MicroRNA Profiling of the Inflammatory Response after Early and Late Asthmatic Reaction
Abstract
1. Introduction
2. Results
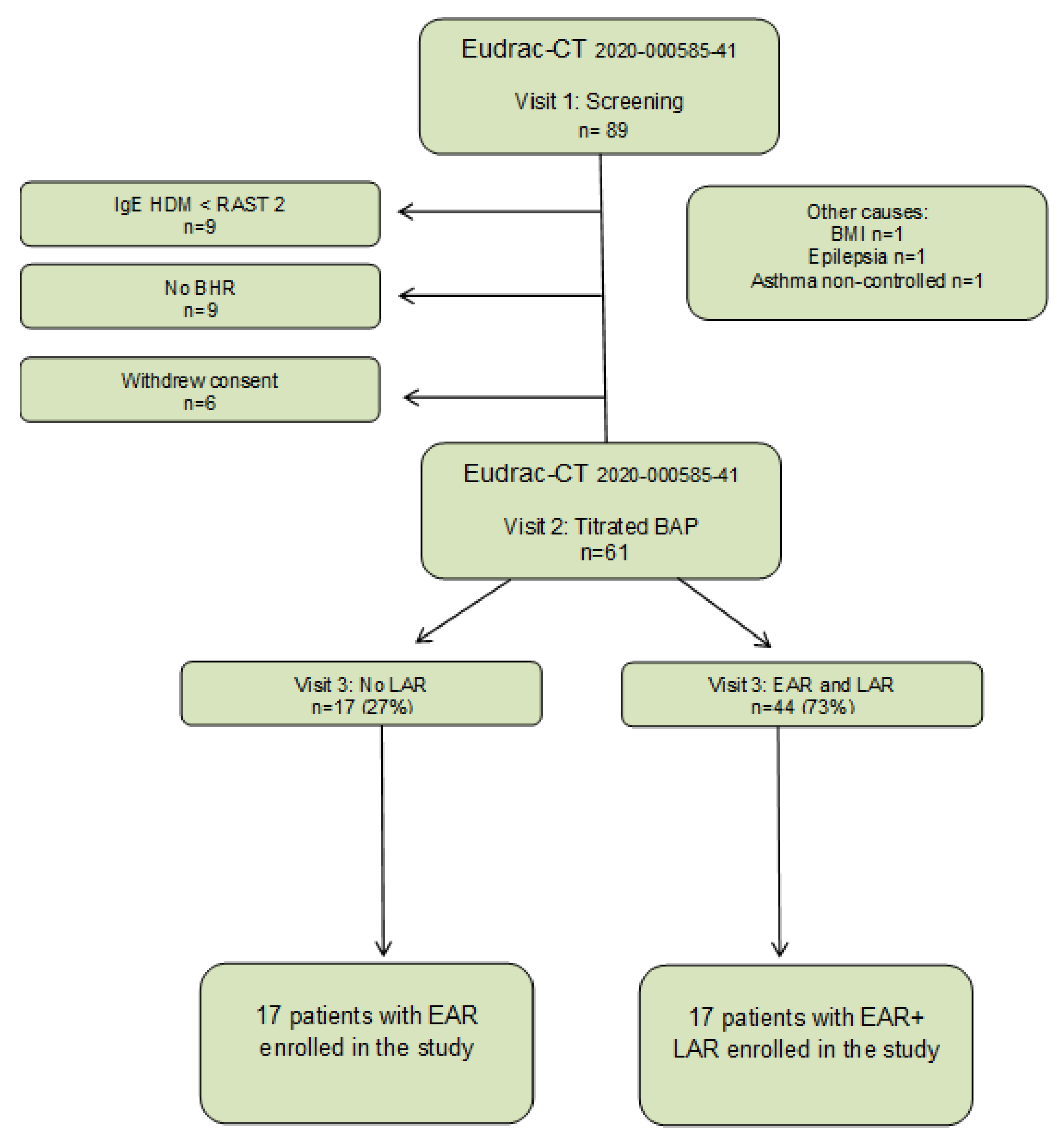
2.1. Bronchial Allergen Provocation
2.2. miRNAs
2.2.1. Influence of BAP on miRNA Expression
2.2.2. KEGG-Pathway and Target Analysis
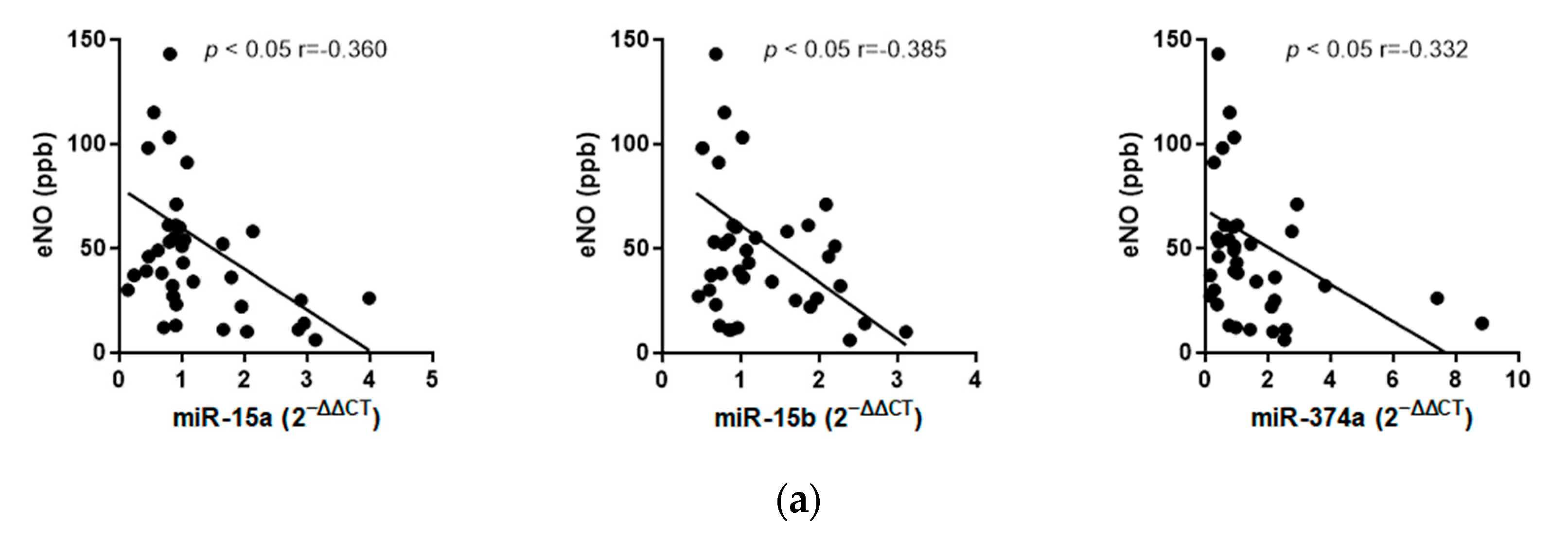
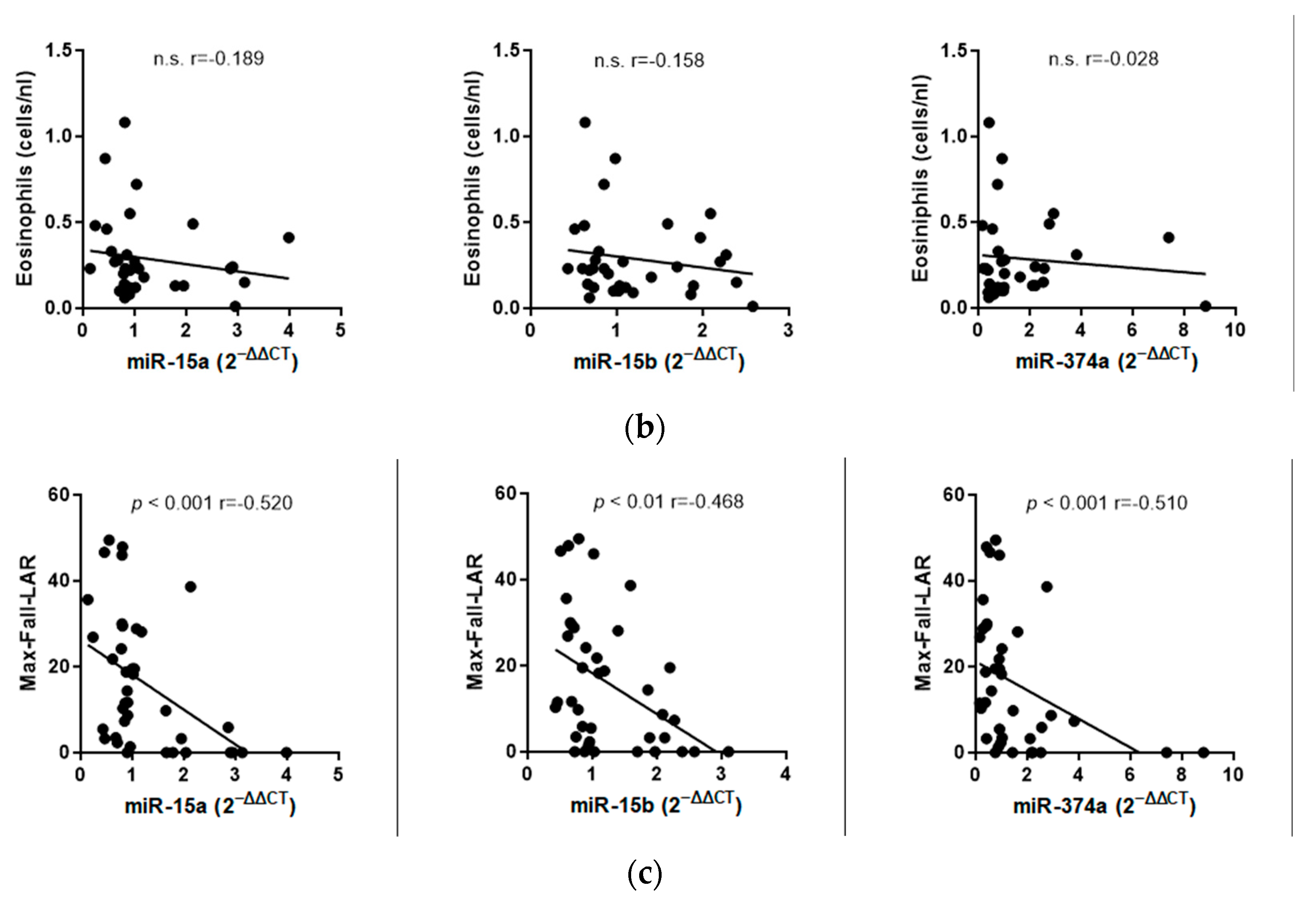
2.2.3. Correlation Analyses
3. Discussion
4. Materials and Methods
4.1. Study Design
- Written informed consent from study participants;
- Age between 18 and 65 years;
- Known sensitization to HDM (RAST class > 2);
- Bronchial hyperreactivity (BHR) measured by methacholine challenge PD 20 FEV1 < 2 mg methacholine;
- BMI < 30.
- Age < 18 years;
- Lung function FVC < 80% and FEV1 < 75%;
- Other chronic diseases or infections (e.g., HIV, TB);
- Pregnancy;
- Treatment with inhaled and systemic corticosteroids;
- Known alcohol, drug, and/or medication abuse;
- Inability to understand the scope of the study.
