (p110a) of PI3K, have been demonstrated in 28% to 46% of patients with HR+/(HER2−) advanced breast cancers [21]. In our cohort database study, PIK3CA mutations were present in 38% (278/728) of all tested samples, in 43% of samples with the HR-/HER2+ subtype and in 42% of samples with HR+/HER2-post-menopausal status. Notably, when patients were treated with CDK4/6 inhibitors, the median time to treatment failure was 12 months (95% CI: 7–21 months) in the PIK3CA mutation group, but this increased to 16 months (95% CI: 11–23 months) in the PIK3CA wild-type group [22]. Thus, the PIK3CA mutations were associated with a reduced sensitivity to CDK4/6 inhibitor, and this is compatible with earlier findings [23,24].
There is consensus that PIK3CA gene mutation often results in hyperactivation of the PI3K-rapamycin (mTOR) pathway. The results obtained from the BOLERO-1/-3 study suggest that patients having tumors with a PIK3CA mutation or a hyperactive PI3K pathway derive PFS benefit from treatment with everolimus compared to those without such a mutation [25]. The most common mutations of PIK3CA in Taiwanese female breast cancers are in exon 20 (the H1047R mutation), in exon 9 (the E545K mutation), and in exon 9 (the E542K mutation) with frequencies of 41.6%, 18.9%, and 10.3%, respectively [22]. Our present results show that there is a significantly higher frequency of 17.62% (65/369) of PIK3CA_SNA in the HER2-low group compared to the HER2-0 group, which has a frequency of 9.35% (23/246) (p = 0.006, Chi-Square test) (Table 4). This suggests that early detection of PIK3CA mutations is important and might help therapeutic decision making in those patients with a HER2-low tumor. Although there was a higher frequency of PIK3CA_CNA (2.85%) in the HER2-0 group than in the HER2-low group, which has a frequency of 0.54%, the clinical significance of this remains to be elucidated due to the limited sample size.
TNBC (ER-, PR-, and HER2-) tumors are characterized by their clinicopathological features, such as their occurrence in younger women, their aggressive nature (a higher tumor grade and a higher Ki67 percentage), and a higher association with metastasis to distant organs. In patients with HER2-low tumors, it is expected that TNBC should be included. In the present study, our findings show that there is a significantly higher percentage of tumors grade III and Ki-67 ≥ 30% among the HER2-0 group compared to the HER2-low group (Table 2) (p < 0.05, Fisher’s exact test), which is compatible with our findings that there is a higher percentage (29.7%) of TNBC in the HER2-0 group compared to 12.2% in the HER2-low group (p < 0.0001, Fisher’s exact test). Based on our NGS analysis, there is also a significantly higher frequency of 28.17% (140/497) for ERBB2_SNA in the luminal-like group compared to the TNBC group, which has a frequency of 16.95% (20/118) (p = 0.014, Fisher’s exact test) (Table 6). Although previous studies have demonstrated that G to A mutation at amino acid codon 655 of the human erbB-2/HER2 gene is a new allele polymorphism of the ERBB2 gene [26,27], the exact role of such mutation as a therapeutic target in luminal-like breast cancer remains further clinical elucidation.
In summary, HER2-negative breast cancer is able to be further divided into the HER2-0 and HER2-low groups. There are different clinical manifestations between these two groups. There is a significantly higher frequency of 17.62% (65/369) of PIK3CA_SNA in the HER2-low group compared to the HER2-0, with a frequency of 9.35% (23/246). When the called gene alterations are compared between the TNBC and luminal-like groups, there is a significantly high frequency of 28.17% (140/497) for ERBB2_SNA in the luminal-like group compared to the TNBC group, which has a frequency of 16.95% (20/118).
4. Materials and Methods
4.1. Study Population
Under the approval of the Institutional Review Board of Taipei Veterans General Hospital (# 2023-06-025BC), we followed the same protocol as the VGH-TAYLOR, which involved a comprehensive precision medicine investigation of the heterogeneity of Taiwanese breast cancer patients (Clinical trial registration: NCT04626440 (ClinicalTrials.gov)) [28]. In short, the study comprised a broad clinical spectrum of breast cancers, namely, Group 1, those planned to receive first-line surgery followed by adjuvant therapy or having early relapse within 3 years; Group 2, those planned to receive first-line neoadjuvant therapy followed by surgery; and Group 3, those planned to receive treatment for de novo stage IV or stage IV disease with recurrence beyond 3 years. Three years (Jan. 2018–Jan. 2020) of enrollment and four years of follow-up after enrollment were included. All patients received treatment following the contemporary practice guidelines of the Comprehensive Breast Health Center at Taipei Veterans General Hospital, which is based on the NCCN and St. Gallen guidelines.
