Genetic Analysis of Partially Resistant and Susceptible Chickpea Cultivars in Response to Ascochyta rabiei Infection
Abstract
1. Introduction
2. Result and Discussion
2.1. Phenotypic Differences of Chickpea Cultivars in Response to Ascochyta Blight
2.2. Genome-Wide Transcriptome Sequencing
2.3. Differentially Expressed Genes in Response to A. rabiei
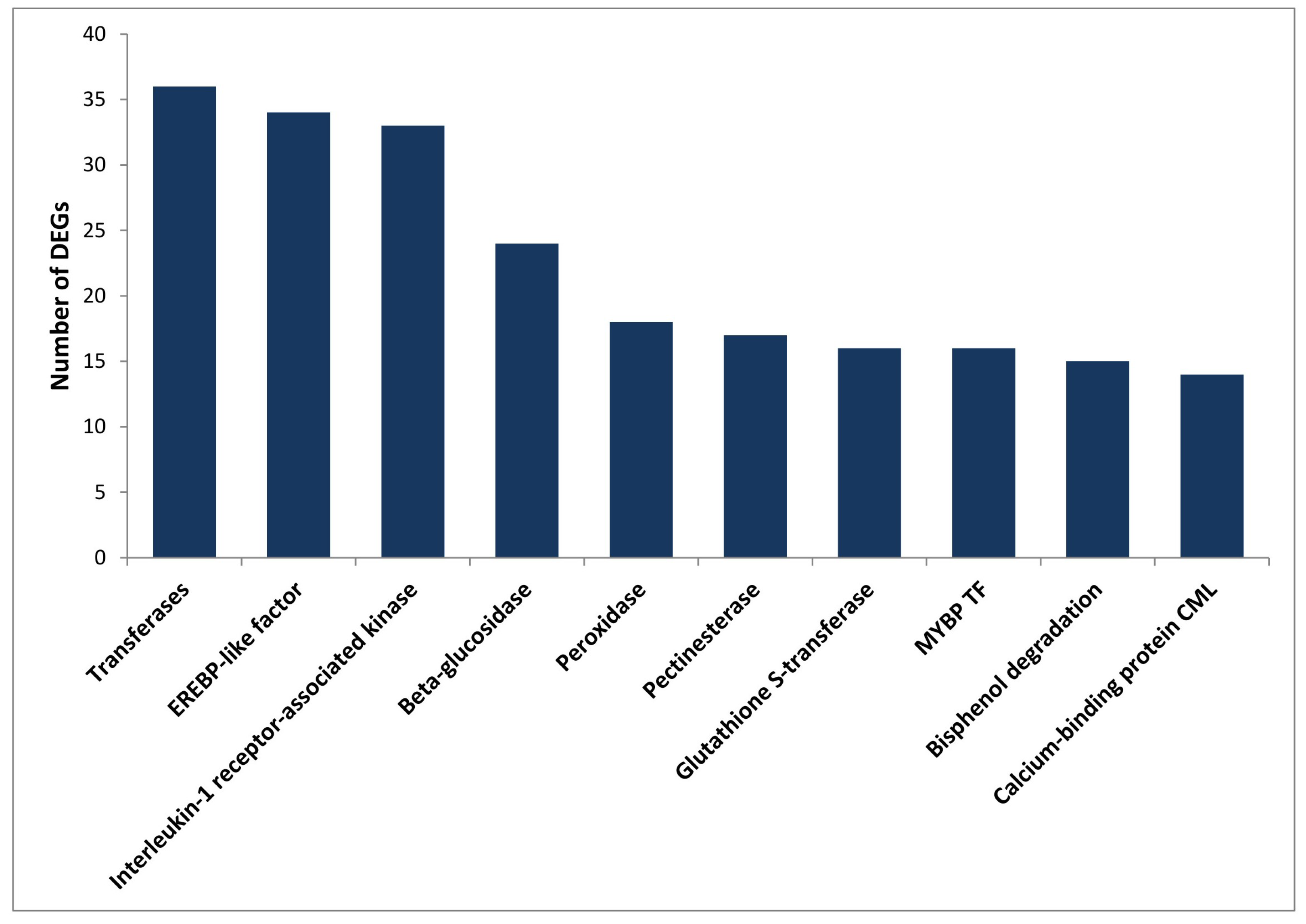
2.4. Differentially Expressed Genes in Partially Resistant Chickpea Cultivars
2.5. Time-Dependent DEGs in Response to A. rabiei
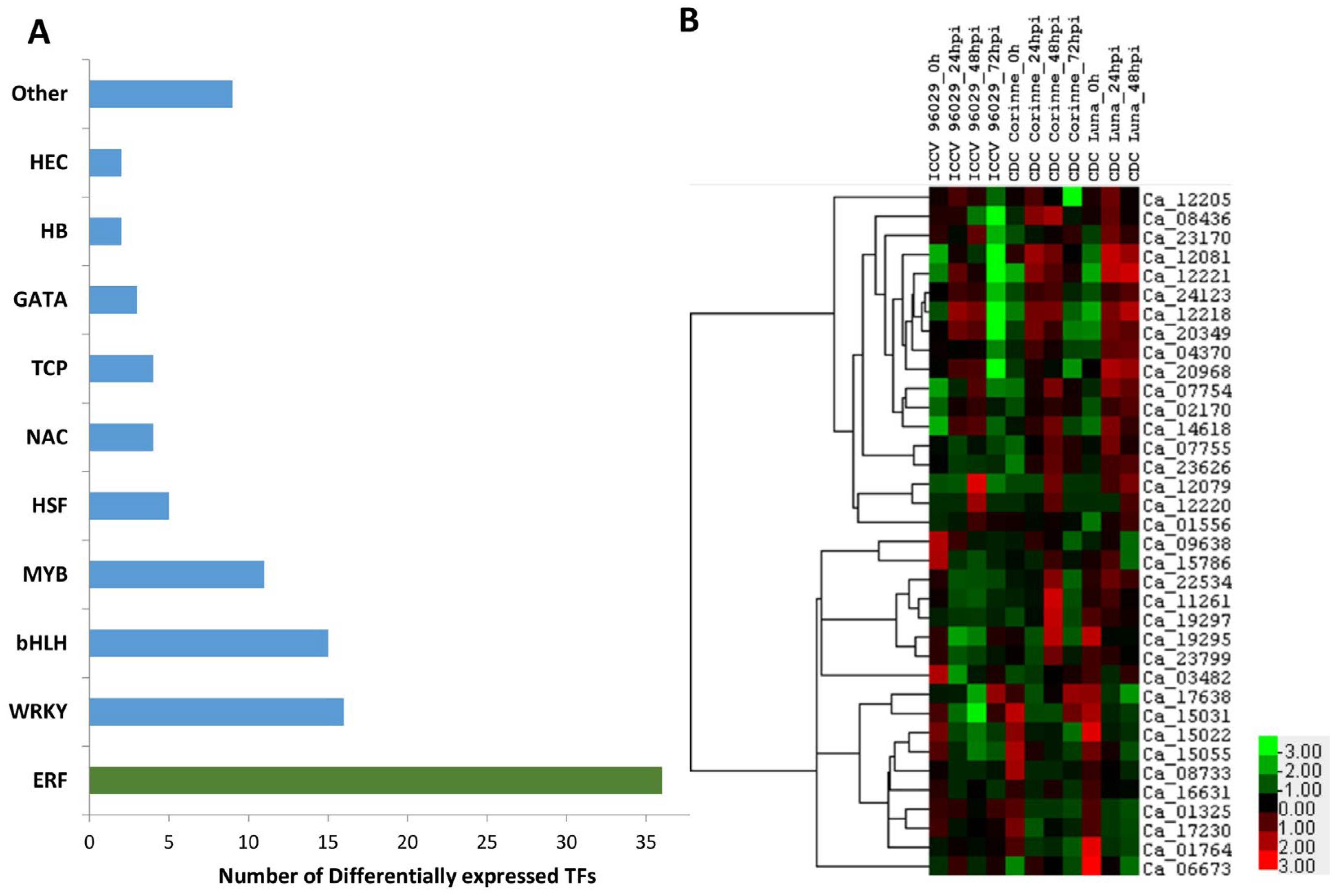
2.6. Transcription Factors Regulation in Response to A. rabiei Infection
2.7. Pathogen Recognition
2.7.1. Differentially Expressed Receptor-like Protein Kinases (RLKs)
2.7.2. Calcium-Binding Protein (CML)
2.8. Pathogenesis-Related Proteins
2.8.1. Peroxidase
2.8.2. Glutathione S-Transferase
2.8.3. Chitinase
2.9. Phenylpropanoid Pathway
2.10. Cytochrome P450 (CYP)
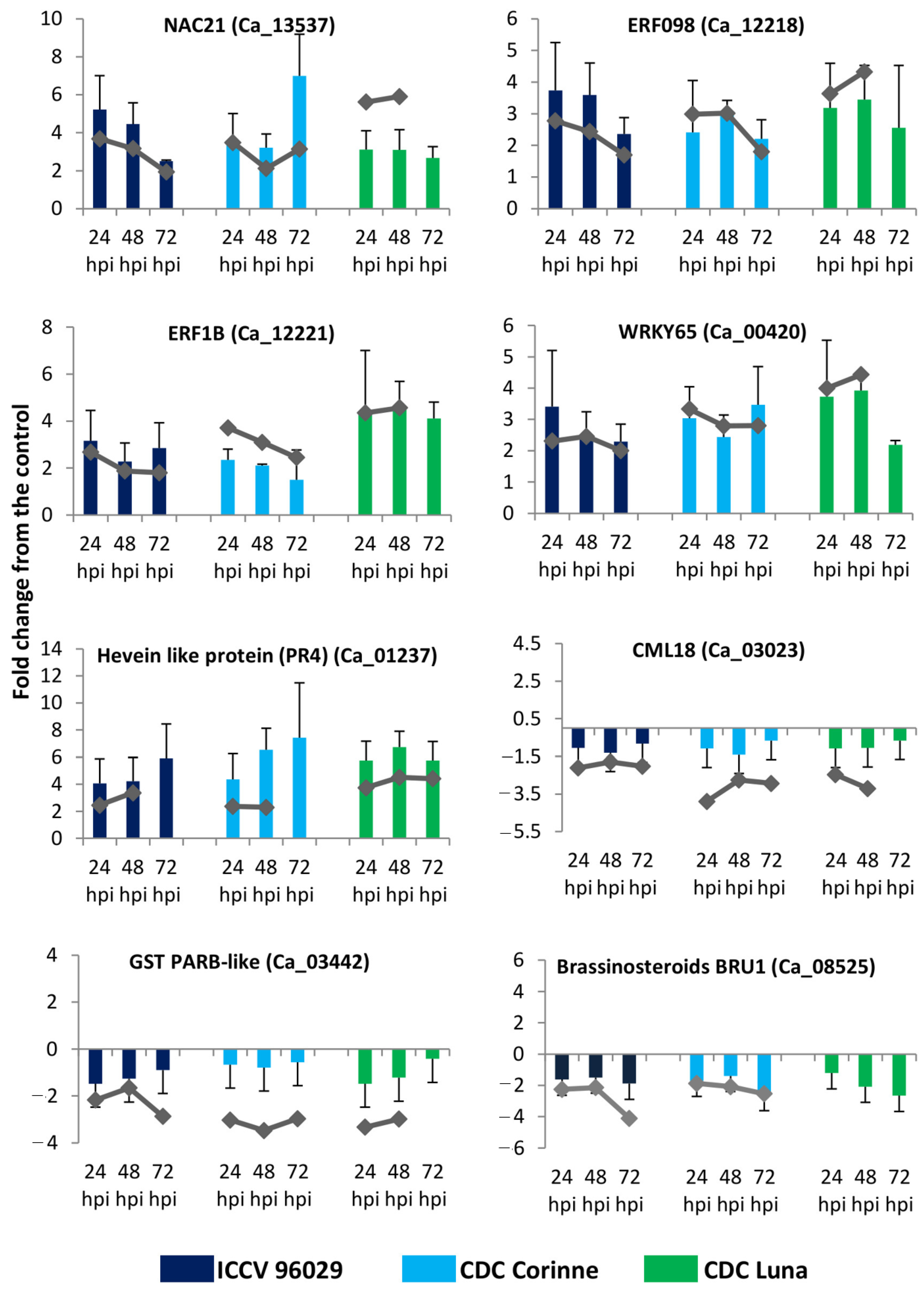
2.11. RT-qPCR Validation of Differentially Expressed Genes in Response to A. rabiei Infection
3. Materials and Methods
3.1. Plant Materials
3.2. A. rabiei Inoculations and Sample Collection for RNA-Seq
3.3. RNA-Seq
3.4. RNA-Seq Data Analysis
3.5. Downstream Data Analysis
3.6. Validation of RNA-Seq Analysis by qPCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaur, P.M.; Singh, M.K.; Samineni, S.; Sajja, S.B.; Jukanti, A.K.; Kamatam, S.; Varshney, R.K. Inheritance of protein content and its relationships with seed size, grain yield and other traits in chickpea. Euphytica 2016, 209, 253–260. [Google Scholar] [CrossRef]
- Sharma, M.; Ghosh, R. An Update on Genetic Resistance of Chickpea to Ascochyta Blight. Agronomy 2016, 6, 18. [Google Scholar] [CrossRef]
- Pandey, B.K.; Singh, U.S.; Chaube, H.S. Mode of Infection of Ascochyta Blight of Chickpea Caused by Ascochyta rabiei. J. Phytopathol. 1987, 119, 88–93. [Google Scholar] [CrossRef]
- Höhl, B.; Pfautsch, M.; Barz, W. Histology of Disease Development in Resistant and Susceptible Cultivars of Chickpea (Cicer arietinum L.) Inoculated with Spores of Ascochyta rabiei. J. Phytopathol. 1990, 129, 31–34. [Google Scholar] [CrossRef]
- Ilarslan, H.; Dolar, F.S. Histological and Ultrastructural Changes in Leaves and Stems of Resistant and Susceptible Chickpea Cultivars to Ascochyta rabiei. J. Phytopathol. 2002, 150, 340–348. [Google Scholar] [CrossRef]
- White, D.; Chen, W. Towards identifying pathogenic determinants of the chickpea pathogen Ascochyta rabiei. Eur. J. Plant Pathol. 2007, 119, 3–12. [Google Scholar] [CrossRef]
- Chongo, G.; Gossen, B. Effect of plant age on resistance to Ascochyta rabiei in chickpea. Can. J. Plant Pathol. 2001, 23, 358–363. [Google Scholar] [CrossRef]
