A Molecular Dynamics Simulation of Complexes of Fullerenes and Lysine-Based Peptide Dendrimers with and without Glycine Spacers
Abstract
1. Introduction
2. Results and Discussion
2.1. Formation of a Dendrimer–Fullerene Complex
2.2. Analysis of the Structure and Properties of Dendrimer–Fullerene Complexes Snapshots, Shapes, Sizes, and Distances between Dendrimer and Fullerene in Complexes
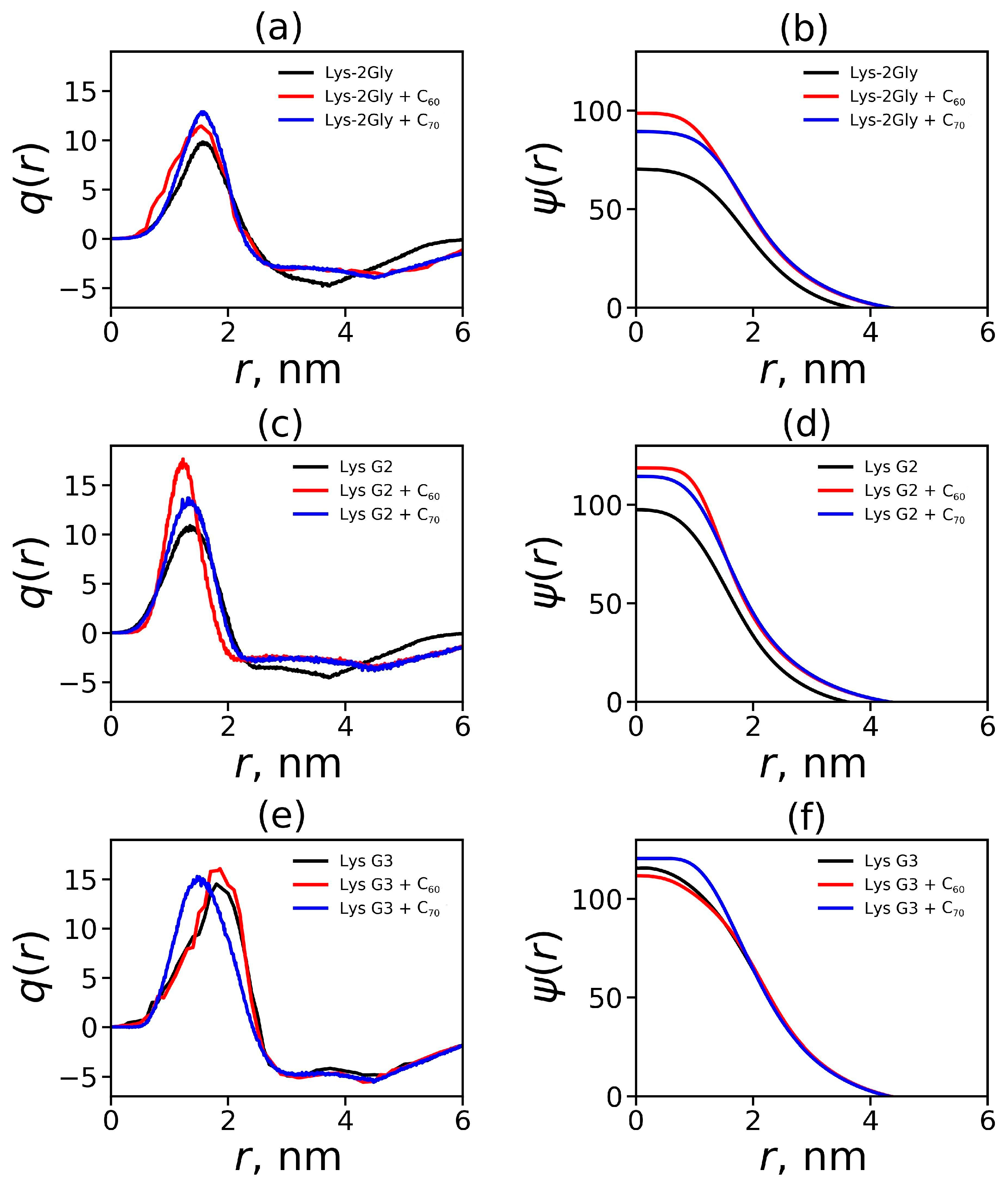
2.3. Radial Distribution Functions
2.4. Electrostatic Properties of the Dendrimer–Fullerene Complex and Individual Dendrimer
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- González Corrales, D.; Fernández Rojas, N.; Solís Vindas, G.; Santamaría Muñoz, M.; Chavarría Rojas, M.; Matarrita Brenes, D.; Rojas Salas, M.F.; Madrigal Redondo, G. Dendrimers and Their Applications. J. Drug Deliv. Ther. 2022, 12, 151–158. [Google Scholar] [CrossRef]
- Ziemba, B.; Borowiec, M.; Franiak-Pietryga, I. There and Back Again: A Dendrimer’s Tale. Drug Chem. Toxicol. 2022, 45, 2169–2184. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.; Alam, M.; Saxena, K. Dendrimers: A Class of Polymers in the Nanotechnology for the Delivery of Active Pharmaceuticals. Curr. Pharm. Des. 2009, 15, 2958–2969. [Google Scholar] [CrossRef] [PubMed]
- D’Emanuele, A.; Attwood, D. Dendrimer-Drug Interactions. Adv. Drug Deliv. Rev. 2005, 57, 2147–2162. [Google Scholar] [CrossRef] [PubMed]
- Madaan, K.; Kumar, S.; Poonia, N.; Lather, V.; Pandita, D. Dendrimers in Drug Delivery and Targeting: Drug-Dendrimer Interactions and Toxicity Issues. J. Pharm. Bioallied Sci. 2014, 6, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.M.; Cullis, P.R. Drug Delivery Systems: Entering the Mainstream. Science 2004, 303, 1818–1822. [Google Scholar] [CrossRef]
- Sosnik, A.; Carcaboso, A.; Chiappetta, D. Polymeric Nanocarriers: New Endeavors for the Optimization of the Technological Aspects of Drugs. Recent Pat. Biomed. Eng. 2008, 1, 43–59. [Google Scholar] [CrossRef]
- Tyshenko, M.G. Medical Nanotechnology Using Genetic Material and the Need for Precaution in Design and Risk Assessments. Int. J. Nanotechnol. 2008, 5, 116–123. [Google Scholar] [CrossRef]
- Veldhoen, S.; Laufer, S.; Restle, T. Recent Developments in Peptide-Based Nucleic Acid Delivery. Int. J. Mol. Sci. 2008, 9, 1276–1320. [Google Scholar] [CrossRef]
- Cheng, Y.; Xu, T. The Effect of Dendrimers on the Pharmacodynamic and Pharmacokinetic Behaviors of Non-Covalently or Covalently Attached Drugs. Eur. J. Med. Chem. 2008, 43, 2291–2297. [Google Scholar] [CrossRef]
- Karande, P.; Trasatti, J.P.; Chandra, D. Novel Approaches for the Delivery of Biologics to the Central Nervous System. In Novel Approaches and Strategies for Biologics, Vaccines and Cancer Therapies; Elsevier: Amsterdam, The Netherlands, 2015; pp. 59–88. [Google Scholar]