4.2. Skin Prick Test
4.3. Measurement of Exhaled NO
4.4. Pulmonary Function Testing
4.5. Bronchial Allergen Provocation
4.6. Laboratoy Measurements
4.7. miRNA and Next-Generation Sequencing
4.8. miRNA Validation by Taqman qPCR
4.9. KEGG-Pathway and Target Analysis
4.10. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schulze, J.; Agache, I.; Eguiluz-Gracia, I.; Trischler, J.; Zielen, S. Medical algorithm: Diagnosis and treatment of house dust mite-driven allergic asthma. Allergy 2023, 78, 1397–1399. [Google Scholar] [CrossRef] [PubMed]
- Schatz, M.; Rosenwasser, L. The allergic asthma phenotype. J. Allergy Clin. Immunol. Pract. 2014, 2, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Boulet, L.P.; Cartier, A.; Thomson, N.C.; Roberts, R.S.; Dolovich, J.; Hargreave, F.E. Asthma and increases in nonallergic bronchial responsiveness from seasonal pollen exposure. J. Allergy Clin. Immunol. 1983, 71, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. New concepts in the pathogenesis of bronchial hyperresponsiveness and asthma. J. Allergy Clin. Immunol. 1989, 83, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Gauvreau, G.M.; El-Gammal, A.I.; O’Byrne, P.M. Allergen-induced airway responses. Eur. Respir. J. 2015, 46, 819. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.; Voss, S.; Zissler, U.; Rose, M.A.; Zielen, S.; Schubert, R. Airway responses and inflammation in subjects with asthma after four days of repeated high-single-dose allergen challenge. Respir. Res. 2012, 13, 78. [Google Scholar] [CrossRef] [PubMed]
- O’Hollaren, M.T.; Yunginger, J.W.; Offord, K.P.; Somers, M.J.; O’Connell, E.J.; Ballard, D.J.; Sachs, M.I. Exposure to an aeroallergen as a possible precipitating factor in respiratory arrest in young patients with asthma. N. Engl. J. Med. 1991, 324, 359–363. [Google Scholar] [CrossRef]
- Salo, P.M.; Arbes, S.J.; Sever, M.; Jaramillo, R.; Cohn, R.D.; London, S.J.; Zeldin, D.C. Exposure to Alternaria alternata in US homes is associated with asthma symptoms. J. Allergy Clin. Immunol. 2006, 118, 892–898. [Google Scholar] [CrossRef]
- Harun, N.-S.; Lachapelle, P.; Douglass, J. Thunderstorm-triggered asthma: What we know so far. J. Asthma Allergy 2019, 12, 101–108. [Google Scholar] [CrossRef]
- Antó, J.M.; Sunyer, J.; Rodriguez-Roisin, R.; Suarez-Cervera, M.; Vazquez, L. Community outbreaks of asthma associated with inhalation of soybean dust. Toxicoepidemiological Committee. N. Engl. J. Med. 1989, 320, 1097–1102. [Google Scholar] [CrossRef]
- Boulet, L.-P.; Gauvreau, G.; Boulay, M.-E.; O’Byrne, P.; Cockcroft, D.W. The allergen bronchoprovocation model: An important tool for the investigation of new asthma anti-inflammatory therapies. Allergy 2007, 62, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Tsai, M.; Piliponsky, A.M. The development of allergic inflammation. Nature 2008, 454, 445–454. [Google Scholar] [CrossRef]
- Locksley, R.M. Asthma and allergic inflammation. Cell 2010, 140, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Elieh Ali Komi, D.; Bjermer, L. Mast Cell-Mediated Orchestration of the Immune Responses in Human Allergic Asthma: Current Insights. Clin. Rev. Allergy Immunol. 2019, 56, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jarjour, N.N.; Busse, W.W.; Kelly, E.A.B. Enhanced generation of helper T type 1 and 2 chemokines in allergen-induced asthma. Am. J. Respir. Crit. Care Med. 2004, 169, 1118–1124. [Google Scholar] [CrossRef] [PubMed]
- Thunberg, S.; Gafvelin, G.; Nord, M.; Grönneberg, R.; Grunewald, J.; Eklund, A.; van Hage, M. Allergen provocation increases TH2-cytokines and FOXP3 expression in the asthmatic lung. Allergy 2010, 65, 311–318. [Google Scholar] [CrossRef] [PubMed]
- De Monchy, J.G.; Kauffman, H.F.; Venge, P.; Koëter, G.H.; Jansen, H.M.; Sluiter, H.J.; De Vries, K. Bronchoalveolar eosinophilia during allergen-induced late asthmatic reactions. Am. Rev. Respir. Dis. 1985, 131, 373–376. [Google Scholar]
- Cartier, A.; Thomson, N.C.; Frith, P.A.; Roberts, R.; Hargreave, F.E. Allergen-induced increase in bronchial responsiveness to histamine: Relationship to the late asthmatic response and change in airway caliber. J. Allergy Clin. Immunol. 1982, 70, 170–177. [Google Scholar] [CrossRef]
- Chapman, D.G.; Irvin, C.G. Mechanisms of airway hyper-responsiveness in asthma: The past, present and yet to come. Clin. Exp. Allergy 2015, 45, 706–719. [Google Scholar] [CrossRef]
- Cockcroft, D.W. Airway hyperresponsiveness and late asthmatic responses. Chest 1988, 94, 178–180. [Google Scholar] [CrossRef]
- Holt, P.G.; Sly, P.D. Th2 cytokines in the asthma late-phase response. Lancet 2007, 370, 1396–1398. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, J.; Jeffery, P.K.; Busse, W.W.; Johnson, M.; Vignola, A.M. Asthma. From bronchoconstriction to airways inflammation and remodeling. Am. J. Respir. Crit. Care Med. 2000, 161, 1720–1745. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Shannon, C.P.; Kim, Y.W.; Yang, C.X.; Balshaw, R.; Cohen Freue, G.V.; Gauvreau, G.M.; FitzGerald, J.M.; Boulet, L.-P.; O’Byrne, P.M.; et al. Novel Blood-based Transcriptional Biomarker Panels Predict the Late-Phase Asthmatic Response. Am. J. Respir. Crit. Care Med. 2018, 197, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Cañas, J.A.; Rodrigo-Muñoz, J.M.; Sastre, B.; Gil-Martinez, M.; Redondo, N.; Del Pozo, V. MicroRNAs as Potential Regulators of Immune Response Networks in Asthma and Chronic Obstructive Pulmonary Disease. Front. Immunol. 2020, 11, 608666. [Google Scholar] [CrossRef] [PubMed]
- Ntontsi, P.; Photiades, A.; Zervas, E.; Xanthou, G.; Samitas, K. Genetics and Epigenetics in Asthma. Int. J. Mol. Sci. 2021, 22, 2412. [Google Scholar] [CrossRef]
- Bélanger, É.; Laprise, C. Could the Epigenetics of Eosinophils in Asthma and Allergy Solve Parts of the Puzzle? Int. J. Mol. Sci. 2021, 22, 8921. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Duecker, R.P.; Adam, E.H.; Wirtz, S.; Gronau, L.; Khodamoradi, Y.; Eberhardt, F.J.; Donath, H.; Gutmann, D.; Vehreschild, M.J.G.T.; Zacharowski, K.; et al. The MiR-320 Family Is Strongly Downregulated in Patients with COVID-19 Induced Severe Respiratory Failure. Int. J. Mol. Sci. 2021, 22, 10351. [Google Scholar] [CrossRef]