- Iruela, M.; Rubio, J.; Barro, F.; Cubero, J.I.; Millán, T.; Gil, J. Detection of two quantitative trait loci for resistance to ascochyta blight in an intra-specific cross of chickpea (Cicer arietinum L.): Development of SCAR markers associated with resistance. Theor. Appl. Genet. 2005, 112, 278–287. [Google Scholar] [CrossRef]
- Lichtenzveig, J.; Bonfil, D.J.; Zhang, H.-B.; Shtienberg, D.; Abbo, S. Mapping quantitative trait loci in chickpea associated with time to flowering and resistance to Didymella rabiei the causal agent of Ascochyta blight. Theor. Appl. Genet. 2006, 113, 1357–1369. [Google Scholar] [CrossRef]
- Tar’an, B.; Warkentin, T.D.; Tullu, A.; Vandenberg, A. Genetic mapping of ascochyta blight resistance in chickpea (Cicer arietinum L.) using a simple sequence repeat linkage map. Genome 2007, 50, 26–34. [Google Scholar] [CrossRef]
- Taran, B.; Warkentin, T.D.; Vandenberg, A. Fast track genetic improvement of ascochyta blight resistance and double podding in chickpea by marker-assisted backcrossing. Theor. Appl. Genet. 2013, 126, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Daba, K.; Deokar, A.; Banniza, S.; Warkentin, T.D.; Tar’an, B. QTL mapping of early flowering and resistance to ascochyta blight in chickpea. Genome 2016, 59, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Madrid, E.; Rajesh, P.N.; Rubio, J.; Gil, J.; Millan, T.; Chen, W. Characterization and genetic analysis of an EIN4-like sequence (CaETR-1) located in QTL (AR1) implicated in ascochyta blight resistance in chickpea. Plant Cell Rep. 2012, 31, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Madrid, E.; Seoane, P.; Claros, M.G.; Barro, F.; Rubio, J.; Gil, J.; Millán, T. Genetic and physical mapping of the QTLAR3 controlling blight resistance in chickpea (Cicer arietinum L). Euphytica 2014, 198, 69–78. [Google Scholar] [CrossRef]
- Varshney, R.K.; Mir, R.R.; Bhatia, S.; Thudi, M.; Hu, Y.; Azam, S.; Zhang, Y.; Jaganathan, D.; You, F.M.; Gao, J.; et al. Integrated physical, genetic and genome map of chickpea (Cicer arietinum L.). Funct. Integr. Genom. 2014, 14, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Deokar, A.; Sagi, M.; Daba, K.; Tar’An, B. QTL sequencing strategy to map genomic regions associated with resistance to ascochyta blight in chickpea. Plant Biotechnol. J. 2018, 17, 275–288. [Google Scholar] [CrossRef]
- Deokar, A.; Sagi, M.; Tar’an, B. Genome-wide SNP discovery for development of high-density genetic map and QTL mapping of ascochyta blight resistance in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2019, 132, 1861–1872. [Google Scholar] [CrossRef]
- Zhang, Z.-F.; Li, Y.-Y.; Xiao, B.-Z. Comparative transcriptome analysis highlights the crucial roles of photosynthetic system in drought stress adaptation in upland rice. Sci. Rep. 2016, 6, 19349. [Google Scholar] [CrossRef]