4.2. Pathology Review
IHC staining to detect ER (clone 6F11; Leica Biosystems, Newcastle, UK; 1:100), PR (clone 16; Leica Biosystems, Newcastle, UK; 1:150), and HER2 (Ventana PATHWAY anti-HER2/neu 4B5 rabbit monoclonal antibody), were evaluated by experienced pathologists from our institute. The positivity of ER and PR was defined as ≥1% of tumor cells exhibiting nuclear staining. HER2 IHC positivity (score 3+) was defined via complete intense membrane staining in >10% of tumor cells. Reflex in situ hybridization (ISH) testing via fluorescence ISH (PathVysion HER2 DNA Probe Kit; Abbott Laboratories, Des Plaines, IL, USA) was performed for cases that gave equivocal HER2 IHC results (score 2+). Patients with an average HER2 copy number of ≥6 signals/cell, or ≥4 signals/cell and a HER2 ISH ratio (HER2 gene signals to chromosome 17 centromere signals) of ≥2 were regarded as ISH-positive by the 2018 American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) guidelines. In the present study, HER2-negative patients were further categorized into two subgroups: the HER2-0 (IHC score 0) group and the HER2-low (IHC score 1+/2+ and FISH-negative) groups.
4.3. Sample Preparation
Sample preparation was carried out to allow next-generation sequencing (NGS) targeted sequencing of the fresh-frozen paraffin-embedded (FFPE) samples. The preparation of the FFPE section was performed at the clinical site following the standard procedures. The hematoxylin and eosin (H&E) staining was performed and interpreted under the guidance of a certified pathologist (Dr. Hsu, CY). Approximately 7 unstained sections of tumor FFPE tissues per subject were retrieved; at least one unstained section was prepared for H&E staining, and 6 unstained sections were prepared for TMO comprehensive assay.
4.4. DNA Extraction
The extraction of DNA was conducted in the central laboratory according to the appropriate laboratory manuals. In short, DNA extraction from FFPE or tissue cores wassubjected to xylene treatment and rehydrated using a series of ethanol washes, followed by the removal of proteins (nucleases) via proteinase K digestion. Nucleic acids were purified from the tissue lysate, and this was followed by phenol extractions, and RNase A was added to eliminate RNA contamination. The addition of sodium acetate and isopropanol precipitated DNA, and high-speed centrifugation was used to pellet the DNA, followed by the salt out process and washing with 70% ethanol and by centrifugation to re-pellet the DNA and stored at −20 °C for further use. Purified DNA can subsequently be used in downstream applications, which include PCR, array comparative genomic hybridization 4 (array CGH), methylated DNA Immunoprecipitation (MeDIP), and sequencing, allowing for an integrative analysis of tissue/tumor samples. Additional sections of FFPE samples of individual subjects might be pursued if a sample failed the nucleic acid quality check. The criteria for the DNA quality check followed the manual of the TMO assay requirement (see below).
4.5. OncomineTM Comprehensive Assay (TMO Comprehensive Assay)
Targeted NGS experiments were performed using the TMO comprehensive assayv3 from FFPE tissues (Thermo Fisher Scientific, Waltham, MA, USA) in order to detect thousands of variants across 161 genes that are relevant to cancer. The TMO comprehensive assay was performed using the collected FFPE tissue samples, and the analyses of the TMO comprehensive assays included a range of specific genes and various types of mutation within these genes, including frameshift, missense, synonymous, single nucleotide alteration (SNA), insertion/deletion (Indel), structure alteration (SA) and copy number alteration (CNA); each analysis was for an individual subject.