- Davis, J.S.; Sun, M.; Kho, A.T.; Moore, K.G.; Sylvia, J.M.; Weiss, S.T.; Lu, Q.; Tantisira, K.G. Circulating microRNAs and association with methacholine PC20 in the Childhood Asthma Management Program (CAMP) cohort. PLoS ONE 2017, 12, e0180329. [Google Scholar] [CrossRef]
- Kho, A.T.; McGeachie, M.J.; Moore, K.G.; Sylvia, J.M.; Weiss, S.T.; Tantisira, K.G. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir. Res. 2018, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Pinkerton, M.; Chinchilli, V.; Banta, E.; Craig, T.; August, A.; Bascom, R.; Cantorna, M.; Harvill, E.; Ishmael, F.T. Differential expression of microRNAs in exhaled breath condensates of patients with asthma, patients with chronic obstructive pulmonary disease, and healthy adults. J. Allergy Clin. Immunol. 2013, 132, 217–219. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.; Rosewich, M.; Dressler, M.; Riemer, C.; Rose, M.A.; Zielen, S. Bronchial allergen challenge using the Medicaid dosimeter. Int. Arch. Allergy Immunol. 2012, 157, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Agache, I.; Antolin-Amerigo, D.; de Blay, F.; Boccabella, C.; Caruso, C.; Chanez, P.; Couto, M.; Covar, R.; Doan, S.; Fauquert, J.; et al. EAACI position paper on the clinical use of the bronchial allergen challenge: Unmet needs and research priorities. Allergy 2022, 77, 1667–1684. [Google Scholar] [CrossRef] [PubMed]
- Kharitonov, S.A.; O’Connor, B.J.; Evans, D.J.; Barnes, P.J. Allergen-induced late asthmatic reactions are associated with elevation of exhaled nitric oxide. Am. J. Respir. Crit. Care Med. 1995, 151, 1894–1899. [Google Scholar] [CrossRef] [PubMed]
- Durham, S.R. The significance of late responses in asthma. Clin. Exp. Allergy 1991, 21, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Cieslewicz, G.; Tomkinson, A.; Adler, A.; Duez, C.; Schwarze, J.; Takeda, K.; Larson, K.A.; Lee, J.J.; Irvin, C.G.; Gelfand, E.W. The late, but not early, asthmatic response is dependent on IL-5 and correlates with eosinophil infiltration. J. Clin. Investig. 1999, 104, 301–308. [Google Scholar] [CrossRef]
- Lopuhaä, C.E.; Out, T.A.; Jansen, H.M.; Aalberse, R.C.; van der Zee, J.S. Allergen-induced bronchial inflammation in house dust mite-allergic patients with or without asthma. Clin. Exp. Allergy 2002, 32, 1720–1727. [Google Scholar] [CrossRef]
- Dente, F.L.; Bacci, E.; Bartoli, M.L.; Cianchetti, S.; Di Franco, A.; Costa, F.; Vagaggini, B.; Paggiaro, P.L. Magnitude of late asthmatic response to allergen in relation to baseline and allergen-induced sputum eosinophilia in mild asthmatic patients. Ann. Allergy Asthma Immunol. 2008, 100, 457–462. [Google Scholar] [CrossRef]
- Pelikan, Z. Effects of inhaled corticosteroids on the dual asthmatic response. Allergy Asthma Proc. 2013, 34, e47–e58. [Google Scholar] [CrossRef]
- Leckie, M.J.; Brinke, A.T.; Khan, J.; Diamant, Z.; O’Connor, B.J.; Walls, C.M.; Mathur, A.K.; Cowley, H.C.; Chung, K.F.; Djukanovic, R.; et al. Effects of an interleukin-5 blocking monoclonal antibody on eosinophils, airway hyper-responsiveness, and the late asthmatic response. Lancet 2000, 356, 2144–2148. [Google Scholar] [CrossRef] [PubMed]
- Trischler, J.; Lieb, A.; Arnold, M.; Schulze, J.; Rosewich, M.; Schubert, R.; Bottoli, I.; Zielen, S. Omalizumab effectively protects against early and late allergic responses in asthma after 4 weeks. Allergy 2017, 72, 1912–1915. [Google Scholar] [CrossRef] [PubMed]
- Weidner, J.; Bartel, S.; Kılıç, A.; Zissler, U.M.; Renz, H.; Schwarze, J.; Schmidt-Weber, C.B.; Maes, T.; Rebane, A.; Krauss-Etschmann, S.; et al. Spotlight on microRNAs in allergy and asthma. Allergy 2021, 76, 1661–1678. [Google Scholar] [CrossRef] [PubMed]
- Kho, A.T.; Sharma, S.; Davis, J.S.; Spina, J.; Howard, D.; McEnroy, K.; Moore, K.; Sylvia, J.; Qiu, W.; Weiss, S.T.; et al. Circulating MicroRNAs: Association with Lung Function in Asthma. PLoS ONE 2016, 11, e0157998. [Google Scholar] [CrossRef] [PubMed]
- Taka, S.; Tzani-Tzanopoulou, P.; Wanstall, H.; Papadopoulos, N.G. MicroRNAs in Asthma and Respiratory Infections: Identifying Common Pathways. Allergy Asthma Immunol. Res. 2020, 12, 4–23. [Google Scholar] [CrossRef] [PubMed]
- Hirai, K.; Shirai, T.; Shimoshikiryo, T.; Ueda, M.; Gon, Y.; Maruoka, S.; Itoh, K. Circulating microRNA-15b-5p as a biomarker for asthma-COPD overlap. Allergy 2021, 76, 766–774. [Google Scholar] [CrossRef]
- Liu, X.; Wang, L.; Li, H.; Lu, X.; Hu, Y.; Yang, X.; Huang, C.; Gu, D. Coactivator-associated arginine methyltransferase 1 targeted by miR-15a regulates inflammation in acute coronary syndrome. Atherosclerosis 2014, 233, 349–356. [Google Scholar] [CrossRef]
- Taheri, M.; Barth, D.A.; Kargl, J.; Rezaei, O.; Ghafouri-Fard, S.; Pichler, M. Emerging Role of Non-Coding RNAs in Regulation of T-Lymphocyte Function. Front. Immunol. 2021, 12, 756042. [Google Scholar] [CrossRef]
- Wei, Y.; Han, B.; Dai, W.; Guo, S.; Zhang, C.; Zhao, L.; Gao, Y.; Jiang, Y.; Kong, X. Exposure to ozone impacted Th1/Th2 imbalance of CD(4+) T cells and apoptosis of ASMCs underlying asthmatic progression by activating lncRNA PVT1-miR-15a-5p/miR-29c-3p signaling. Aging 2020, 12, 25229–25255. [Google Scholar] [CrossRef]
- Doumatey, A.P.; He, W.J.; Gaye, A.; Lei, L.; Zhou, J.; Gibbons, G.H.; Adeyemo, A.; Rotimi, C.N. Circulating MiR-374a-5p is a potential modulator of the inflammatory process in obesity. Sci. Rep. 2018, 8, 7680. [Google Scholar] [CrossRef]
- Shen, J.; Ma, X. miR-374a-5p alleviates sepsis-induced acute lung injury by targeting ZEB1 via the p38 MAPK pathway. Exp. Ther. Med. 2022, 24, 564. [Google Scholar] [CrossRef] [PubMed]
- Weidner, J.; Ekerljung, L.; Malmhäll, C.; Miron, N.; Rådinger, M. Circulating microRNAs correlate to clinical parameters in individuals with allergic and non-allergic asthma. Respir. Res. 2020, 21, 107. [Google Scholar] [CrossRef] [PubMed]