- Surekha, B.; Kommana, N.S.; Dubey, S.K.; Kumar, A.V.P.; Shukla, R.; Kesharwani, P. PAMAM Dendrimer as a Talented Multifunctional Biomimetic Nanocarrier for Cancer Diagnosis and Therapy. Colloids Surf. B Biointerfaces 2021, 204, 111837. [Google Scholar] [CrossRef] [PubMed]
- Janaszewska, A.; Lazniewska, J.; Trzepiński, P.; Marcinkowska, M.; Klajnert-Maculewicz, B. Cytotoxicity of Dendrimers. Biomolecules 2019, 9, 330. [Google Scholar] [CrossRef] [PubMed]
- Chis, A.A.; Dobrea, C.; Morgovan, C.; Arseniu, A.M.; Rus, L.L.; Butuca, A.; Juncan, A.M.; Totan, M.; Vonica-Tincu, A.L.; Cormos, G.; et al. Applications and Limitations of Dendrimers in Biomedicine. Molecules 2020, 25, 3982. [Google Scholar] [CrossRef] [PubMed]
- Luong, D.; Kesharwani, P.; Deshmukh, R.; Mohd Amin, M.C.I.; Gupta, U.; Greish, K.; Iyer, A.K. PEGylated PAMAM Dendrimers: Enhancing Efficacy and Mitigating Toxicity for Effective Anticancer Drug and Gene Delivery. Acta Biomater. 2016, 43, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Kolhatkar, R.B.; Kitchens, K.M.; Swaan, P.W.; Ghandehari, H. Surface Acetylation of Polyamidoamine (PAMAM) Dendrimers Decreases Cytotoxicity While Maintaining Membrane Permeability. Bioconjug. Chem. 2007, 18, 2054–2060. [Google Scholar] [CrossRef]
- Carvalho, M.R.; Carvalho, C.R.; Maia, F.R.; Caballero, D.; Kundu, S.C.; Reis, R.L.; Oliveira, J.M. Peptide-Modified Dendrimer Nanoparticles for Targeted Therapy of Colorectal Cancer. Adv. Ther. 2019, 2, 1900132. [Google Scholar] [CrossRef]
- Kharwade, R.; More, S.; Warokar, A.; Agrawal, P.; Mahajan, N. Starburst Pamam Dendrimers: Synthetic Approaches, Surface Modifications, and Biomedical Applications. Arab. J. Chem. 2020, 13, 6009–6039. [Google Scholar] [CrossRef]
- Santos, S.S.; Gonzaga, R.V.; Silva, J.V.; Savino, D.F.; Prieto, D.; Shikay, J.M.; Silva, R.S.; Paulo, L.H.A.; Ferreira, E.I.; Giarolla, J. Peptide Dendrimers: Drug/Gene Delivery and Other Approaches. Can. J. Chem. 2017, 95, 907–916. [Google Scholar] [CrossRef]
- Sapra, R.; Verma, R.P.; Maurya, G.P.; Dhawan, S.; Babu, J.; Haridas, V. Designer Peptide and Protein Dendrimers: A Cross-Sectional Analysis. Chem. Rev. 2019, 119, 11391–11441. [Google Scholar] [CrossRef]
- Sadler, K.; Tam, J.P. Peptide Dendrimers: Applications and Synthesis. Rev. Mol. Biotechnol. 2002, 90, 195–229. [Google Scholar] [CrossRef]
- Singh, S.K.; Sharma, V.K. Dendrimers: A Class of Polymer in the Nanotechnology for Drug Delivery. In Nanomedicine for Drug Delivery and Therapeutics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 373–409. ISBN 9781118414095. [Google Scholar]