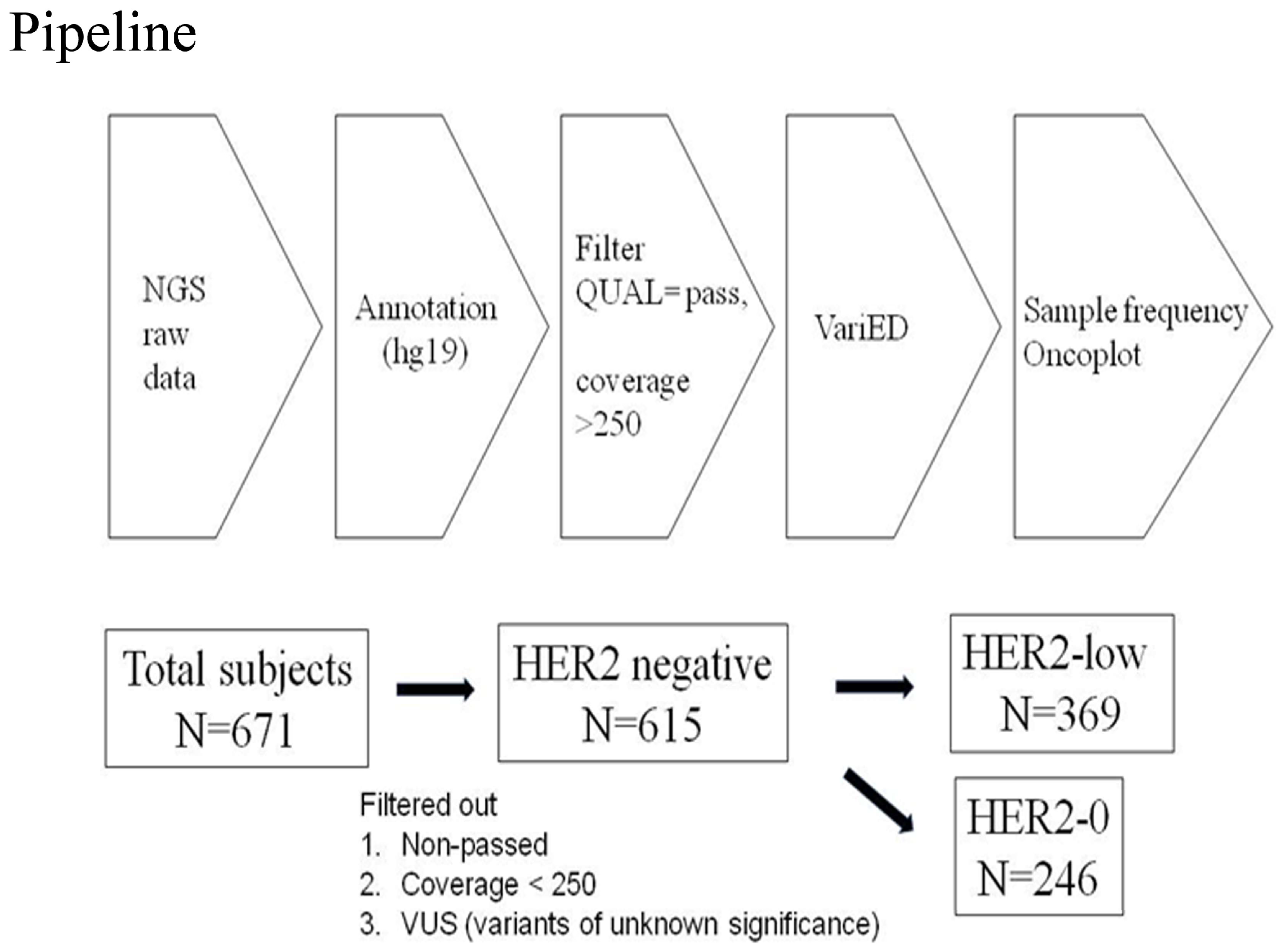
Amplicon libraries were constructed using multiplex PCR primers after the preparation of DNA from the FFPE samples. Sequencing was performed using Ion Gene Studio S5 System and Ion 540 Chips. Raw data processing, alignment, and variant calling were performed using v3–w3.2–DNA and Fusions–Single Sample version 5.10, with the variant calling by the Torrent Variant Caller plug-in. Further management involved Ion ReporterTM Software with the workflow “Oncomine Comprehensive v3–w3.2–DNA and Fusions–Single Sample” version 5.10 being selected and the filter chain “Oncomine Variants” version 5.10 being applied [7]. The pipeline of next-generation sequencing analysis in this study is described in Figure 4.