- Hossny, E.; El-Awady, H.; Bakr, S.; Labib, A. Vascular endothelial growth factor overexpression in induced sputum of children with bronchial asthma. Pediatr. Allergy Immunol. 2009, 20, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Inoue, Y.; Shimojo, N.; Yamaide, F.; Morita, Y.; Arima, T.; Tomiita, M.; Kohno, Y. Lower levels of hsa-mir-15a, which decreases VEGFA, in the CD4+ T cells of pediatric patients with asthma. J. Allergy Clin. Immunol. 2013, 132, 1224–1227.e12. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qiao, J. Protein-protein interaction network analysis and identifying regulation microRNAs in asthmatic children. Allergol. Immunopathol. 2015, 43, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Kierbiedź-Guzik, N.; Sozańska, B. miRNAs as Modern Biomarkers in Asthma Therapy. Int. J. Mol. Sci. 2023, 24, 11499. [Google Scholar] [CrossRef] [PubMed]
- Milger, K.; Götschke, J.; Krause, L.; Nathan, P.; Alessandrini, F.; Tufman, A.; Fischer, R.; Bartel, S.; Theis, F.J.; Behr, J.; et al. Identification of a plasma miRNA biomarker signature for allergic asthma: A translational approach. Allergy 2017, 72, 1962–1971. [Google Scholar] [CrossRef]
- Coskunpinar, E.; Akcesme, B.; Tas, S.K.; Aynaci, A. Investigation of miRNAs that are effective in the pathogenesis of asthma. J. Asthma 2023, 60, 2145–2152. [Google Scholar] [CrossRef]
- Hur, G.Y.; Broide, D.H. Genes and Pathways Regulating Decline in Lung Function and Airway Remodeling in Asthma. Allergy Asthma Immunol. Res. 2019, 11, 604–621. [Google Scholar] [CrossRef]
- Fang, L.; Roth, M.; S’ng, C.T.; Tamm, M.; Han, B.; Hoang, B.X. Zinc salicylate reduces airway smooth muscle cells remodelling by blocking mTOR and activating p21((Waf1/Cip1)). J. Nutr. Biochem. 2021, 89, 108563. [Google Scholar] [CrossRef]
- Qiao, L.; Xu, Y.; Liu, X.; Xie, J.; Wang, J.; Du, C.; Zhang, J.; Ni, W.; Chen, S. Role of protein kinase C alpha and cyclin D1 in the proliferation of airway smooth muscle in asthmatic rats. Chin. Med. J. 2008, 121, 2070–2076. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-H.; Chiu, K.-L.; Hsia, T.-C.; Shen, T.-C.; Chen, L.-H.; Yu, C.-C.; Mong, M.-C.; Chang, W.-S.; Tsai, C.-W.; Bau, D.-T. Significant Association of Cyclin D1 Promoter Genotypes With Asthma Susceptibility in Taiwan. Vivo 2021, 35, 2041–2046. [Google Scholar] [CrossRef] [PubMed]
- Noori, M.S.; Bhatt, P.M.; Courreges, M.C.; Ghazanfari, D.; Cuckler, C.; Orac, C.M.; McMills, M.C.; Schwartz, F.L.; Deosarkar, S.P.; Bergmeier, S.C.; et al. Identification of a novel selective and potent inhibitor of glycogen synthase kinase-3. Am. J. Physiol. Cell Physiol. 2019, 317, C1289–C1303. [Google Scholar] [CrossRef] [PubMed]
- Bentley, J.K.; Deng, H.; Linn, M.J.; Lei, J.; Dokshin, G.A.; Fingar, D.C.; Bitar, K.N.; Henderson, W.R.; Hershenson, M.B.; Prakash, Y.S.; et al. Airway smooth muscle hyperplasia and hypertrophy correlate with glycogen synthase kinase-3(beta) phosphorylation in a mouse model of asthma. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 296, L176–L184. [Google Scholar] [CrossRef]
- Bao, Z.; Lim, S.; Liao, W.; Lin, Y.; Thiemermann, C.; Leung, B.P.; Wong, W.S.F. Glycogen synthase kinase-3beta inhibition attenuates asthma in mice. Am. J. Respir. Crit. Care Med. 2007, 176, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Cui, J.; Zhang, Y.; Zheng, Z.; Meng, J.; Du, J. MicroRNA-15b-5p inhibits tumor necrosis factor alpha-induced proliferation, migration, and extracellular matrix production of airway smooth muscle cells via targeting yes-associated protein 1. Bioengineered 2022, 13, 5396–5406. [Google Scholar] [CrossRef]
- Li, Z.; Liu, L.; Du, C.; Yu, Z.; Yang, Y.; Xu, J.; Wei, X.; Zhan, F.; Lai, Y.; Qiu, L.; et al. Therapeutic effects of oligo-single-stranded DNA mimicking of hsa-miR-15a-5p on multiple myeloma. Cancer Gene Ther. 2020, 27, 869–877. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, J.; Zhao, M.; Luo, X.; Liang, Z.; Zhen, Y.; Fu, Q.; Deng, X.; Lin, X.; Li, L.; et al. microRNA-374a suppresses colon cancer progression by directly reducing CCND1 to inactivate the PI3K/AKT pathway. Oncotarget 2016, 7, 41306–41319. [Google Scholar] [CrossRef]
- Sun, J.; Yan, P.; Chen, Y.; Chen, Y.; Yang, J.; Xu, G.; Mao, H.; Qiu, Y. MicroRNA-26b inhibits cell proliferation and cytokine secretion in human RASF cells via the Wnt/GSK-3β/β-catenin pathway. Diagn. Pathol. 2015, 10, 72. [Google Scholar] [CrossRef]
- Kabesch, M.; Tost, J. Recent findings in the genetics and epigenetics of asthma and allergy. Semin. Immunopathol. 2020, 42, 43–60. [Google Scholar] [CrossRef]
- Solberg, O.D.; Ostrin, E.J.; Love, M.I.; Peng, J.C.; Bhakta, N.R.; Hou, L.; Nguyen, C.; Solon, M.; Nguyen, C.; Barczak, A.J.; et al. Airway epithelial miRNA expression is altered in asthma. Am. J. Respir. Crit. Care Med. 2012, 186, 965–974. [Google Scholar] [CrossRef]
- Elbehidy, R.M.; Youssef, D.M.; El-Shal, A.S.; Shalaby, S.M.; Sherbiny, H.S.; Sherief, L.M.; Akeel, N.E. MicroRNA-21 as a novel biomarker in diagnosis and response to therapy in asthmatic children. Mol. Immunol. 2016, 71, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Zielen, S.; Lieb, A.; De La Motte, S.; Wagner, F.; de Monchy, J.; Fuhr, R.; Munzu, C.; Koehne-Voss, S.; Rivière, G.-J.; Kaiser, G.; et al. Omalizumab protects against allergen-induced bronchoconstriction in allergic (immunoglobulin E-mediated) asthma. Int. Arch. Allergy Immunol. 2013, 160, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Slutsky, A.S.; Drazen, J.M.; Silkoff, P.E.; Gaston, B.M.; Holden, W.; Romero, F.A.; Alving, K.; Baraldi, E.; Barnes, P.J.; Bratton, D.; et al. Recommendations for standardized procedures for the on-line and off-line measurement of exhaled lower respiratory nitric oxide and nasal nitric oxide in adults and children-1999. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, July 1999. Am. J. Respir. Crit. Care Med. 1999, 160, 2104–2117. [Google Scholar] [CrossRef]
- Fussbroich, D.; Kohnle, C.; Schwenger, T.; Driessler, C.; Dücker, R.P.; Eickmeier, O.; Gottwald, G.; Jerkic, S.P.; Zielen, S.; Kreyenberg, H.; et al. A combination of LCPUFAs regulates the expression of miRNA-146a-5p in a murine asthma model and human alveolar cells. Prostaglandins Other Lipid Mediat. 2020, 147, 106378. [Google Scholar] [CrossRef]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 5 September 2023).