- Martinho, N.; Silva, L.C.; Florindo, H.F.; Brocchini, S.; Zloh, M.; Barata, T.S. Rational Design of Novel, Fluorescent, Tagged Glutamic Acid Dendrimers with Different Terminal Groups and in Silico Analysis of Their Properties. Int. J. Nanomed. 2017, 12, 7053–7073. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.; Bugno, J.; Lee, S.; Hong, S. Dendrimer-based Nanocarriers: A Versatile Platform for Drug Delivery. WIREs Nanomed. Nanobiotechnol. 2017, 9, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Palmerston Mendes, L.; Pan, J.; Torchilin, V. Dendrimers as Nanocarriers for Nucleic Acid and Drug Delivery in Cancer Therapy. Molecules 2017, 22, 1401. [Google Scholar] [CrossRef] [PubMed]
- Kuang, T.; Fu, D.; Chang, L.; Yang, Z.; Chen, Z.; Jin, L.; Chen, F.; Peng, X. Recent Progress in Dendrimer-Based Gene Delivery Systems. Curr. Org. Chem. 2016, 20, 1820–1826. [Google Scholar] [CrossRef]
- Pompilio, A.; Geminiani, C.; Mantini, P.; Siriwardena, T.N.; Di Bonaventura, I.; Reymond, J.L.; Di Bonaventura, G. Peptide Dendrimers as “Lead Compounds” for the Treatment of Chronic Lung Infections by Pseudomonas Aeruginosa in Cystic Fibrosis Patients: In Vitro and in Vivo Studies. Infect. Drug Resist. 2018, 11, 1767–1782. [Google Scholar] [CrossRef] [PubMed]
- Ben Jeddou, F.; Falconnet, L.; Luscher, A.; Siriwardena, T.; Reymond, J.-L.; van Delden, C.; Köhler, T. Adaptive and Mutational Responses to Peptide Dendrimer Antimicrobials in Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2020, 64, e02040-19. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Jordan, O.; Hanawa, T.; Borchard, G.; Patrulea, V. Are Antimicrobial Peptide Dendrimers an Escape from ESKAPE? Adv. Wound Care 2020, 9, 378–395. [Google Scholar] [CrossRef]
- Cañas-Arranz, R.; de León, P.; Forner, M.; Defaus, S.; Bustos, M.J.; Torres, E.; Andreu, D.; Blanco, E.; Sobrino, F. Immunogenicity of a Dendrimer B2T Peptide Harboring a T-Cell Epitope From FMDV Non-Structural Protein 3D. Front. Vet. Sci. 2020, 7, 498. [Google Scholar] [CrossRef]
- Mhlwatika, Z.; Aderibigbe, B. Application of Dendrimers for the Treatment of Infectious Diseases. Molecules 2018, 23, 2205. [Google Scholar] [CrossRef]
- Kandeel, M.; Al-Taher, A.; Park, B.K.; Kwon, H.; Al-Nazawi, M. A Pilot Study of the Antiviral Activity of Anionic and Cationic Polyamidoamine Dendrimers against the Middle East Respiratory Syndrome Coronavirus. J. Med. Virol. 2020, 92, 1665–1670. [Google Scholar] [CrossRef]
- Crespo, L.; Sanclimens, G.; Pons, M.; Giralt, E.; Royo, M.; Albericio, F. Peptide and Amide Bond-Containing Dendrimers. Chem. Rev. 2005, 105, 1663–1681. [Google Scholar] [CrossRef] [PubMed]
- Johansson, E.M.V.; Crusz, S.A.; Kolomiets, E.; Buts, L.; Kadam, R.U.; Cacciarini, M.; Bartels, K.-M.; Diggle, S.P.; Cámara, M.; Williams, P.; et al. Inhibition and Dispersion of Pseudomonas Aeruginosa Biofilms by Glycopeptide Dendrimers Targeting the Fucose-Specific Lectin LecB. Chem. Biol. 2008, 15, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Gorain, B.; Choudhury, H.; Pandey, M.; Mohd Amin, M.C.I.; Singh, B.; Gupta, U.; Kesharwani, P. Dendrimers as Effective Carriers for the Treatment of Brain Tumor. In Nanotechnology-Based Targeted Drug Delivery Systems for Brain Tumors; Elsevier: Amsterdam, The Netherlands, 2018; pp. 267–305. [Google Scholar]