- Bhardwaj, A.R.; Joshi, G.; Kukreja, B.; Malik, V.; Arora, P.; Pandey, R.; Shukla, R.N.; Bankar, K.G.; Katiyar-Agarwal, S.; Goel, S.; et al. Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 2015, 15, 9. [Google Scholar] [CrossRef]
- Goyer, A.; Hamlin, L.; Crosslin, J.M.; Buchanan, A.; Chang, J.H. RNA-Seq analysis of resistant and susceptible potato varieties during the early stages of potato virus Y infection. BMC Genom. 2015, 16, 1–13. [Google Scholar] [CrossRef]
- Kamber, T.; Buchmann, J.P.; Pothier, J.F.; Smits, T.H.M.; Wicker, T.; Duffy, B. Fire blight disease reactome: RNA-seq transcriptional profile of apple host plant defense responses to Erwinia amylovora pathogen infection. Sci. Rep. 2016, 6, 21600. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; Oono, Y.; Kanamori, H.; Matsumoto, T.; Itoh, T.; Minami, E. Simultaneous RNA-Seq Analysis of a Mixed Transcriptome of Rice and Blast Fungus Interaction. PLoS ONE 2012, 7, e49423. [Google Scholar] [CrossRef] [PubMed]
- Coram, T.E.; Pang, E.C.K. Expression profiling of chickpea genes differentially regulated during a resistance response to Ascochyta rabiei. Plant Biotechnol. J. 2006, 4, 647–666. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, P.; Cheruku, J.R.; Kumar, K.; Yadav, S.; Singh, A.; Kumari, P.; Dube SCUpadhyaya, K.C.; Verma, P.K. Differential transcript accumulation in chickpea during early phases of compatible interaction with a necrotrophic fungus Ascochyta rabiei. Mol. Biol. Rep. 2012, 39, 4635–4646. [Google Scholar] [CrossRef]
- Leo, A.E.; Linde, C.C.; Ford, R. Defence gene expression profiling to Ascochyta rabiei aggressiveness in chickpea. Theor. Appl. Genet. 2016, 129, 1333–1345. [Google Scholar] [CrossRef]
- Sagi, M.S.; Deokar, A.A.; Tar’an, B. Genetic Analysis of NBS-LRR Gene Family in Chickpea and Their Expression Profiles in Response to Ascochyta Blight Infection. Front. Plant Sci. 2017, 8, 838. [Google Scholar] [CrossRef]
- Fondevilla, S.; Krezdorn, N.; Rotter, B.; Kahl, G.; Winter, P. In planta Identification of Putative Pathogenicity Factors from the Chickpea Pathogen Ascochyta rabiei by De novo Transcriptome Sequencing Using RNA-Seq and Massive Analysis of cDNA Ends. Front. Microbiol. 2015, 6, 1329. [Google Scholar] [CrossRef]
- Du, H.; Wang, Y.; Yang, J.; Yang, W. Comparative Transcriptome Analysis of Resistant and Susceptible Tomato Lines in Response to Infection by Xanthomonas perforans Race T3. Front. Plant Sci. 2015, 6, 1173. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, Q.; Yang, Q.; Liu, H.; Li, Q.; Yi, X.; Cheng, Y.; Guo, L.; Fan, C.; Zhou, Y. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 2016, 6, 19007. [Google Scholar] [CrossRef]