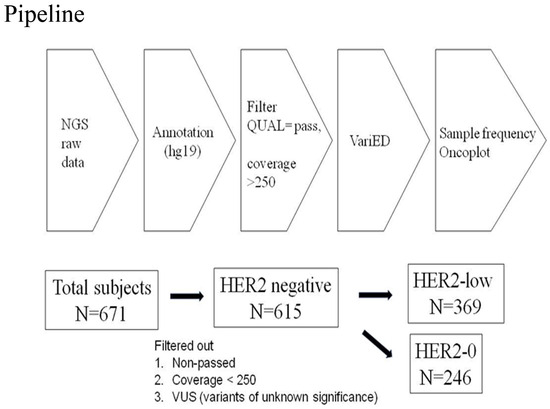
Figure 4.
Pipeline of next-generation sequencing analysis in this study. From 2018-1 to 2020-1, a total of 671 patients with HER2-negative tumors were included. The raw data of NGS were annotated usinghg19 human genome reference and VariED database. The statistical significance of sample frequency of mutations was selected between HER2-0 andHER2-low groups with a p value < 0.05 via Chi-square test or Fisher’s exact test. Among 671 tumors receiving NGS target sequencing, those data with non-pass quality, coverage < 250, and variants with unknown significance were excluded. Finally, 615 patients with HER2-negative tumors were categorized into HER2-0 (n = 246) and HER2-low (n = 369) groups. The definition of HER2-0 and HER2-low was HER2-0, immunohistochemistry score (IHC) 0, and HER2-low, IHC score 1+/2+, and FISH-negative, respectively.
4.6. Statistical Methods
Descriptive statistical analysis was applied to compare baseline characteristics among the risk assessment and clinicopathological features. Categorical data were summarized in counts and percentages. The Chi-Square test or Fisher’s exact test was used to compare the distributions of categorical variables. A two-sided p-value of <0.05 was considered statistically significant. All statistical analyses were performed using Python software (version 3.9.6. Python Software Foundation, Wilmington, Del; open source; https://www.python.org/ (accessed on 8 July 2023))
5. Conclusions
We conclude that there are different clinical manifestations present in the HER2-0 group and the HER2-low group. There is also a significantly higher frequency of PIK3CA_SNA in the HER2-low group than in the HER2-0 group. It is suggested that the early detection of the presence of a PIK3CA mutation is important and might help therapeutic decision making in patients with HER2-low tumors.
Supplementary Materials
The supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms25021318/s1.
Author Contributions
J.-H.C. and Y.-F.T. designed the study. Y.-F.T. drafted the manuscript. C.-Y.H. reviewed pathological slides. C.-C.H., J.-I.L., C.-Y.L., T.-C.C., Y.-S.L., Y.-F.T., C.-J.F., P.-J.L. and L.-M.T. collected the subject’s data. J.-H.C., C.-Y.L. and L.-M.T. supervised the research. C.-Y.L. and J.-H.C. took full responsibility for execution and finalized the submitted manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Taipei Veterans General Hospital (V112DHA0100359) and the Melissa Lee Cancer Foundation (MLCF_V112_A11204).
Institutional Review Board Statement
This work has been approved by the Institutional Review Board of Taipei Veterans General Hospital (2023-06-025BC).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Acknowledgments
The establishment of this database was exclusively sponsored by the YongLinHeathcare Foundation under clinical study protocol (# QCR18002). The authors would like to thank Morris Chang (ABMRD002) and Shih-Hsiang Chou for their data collection and running the statistical analysis.
Conflicts of Interest
The authors confirm that they have read BioMed Central’s guidance on competing interests and have included a statement indicating that none of the authors have any competing interests in the manuscript.