- Duecker, R.P.; Messa, I.D.M.; Jerkic, S.; Kochems, A.; Gottwald, G.; Moreno-Galdó, A.; Rosewich, M.; Gronau, L.; Zielen, S.; Geburtig-Chiocchetti, A.; et al. Epigenetic regulation of inflammation by microRNAs in post-infectious bronchiolitis obliterans. Clin. Transl. Immunol. 2022, 11, e1376. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]

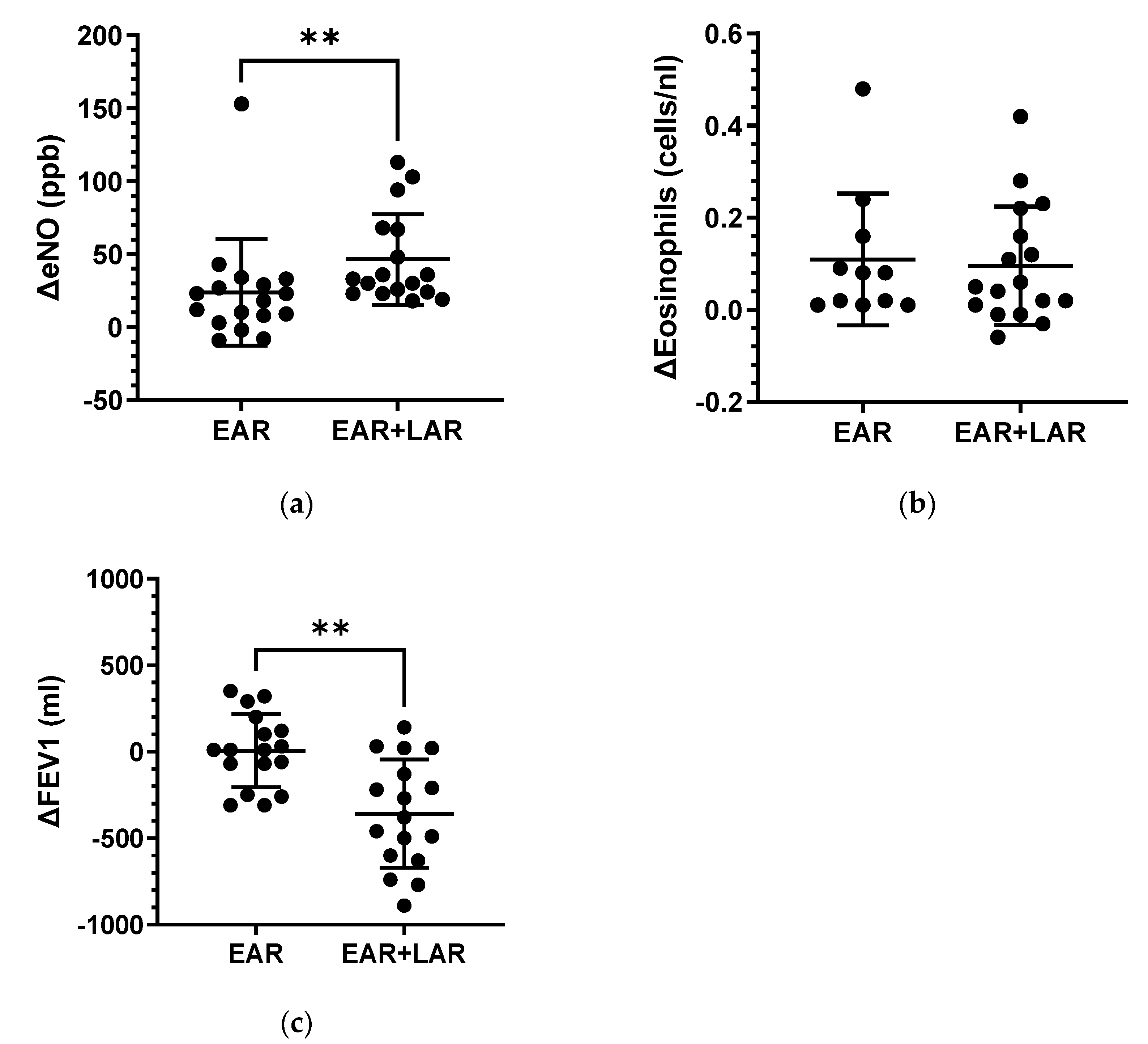
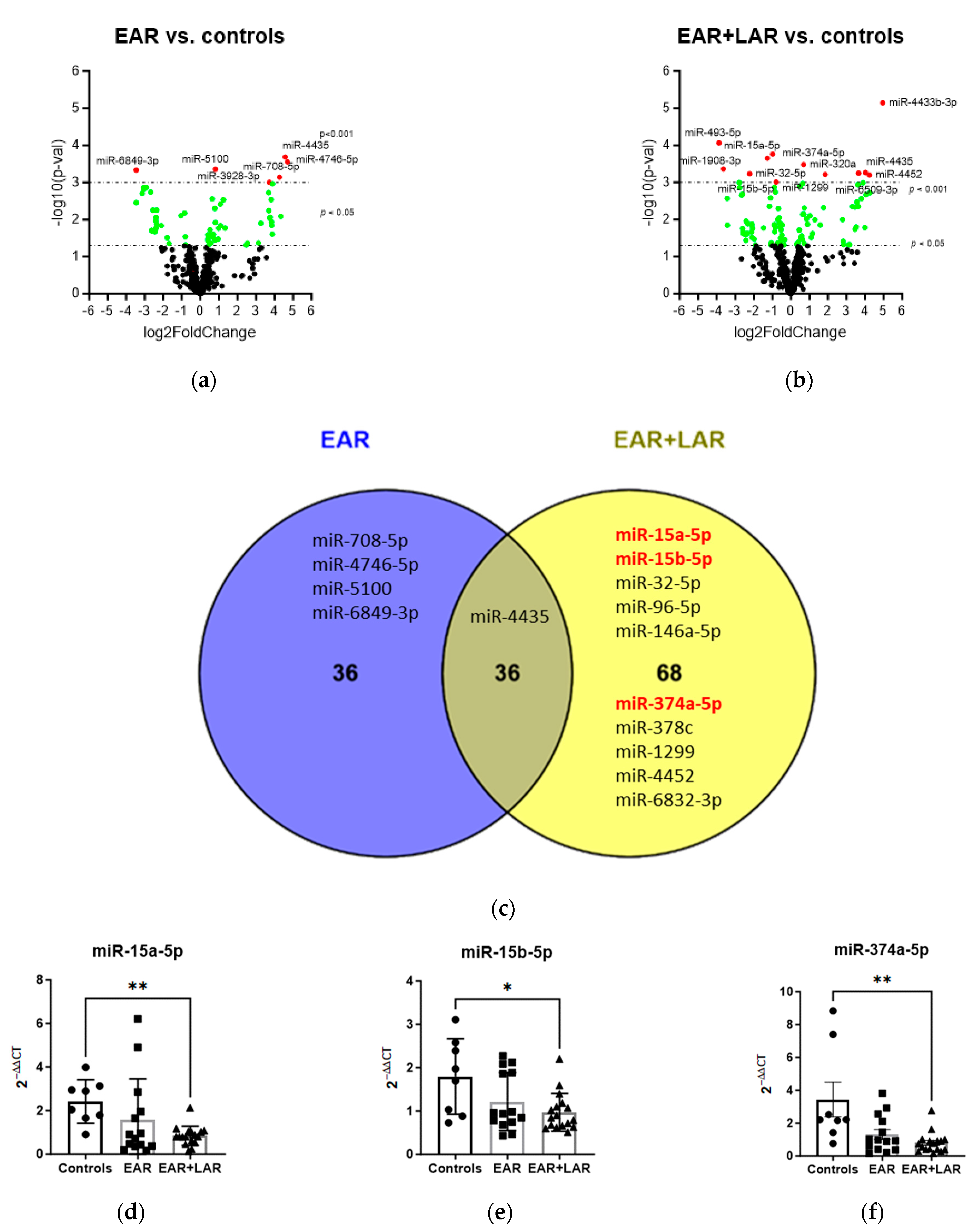
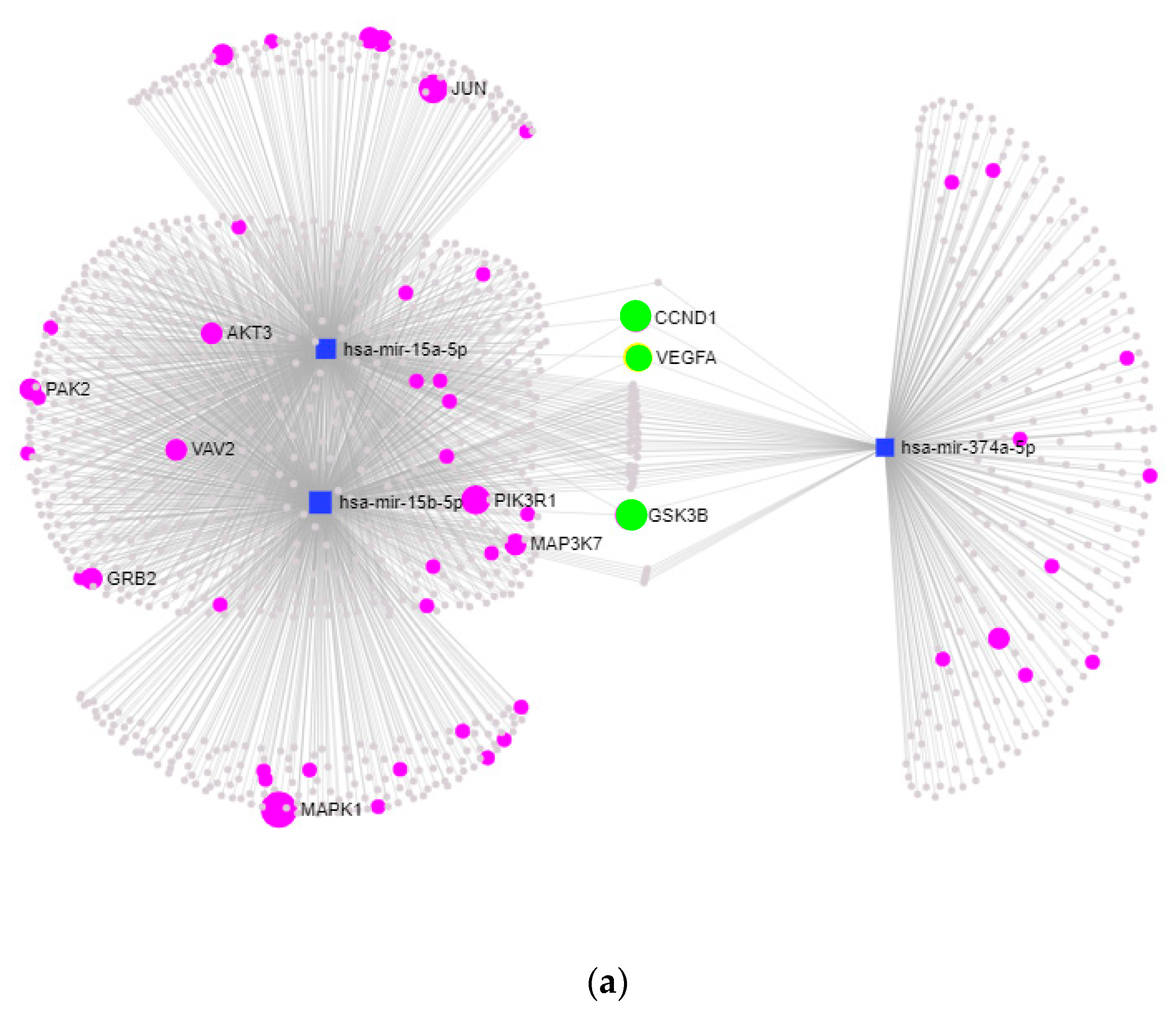
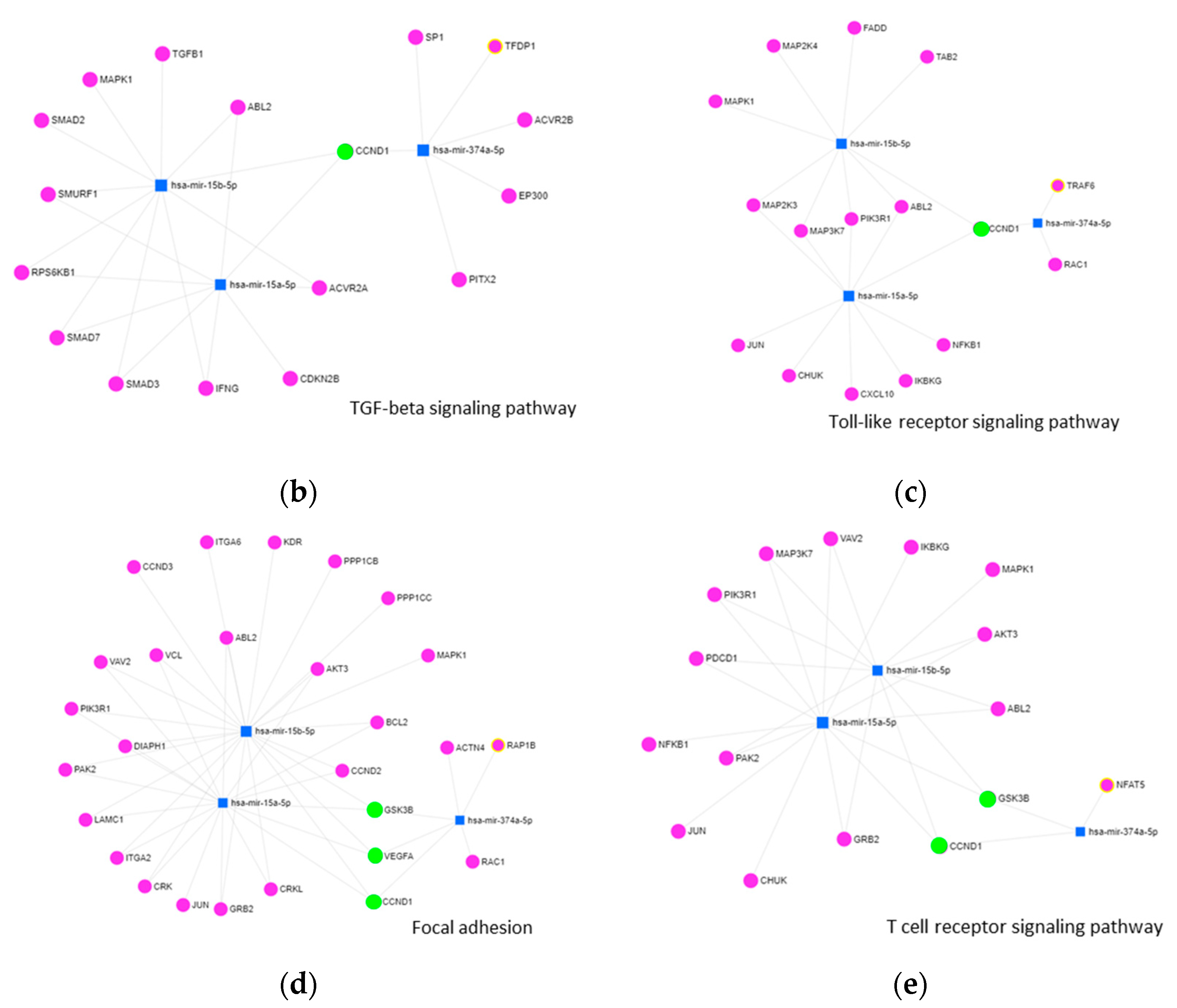
| Controls | EAR/ No LAR | EAR Plus LAR | |
|---|---|---|---|
| Numbers | n = 8 | n = 17 | n = 17 |
| Age (years) | 25 (18–35) | 27 (18–38) | 24 (19–32) |
| Sex male/female | 3/5 | 12/5 | 6/11 |
| Asthma control test (ACT) | - | 23 (16–25) | 23 (15–25) |
| eNO (ppb) | 13.5 (6–36) | 18 (8–74) | 21 (7–104) |
| FEV1 (%) | 98.5 (90.0–107.6) | 93.4 (79.2–131.4) | 97.9 (82.6–115) |