- Nehra, K.; Chugh, L.; Dhillon, S.; Singh, R. Induction, Purification and Characterization of Chitinases from Chickpea (Cicer arietinum L.) Leaves and Pods Infected with Ascochyta rabiei. J. Plant Physiol. 1994, 144, 7–11. [Google Scholar] [CrossRef]
- Cletus, J.; Balasubramanian, V.; Vashisht, D.; Sakthivel, N. Transgenic expression of plant chitinases to enhance disease resistance. Biotechol. Lett. 2013, 35, 1719–1732. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O.K. Secretome Analysis Reveals an Arabidopsis Lipase Involved in Defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional Analysis of the Arabidopsis PAL Gene Family in Plant Growth, Development, and Response to Environmental Stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Anbessa, Y.; Taran, B.; Warkentin, T.D.; Tullu, A.; Vandenberg, A. Genetic analyses and conservation of QTL for ascochyta blight resistance in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2009, 119, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Deokar, A.A.; Kondawar, V.; Kohli, D.; Aslam, M.; Jain, P.K.; Karuppayil, S.M.; Varshney, R.K.; Srinivasan, R. The CarERF genes in chickpea (Cicer arietinum L.) and the identification of CarERF116 as abiotic stress responsive transcription factor. Funct. Integr. Genom. 2014, 15, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Heyman, J.; Canher, B.; Bisht, A.; Christiaens, F.; De Veylder, L. Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J. Cell Sci. 2018, 131, jcs208215. [Google Scholar] [CrossRef] [PubMed]
- McGrath, K.C.; Dombrecht, B.; Manners, J.M.; Schenk, P.M.; Edgar, C.I.; Maclean, D.J.; Scheible, W.R.; Udvardi, M.K.; Kazan, K. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005, 139, 949–959. [Google Scholar] [CrossRef]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Molina, A.; Funnell-Harris, D.L.; Sattler, S.E.; O’neill, P.M.; Gries, T.; Tetreault, H.M.; Clemente, T.E.; Zhao, C.; Wang, H.; et al. Ethylene Response Factor 1 Mediates Arabidopsis Resistance to the Soilborne Fungus Fusarium oxysporum. Mol. Plant-Microbe Interact. 2004, 17, 763–770. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002, 29, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Lichtenzveig, J.; Gleason, C.; Oliver, R.P.; Singh, K.B. The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia. Plant Physiol. 2010, 154, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Qi, L.; Liu, X.; Cai, S.; Xu, H.; Huang, R.; Li, J.; Wei, X.; Zhang, Z. The Wheat Ethylene Response Factor Transcription Factor PATHOGEN-INDUCED ERF1 Mediates Host Responses to Both the Necrotrophic Pathogen Rhizoctonia cerealis and Freezing Stresses. Plant Physiol. 2014, 164, 1499–1514. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. Induction of Phytoalexin Biosynthesis: WRKY33 Is a Target of MAPK Signaling. Plant Cell 2011, 23, 1190. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Wang, H.; Meng, J.; Peng, X.; Tang, X.; Zhou, P.; Xiang, J.; Deng, X. Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 2015, 89, 157–171. [Google Scholar] [CrossRef]