Abbreviations
| ADC | antibody–drug conjugate |
| CNA | copy number alteration |
| ER | estrogen receptor |
| HER2 | human epidermal growth factor receptor 2 |
| IHC | immunohistochemistry |
| Indel | insertion/deletion |
| ISH | in situ hybridization |
| NGS | next-generation sequencing |
| pCR | pathological complete response |
| PR | progesterone receptor |
| SA | structure alteration |
| SNA | single nucleotide alteration |
| T-DXd | Trastuzumab deruxtecan |
| VUS | variants of unknown significance |
References
- Tarantino, P.; Curigliano, G.; Tolaney, S.M. Navigating the HER2-Low Paradigm in Breast Oncology: New Standards, Future Horizons. Cancer Discov. 2022, 12, 2026–2030. [Google Scholar] [CrossRef] [PubMed]
- Fehrenbacher, L.; Cecchini, R.S.; Geyer, C.E., Jr.; Rastogi, P.; Costantino, J.P.; Atkins, J.N.; Crown, J.P.; Polikoff, J.; Boileau, J.F.; Provencher, L.; et al. NSABP B-47/NRG Oncology Phase III Randomized Trial Comparing Adjuvant Chemotherapy With or Without Trastuzumab in High-Risk Invasive Breast Cancer Negative for HER2 by FISH and With IHC 1+ or 2. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Schettini, F.; Chic, N.; Brasó-Maristany, F.; Paré, L.; Pascual, T.; Conte, B.; Martínez-Sáez, O.; Adamo, B.; Vidal, M.; Barnadas, E.; et al. Clinical, pathological, and PAM50 gene expression features of HER2-low breast cancer. NPJ Breast Cancer 2021, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, P.; Hamilton, E.; Tolaney, S.M.; Cortes, J.; Morganti, S.; Ferraro, E.; Marra, A.; Viale, G.; Trapani, D.; Cardoso, F.; et al. HER2-Low Breast Cancer: Pathological and Clinical Landscape. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 1951–1962. [Google Scholar] [CrossRef]
- Modi, S.; Jacot, W.; Yamashita, T.; Sohn, J.; Vidal, M.; Tokunaga, E.; Tsurutani, J.; Ueno, N.T.; Prat, A.; Chae, Y.S.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N. Engl. J. Med. 2022, 387, 9–20. [Google Scholar] [CrossRef]
- Huang, C.-C.; Tsai, Y.-F.; Liu, C.-Y.; Chao, T.-C.; Lien, P.-J.; Lin, Y.-S.; Feng, C.-J.; Chiu, J.-H.; Hsu, C.-Y.; Tseng, L.-M. Comprehensive molecular profiling of Taiwanese breast cancers revealed potential therapeutic targets: Prevalence of actionable mutations among 380 targeted sequencing analyses. Res. Sq. 2020, 21, 1–14. [Google Scholar] [CrossRef]
- Huang, C.C.; Tsai, Y.F.; Liu, C.Y.; Lien, P.J.; Lin, Y.S.; Chao, T.C.; Feng, C.J.; Chen, Y.J.; Lai, J.I.; Phan, N.N.; et al. Prevalence of Tumor Genomic Alterations in Homologous Recombination Repair Genes Among Taiwanese Breast Cancers. Ann. Surg. Oncol. 2022, 29, 3578–3590. [Google Scholar] [CrossRef]
- Tarantino, P.; Jin, Q.; Tayob, N.; Jeselsohn, R.M.; Schnitt, S.J.; Vincuilla, J.; Parker, T.; Tyekucheva, S.; Li, T.; Lin, N.U.; et al. Prognostic and Biologic Significance of ERBB2-Low Expression in Early-Stage Breast Cancer. JAMA Oncol. 2022, 8, 1177–1183. [Google Scholar] [CrossRef]
- Modi, S.; Niikura, N.; Yamashita, T.; Jacot, W.; Sohn, J.; Tokunaga, E.; Vidal, M.J.; Park, Y.H.; Lee, K.S.; Chae, Y.; et al. Trastuzumab deruxtecan (T-DXd) vs treatment of physician’s choice (TPC) in patients (pts) with HER2-low, hormone receptor-positive (HR+) unresectable and/or metastatic breast cancer (mBC): Exploratory biomarker analysis of DESTINY-Breast04. J. Clin. Oncol. 2023, 41 (Suppl. S16), 1020. [Google Scholar] [CrossRef]
- Mosele, F.; Deluche, E.; Lusque, A.; Le Bescond, L.; Filleron, T.; Pradat, Y.; Ducoulombier, A.; Pistilli, B.; Bachelot, T.; Viret, F.; et al. Trastuzumab deruxtecan in metastatic breast cancer with variable HER2 expression: The phase 2 DAISY trial. Nat. Med. 2023, 29, 2110–2120. [Google Scholar] [CrossRef] [PubMed]