- Cooper, B.M.; Iegre, J.; O’Donovan, D.H.; Ölwegård Halvarsson, M.; Spring, D.R. Peptides as a Platform for Targeted Therapeutics for Cancer: Peptide–Drug Conjugates (PDCs). Chem. Soc. Rev. 2021, 50, 1480–1494. [Google Scholar] [CrossRef] [PubMed]
- Gorzkiewicz, M.; Kopeć, O.; Janaszewska, A.; Konopka, M.; Pędziwiatr-Werbicka, E.; Tarasenko, I.I.; Bezrodnyi, V.V.; Neelov, I.M.; Klajnert-Maculewicz, B. Poly(Lysine) Dendrimers Form Complexes with SiRNA and Provide Its Efficient Uptake by Myeloid Cells: Model Studies for Therapeutic Nucleic Acid Delivery. Int. J. Mol. Sci. 2020, 21, 3138. [Google Scholar] [CrossRef]
- Sheveleva, N.N.; Markelov, D.A.; Vovk, M.A.; Mikhailova, M.E.; Tarasenko, I.I.; Neelov, I.M.; Lähderanta, E. NMR Studies of Excluded Volume Interactions in Peptide Dendrimers. Sci. Rep. 2018, 8, 8916. [Google Scholar] [CrossRef] [PubMed]
- Sheveleva, N.N.; Markelov, D.A.; Vovk, M.A.; Mikhailova, M.E.; Tarasenko, I.I.; Tolstoy, P.M.; Neelov, I.M.; Lähderanta, E. Lysine-Based Dendrimer with Double Arginine Residues. RSC Adv. 2019, 9, 18018–18026. [Google Scholar] [CrossRef] [PubMed]
- Sheveleva, N.N.; Markelov, D.A.; Vovk, M.A.; Tarasenko, I.I.; Mikhailova, M.E.; Ilyash, M.Y.; Neelov, I.M.; Lahderanta, E. Stable Deuterium Labeling of Histidine-Rich Lysine-Based Dendrimers. Molecules 2019, 24, 2481. [Google Scholar] [CrossRef]
- Sheveleva, N.N.; Bezrodnyi, V.V.; Mikhtaniuk, S.E.; Shavykin, O.V.; Neelov, I.M.; Tarasenko, I.I.; Vovk, M.A.; Mikhailova, M.E.; Penkova, A.V.; Markelov, D.A. Local Orientational Mobility of Collapsed Dendrimers. Macromolecules 2021, 54, 11083–11092. [Google Scholar] [CrossRef]
- Bezrodnyi, V.V.; Mikhtaniuk, S.E.; Shavykin, O.V.; Neelov, I.M.; Sheveleva, N.N.; Markelov, D.A. Size and Structure of Empty and Filled Nanocontainer Based on Peptide Dendrimer with Histidine Spacers at Different PH. Molecules 2021, 26, 6552. [Google Scholar] [CrossRef]
- Santos, A.; Veiga, F.; Figueiras, A. Dendrimers as Pharmaceutical Excipients: Synthesis, Properties, Toxicity and Biomedical Applications. Materials 2020, 13, 65. [Google Scholar] [CrossRef]
- Bai, S.; Thomas, C.; Ahsan, F. Dendrimers as a Carrier for Pulmonary Delivery of Enoxaparin, a Low-Molecular Weight Heparin. J. Pharm. Sci. 2007, 96, 2090–2106. [Google Scholar] [CrossRef] [PubMed]
- Gupta, L.; Sharma, A.K.; Gothwal, A.; Khan, M.S.; Khinchi, M.P.; Qayum, A.; Singh, S.K.; Gupta, U. Dendrimer Encapsulated and Conjugated Delivery of Berberine: A Novel Approach Mitigating Toxicity and Improving in Vivo Pharmacokinetics. Int. J. Pharm. 2017, 528, 88–99. [Google Scholar] [CrossRef]