- Mzid, R.; Marchive, C.; Blancard, D.; Deluc, L.; Barrieu, F.; Corio-Costet, M.; Drira, N.; Hamdi, S.; Lauvergeat, V. Overexpression of VvWRKY2 in tobacco enhances broad resistance to necrotrophic fungal pathogens. Physiol. Plant. 2007, 131, 434–447. [Google Scholar] [CrossRef]
- Mengiste, T.; Chen, X.; Salmeron, J.; Dietrich, R. The Botrytis Susceptible1 Gene Encodes an R2R3MYB Transcription Factor Protein That Is Required for Biotic and Abiotic Stress Responses in Arabidopsis. Plant Cell 2003, 15, 2551–2565. [Google Scholar] [CrossRef]
- Shan, T.; Rong, W.; Xu, H.; Du, L.; Liu, X.; Zhang, Z. The wheat R2R3-MYB transcription factor TaRIM1 participates in resistance response against the pathogen Rhizoctonia cerealis infection through regulating defense genes. Sci. Rep. 2016, 6, 28777. [Google Scholar] [CrossRef] [PubMed]
- Windram, O.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.; Cooke, E.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E.; et al. Arabidopsis Defense against Botrytis cinerea: Chronology and Regulation Deciphered by High-Resolution Temporal Transcriptomic Analysis. Plant Cell 2012, 24, 3530–3557. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kuo, Y.C.; Mishra, S.; Tsai, C.H.; Chien, C.C.; Chen, C.W.; Desclos-Theveniau, M.; Chu, P.W.; Schulze, B.; Chinchilla, D.; et al. The Lectin Receptor Kinase-VI.2 Is Required for Priming and Positively Regulates Arabidopsis Pattern-Triggered Immunity. Plant Cell. 2012, 24, 1256–1270. [Google Scholar] [CrossRef]
- Huang, P.Y.; Yeh, Y.H.; Liu, A.C.; Cheng, C.P.; Zimmerli, L. The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 2014, 79, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-Y.; Wang, G.-L.; Chen, L.-L.; Kim, H.-S.; Pi, L.-Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.-X.; Zhu, L.-H.; et al. A Receptor Kinase-Like Protein Encoded by the Rice Disease Resistance Gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nürnberger, T.; Jones, J.D.G.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, X.-C.; Neece, D.; Ramonell, K.M.; Clough, S.; Kim, S.-Y.; Stacey, M.G.; Stacey, G. A LysM Receptor-Like Kinase Plays a Critical Role in Chitin Signaling and Fungal Resistance in Arabidopsis. Plant Cell 2008, 20, 471–481. [Google Scholar] [CrossRef]