- Razandi, M.; Pedram, A.; Park, S.T.; Levin, E.R. Proximal events in signaling by plasma membrane estrogen receptors. J. Biol. Chem. 2003, 278, 2701–2712. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Massarweh, S.; Osborne, C.K.; Wakeling, A.E.; Ali, S.; Weiss, H.; Schiff, R. Mechanisms of tamoxifen resistance: Increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J. Natl. Cancer Inst. 2004, 96, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Arpino, G.; Wiechmann, L.; Osborne, C.K.; Schiff, R. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: Molecular mechanism and clinical implications for endocrine therapy resistance. Endocr. Rev. 2008, 29, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, M.; Hu, H.; Wang, Y.C.; Fu, X.; Nardone, A.; Herrera, S.; Mao, S.; Contreras, A.; Gutierrez, C.; Wang, T.; et al. Upregulation of ER Signaling as an Adaptive Mechanism of Cell Survival in HER2-Positive Breast Tumors Treated with Anti-HER2 Therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 3995–4003. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, Y.; Warden, C.; Chen, S. Cross-talk between ER and HER2 regulates c-MYC-mediated glutamine metabolism in aromatase inhibitor resistant breast cancer cells. J. Steroid Biochem. Mol. Biol. 2015, 149, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Viganò, L.; Locatelli, A.; Ulisse, A.; Galbardi, B.; Dugo, M.; Tosi, D.; Tacchetti, C.; Daniele, T.; Győrffy, B.; Sica, L.; et al. Modulation of the Estrogen/erbB2 Receptors Cross-talk by CDK4/6 Inhibition Triggers Sustained Senescence in Estrogen Receptor- and ErbB2-positive Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2022, 28, 2167–2179. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.T.; Asthana, S.; Gao, S.P.; Lee, B.H.; Chapman, J.S.; Kandoth, C.; Gao, J.; Socci, N.D.; Solit, D.B.; Olshen, A.B.; et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 2016, 34, 155–163. [Google Scholar] [CrossRef]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef]
- Samuels, Y.; Waldman, T. Oncogenic mutations of PIK3CA in human cancers. Curr. Top. Microbiol. Immunol. 2010, 347, 21–41. [Google Scholar]
- Martínez-Sáez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; González-Farré, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Brasó-Maristany, F.; et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. BCR 2020, 22, 45. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.C.; Tsai, Y.F.; Liu, C.Y.; Lien, P.J.; Lin, Y.S.; Feng, C.J.; Chen, Y.J.; Lai, J.I.; Hsu, C.Y.; Lynn, J.J.; et al. Prevalence of PIK3CA mutations in Taiwanese patients with breast cancer: A retrospective next-generation sequencing database analysis. Front. Oncol. 2023, 13, 1192946. [Google Scholar] [CrossRef] [PubMed]
- Pavithran, K.; Jayamohanan, H.; Jose, W.M.; Soman, S.; Vijaykumar, D.K.; Ariyannur, P.S. 256P PI3K mutation is associated with reduced sensitivity to CDK4/6 inhibitors in metastatic breast cancer. Ann. Oncol. 2021, 32, S471. [Google Scholar] [CrossRef]
- Del Re, M.; Crucitta, S.; Lorenzini, G.; De Angelis, C.; Diodati, L.; Cavallero, D.; Bargagna, I.; Cinacchi, P.; Fratini, B.; Salvadori, B.; et al. PI3K mutations detected in liquid biopsy are associated to reduced sensitivity to CDK4/6 inhibitors in metastatic breast cancer patients. Pharmacol. Res. 2021, 163, 105241. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Hurvitz, S.; Fasolo, A.; Tseng, L.M.; Jerusalem, G.; Wilks, S.; O’Regan, R.; Isaacs, C.; Toi, M.; Burris, H.; et al. Molecular Alterations and Everolimus Efficacy in Human Epidermal Growth Factor Receptor 2-Overexpressing Metastatic Breast Cancers: Combined Exploratory Biomarker Analysis From BOLERO-1 and BOLERO-3. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016, 34, 2115–2124. [Google Scholar] [CrossRef] [PubMed]
- Papewalis, J.; Nikitin, A.; Rajewsky, M.F. G to A polymorphism at amino acid codon 655 of the human erbB-2/HER2 gene. Nucleic Acids Res. 1991, 19, 5452. [Google Scholar] [CrossRef] [PubMed]
- Ehsani, A.; Low, J.; Wallace, R.B.; Wu, A.M. Characterization of a New Allele of the Human ERBB2 Gene by Allele-Specific Competition Hybridization. Genomics 1993, 15, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Huang, C.C.; Tsai, Y.F.; Chao, T.C.; Lien, P.J.; Lin, Y.S.; Feng, C.J.; Chen, J.L.; Chen, Y.J.; Chiu, J.H.; et al. VGH-TAYLOR: Comprehensive precision medicine study protocol on the heterogeneity of Taiwanese breast cancer patients. Future Oncol. 2021, 17, 4057–4069. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).