| Metacholine PD20 (mg) | 1.3 (0.11–2.90) | 0.68 (0.01–2.69) | 0.48 (0.01–1.33) |
| Total-IgE (KU/L) | 298 (57–691) | 156 (28–947) | 148 (4–985) |
| Dpt- specific IgE (KU/L) | 7.5 * (0–13.0) | 6.38 (0.4–21.6) | 15.05 * (1.21–63.3) |
| Dfa-specific IgE (KU/L) | 2.2 * (0.5–16.8) | 7.85 (0.48–37.1) | 18.15 * (0.57–65.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duecker, R.P.; Alemdar, O.; Wimmers, A.; Gronau, L.; Chiocchetti, A.G.; Valesky, E.M.; Donath, H.; Trischler, J.; Blumchen, K.; Zielen, S.; et al. MicroRNA Profiling of the Inflammatory Response after Early and Late Asthmatic Reaction. Int. J. Mol. Sci. 2024, 25, 1356. https://doi.org/10.3390/ijms25021356
Duecker RP, Alemdar O, Wimmers A, Gronau L, Chiocchetti AG, Valesky EM, Donath H, Trischler J, Blumchen K, Zielen S, et al. MicroRNA Profiling of the Inflammatory Response after Early and Late Asthmatic Reaction. International Journal of Molecular Sciences. 2024; 25(2):1356. https://doi.org/10.3390/ijms25021356
Chicago/Turabian StyleDuecker, Ruth P., Oguzhan Alemdar, Andreas Wimmers, Lucia Gronau, Andreas G. Chiocchetti, Eva M. Valesky, Helena Donath, Jordis Trischler, Katharina Blumchen, Stefan Zielen, and et al. 2024. "MicroRNA Profiling of the Inflammatory Response after Early and Late Asthmatic Reaction" International Journal of Molecular Sciences 25, no. 2: 1356. https://doi.org/10.3390/ijms25021356
APA StyleDuecker, R. P., Alemdar, O., Wimmers, A., Gronau, L., Chiocchetti, A. G., Valesky, E. M., Donath, H., Trischler, J., Blumchen, K., Zielen, S., & Schubert, R. (2024). MicroRNA Profiling of the Inflammatory Response after Early and Late Asthmatic Reaction. International Journal of Molecular Sciences, 25(2), 1356. https://doi.org/10.3390/ijms25021356