- Zhang, L.; Kars, I.; Essenstam, B.; Liebrand, T.W.H.; Wagemakers, L.; Elberse, J.; Tagkalaki, P.; Tjoitang, D.; van den Ackerveken, G.; van Kan, J.A.L. Fungal Endopolygalacturonases Are Recognized as Microbe-Associated Molecular Patterns by the Arabidopsis Receptor-Like Protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 2014, 164, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fraiture, M.; Kolb, D.; Loffelhardt, B.; Desaki, Y.; Boutrot, F.F.; Tor, M.; Zipfel, C.; Gust, A.A.; Brunner, F. Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 2013, 25, 4227–4241. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Avni, A. The Receptor for the Fungal Elicitor Ethylene-Inducing Xylanase Is a Member of a Resistance-Like Gene Family in Tomato. Plant Cell 2004, 16, 1604–1615. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Ali, G.S.; Reddy, A. Characterization of a pathogen-induced calmodulin-binding protein: Mapping of four Ca2+-dependent calmodulin-binding domains. Plant Mol. Biol. 2003, 52, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Ranty, B.; Aldon, D.; Galaud, J.P. Plant Calmodulins and Calmodulin-Related Proteins: Multifaceted Relays to Decode Calcium Signals. Plant Signal. Behav. 2006, 1, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Takabatake, R.; Karita, E.; Seo, S.; Mitsuhara, I.; Kuchitsu, K.; Ohashi, Y. Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 2007, 48, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.W.; Lee, D.H.; Hwang, B.K. The Pepper Calmodulin Gene CaCaM1 Is Involved in Reactive Oxygen Species and Nitric Oxide Generation Required for Cell Death and the Defense Response. Mol. Plant-Microbe Interact. 2009, 22, 1389–1400. [Google Scholar] [CrossRef]
- Chassot, C.; Nawrath, C.; Métraux, J. Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 2007, 49, 972–980. [Google Scholar] [CrossRef]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef]
- Anjana, G.; Kini, K.R.; Shetty, H.S.; Prakash, H.S. Changes in peroxidase activity in sunflower during infection by necrotrophic pathogenAlternaria helianthi. Arch. Phytopathol. Plant Prot. 2008, 41, 586–596. [Google Scholar] [CrossRef]
- Kaur, L.; Singh, V.P.; Gupta, A.K. Peroxidase: A marker for Ascochyta blight resistance in chickpea. Arch. Phytopathol. Plant Protect. 2012, 45, 42–46. [Google Scholar] [CrossRef]
- Tognolli, M.; Penel, C.; Greppin, H.; Simon, P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 2002, 288, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Wally, O.; Punja, Z.K. Enhanced disease resistance in transgenic carrot (Daucus carota L.) plants over-expressing a rice cationic peroxidase. Planta 2010, 232, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.D.; Goodwin, P.H.; Hsiang, T. Induction of glutathione S-transferase genes of Nicotiana benthamiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. J. Exp. Bot. 2005, 56, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Hamid, K.; Strange, R.N. Phytotoxicity of solanapyrones A and B produced by the chickpea pathogen Ascochyta rabiei (Pass.) Labr. and the apparent metabolism of solanapyrone A by chickpea tissues. Physiol. Mol. Plant Pathol. 2000, 56, 235–244. [Google Scholar] [CrossRef]
- Almeida, N.F.; Krezdorn, N.; Rotter, B.; Winter, P.; Rubiales, D.; Vaz Patto, M.C. Lathyrus sativus transcriptome resistance response to Ascochyta lathyri investigated by deepSuperSAGE analysis. Front Plant Sci. 2015, 6, 178. [Google Scholar] [CrossRef]
- Fondevilla, S.; Rotter, B.; Krezdorn, N.; Jüngling, R.; Winter, P.; Rubiales, D. Identification of Genes Involved in Resistance to Didymella pinodes in Pea by deepSuperSAGE Transcriptome Profiling. Plant Mol. Biol. Rep. 2013, 32, 258–269. [Google Scholar] [CrossRef][Green Version]
- Catinot, J.; Huang, J.B.; Huang, P.Y.; Tseng, M.Y.; Chen, Y.L.; Gu, S.Y.; Lo, W.S.; Wang, L.C.; Chen, Y.R.; Zimmerli, L. ETHYLENE RE-SPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate—And ethylene-responsive defence genes. Plant Cell Environ. 2015, 38, 2721–2734. [Google Scholar] [CrossRef]
- Kavousi, H.; Marashi, H.; Mozafari, J.; Bagheri, A. Expression of Phenylpropanoid Pathway Genes in Chickpea Defense Against Race 3 of Ascochyta rabiei. Plant Pathol. J. 2009, 8, 127–132. [Google Scholar] [CrossRef]
- Daniel, S.; Barz, W. Elicitor-induced metabolic changes in cell cultures of chickpea (Cicer arietinum L.) cultivars resistant and susceptible to Ascochyta rabiei: II. Differential induction of chalcone-synthase-mRNA activity and analysis of in-vitro-translated protein patterns. Planta 1990, 182, 279–286. [Google Scholar] [CrossRef]
- Oliveira, M.B.; de Andrade, R.V.; Grossi-De-Sá, M.F.; Petrofeza, S. Analysis of genes that are differentially expressed during the Sclerotinia sclerotiorum–Phaseolus vulgaris interaction. Front. Microbiol. 2015, 6, 1162. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.E.; Mengesha, B.; Tang, H.; Mengiste, T.; Bluhm, B.H. Resistance to Botrytis cinerea in Solanum lycopersicoides involves widespread transcriptional reprogramming. BMC Genom. 2014, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a Soybean Cytochrome P450 Family Gene Involved in the Jasmonic Acid and Ethylene Signaling Pathway, Enhances Plant Resistance to Biotic and Abiotic Stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef]
- Delventhal, R.; Falter, C.; Strugala, R.; Zellerhoff, N.; Schaffrath, U. Ectoparasitic growth of Magnaporthe on barley triggers expression of the putative barley wax biosynthesis gene CYP96B22 which is involved in penetration resistance. BMC Plant Biol. 2014, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Taran, B.; Warkentin, T.; Malhotra, R.; Banniza, S.; Vandenberg, A. CDC Luna kabuli chickpea. Can. J. Plant Sci. 2009, 89, 517–518. [Google Scholar] [CrossRef]
- Taran, B.; Warkentin, T.; Banniza, S.; Vandenberg, A. CDC Corinne desi chickpea. Can. J. Plant Sci. 2009, 89, 515–516. [Google Scholar] [CrossRef]
- Chongo, G.; Gossen, B.D.; Buchwaldt, L.; Adhikari, T.; Rimmer, S.R. Genetic Diversity of Ascochyta rabiei in Canada. Plant Dis. 2004, 88, 4–10. [Google Scholar] [CrossRef]
- Armstrong-Cho, C.; Lulsdorf, M.M.; Hashemi, P.; Banniza, S. Characterization of resistance to ascochyta blight of selected wild Cicer germplasm. Botany 2015, 93, 723–734. [Google Scholar] [CrossRef]
- Verma, S.; Gazara, R.K.; Nizam, S.; Parween, S.; Chattopadhyay, D.; Verma, P.K. Draft genome sequencing and secretome analysis of fungal phytopathogen Ascochyta rabiei provides insight into the necrotrophic effector repertoire. Sci. Rep. 2016, 6, 24638. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Time Interval | A. rabiei-Responsive DEGs in ICCV 96029 | A. rabiei-Responsive DEGs in CDC Corinne | A. rabiei-Responsive DEGs in CDC Luna | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated | Down-Regulated | Sub-Total | Up-Regulated | Down-Regulated | Sub-Total | Up-Regulated | Down-Regulated | Sub-Total | |
| 0–24 hpi | 267 | 175 | 442 | 201 | 365 | 566 | 696 | 963 | 1659 |
| 0–48 hpi | 343 | 169 | 512 | 211 | 153 | 364 | 712 | 1240 | 1952 |
| 0–72 hpi | 348 | 186 | 534 | 373 | 218 | 591 | - | - | - |
| Non-redundant DEGs in ICCV 96029 | 1051 | Non-redundant DEGs in CDC Corinne | 1132 | Non-redundant DEGs in CDC Luna | 2219 | ||||
| Non-redundant DEGs in response to A. rabiei in all three cultivars | 3073 | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deokar, A.A.; Sagi, M.; Tar’an, B. Genetic Analysis of Partially Resistant and Susceptible Chickpea Cultivars in Response to Ascochyta rabiei Infection. Int. J. Mol. Sci. 2024, 25, 1360. https://doi.org/10.3390/ijms25021360
Deokar AA, Sagi M, Tar’an B. Genetic Analysis of Partially Resistant and Susceptible Chickpea Cultivars in Response to Ascochyta rabiei Infection. International Journal of Molecular Sciences. 2024; 25(2):1360. https://doi.org/10.3390/ijms25021360
Chicago/Turabian StyleDeokar, Amit A., Mandeep Sagi, and Bunyamin Tar’an. 2024. "Genetic Analysis of Partially Resistant and Susceptible Chickpea Cultivars in Response to Ascochyta rabiei Infection" International Journal of Molecular Sciences 25, no. 2: 1360. https://doi.org/10.3390/ijms25021360
APA StyleDeokar, A. A., Sagi, M., & Tar’an, B. (2024). Genetic Analysis of Partially Resistant and Susceptible Chickpea Cultivars in Response to Ascochyta rabiei Infection. International Journal of Molecular Sciences, 25(2), 1360. https://doi.org/10.3390/ijms25021360