- Madaan, K.; Lather, V.; Pandita, D. Evaluation of Polyamidoamine Dendrimers as Potential Carriers for Quercetin, a Versatile Flavonoid. Drug Deliv. 2016, 23, 254–262. [Google Scholar] [CrossRef]
- Pentek, T.; Newenhouse, E.; O’Brien, B.; Chauhan, A. Development of a Topical Resveratrol Formulation for Commercial Applications Using Dendrimer Nanotechnology. Molecules 2017, 22, 137. [Google Scholar] [CrossRef]
- Falconieri, M.; Adamo, M.; Monasterolo, C.; Bergonzi, M.; Coronnello, M.; Bilia, A. New Dendrimer-Based Nanoparticles Enhance Curcumin Solubility. Planta Med. 2016, 83, 420–425. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, J.; Zheng, Y.; Guo, R.; Wang, S.; Mignani, S.; Caminade, A.M.; Majoral, J.P.; Shi, X. Doxorubicin-Conjugated PAMAM Dendrimers for pH-Responsive Drug Release and Folic Acid-Targeted Cancer Therapy. Pharmaceutics 2018, 10, 162. [Google Scholar] [CrossRef]
- Almuqbil, R.M.; Heyder, R.S.; Bielski, E.R.; Durymanov, M.; Reineke, J.J.; da Rocha, S.R.P. Dendrimer Conjugation Enhances Tumor Penetration and Efficacy of Doxorubicin in Extracellular Matrix-Expressing 3D Lung Cancer Models. Mol. Pharm. 2020, 17, 1648–1662. [Google Scholar] [CrossRef]
- Teow, H.M.; Zhou, Z.; Najlah, M.; Yusof, S.R.; Abbott, N.J.; D’Emanuele, A. Delivery of paclitaxel across cellular barriers using a dendrimer-based nanocarrier. Int. J. Pharm. 2013, 441, 701–711. [Google Scholar] [CrossRef]
- Jain, N.K.; Tare, M.S.; Mishra, V.; Tripathi, P.K. The development, characterization and in vivo antiovarian cancer activity of poly(propylene imine) (PPI)-antibody conjugates containing encapsulated paclitaxel. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, J.; Yu, B.; Zhang, J.; Hu, H.; Cong, H.; Shen, Y. The drug loading behavior of PAMAM dendrimer: Insights from experimental and simulation study. Sci. China Technol. Sci. 2023, 66, 1129–1140. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Rajhi, A.A.; Ali, E.; Chandra, S.; Ahmed, H.H.; Adhab, Z.H.; Alkhayyat, A.S.; Alsalamy, A.; Saadh, M.J. Acetyl-terminated PAMAM dendrimers for pH-sensitive delivery of irinotecan and fluorouracil: A molecular dynamics simulation study. Chem. Phys. Lett. 2023, 832, 140876. [Google Scholar] [CrossRef]
- Wolski, P.; Panczyk, T.; Brzyska, A. Molecular Dynamics Simulations of Carbon Quantum Dots Polyamidoamine Dendrimer Nanocomposites. J. Phys. Chem. C 2023, 127, 16740–16750. [Google Scholar] [CrossRef]
- Shen, Z.-L.; Tian, W.-D.; Chen, K.; Ma, Y.-Q. Molecular dynamics simulation of G-actin interacting with PAMAM dendrimers. J. Mol. Graph. Model. 2018, 84, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Stojceski, F.; Grasso, G.; Pallante, L.; Danani, A. Molecular and Coarse-Grained Modeling to Characterize and Optimize Dendrimer-Based Nanocarriers for Short Interfering RNA Delivery. ACS Omega 2020, 5, 2978–2986. [Google Scholar] [CrossRef] [PubMed]
- Trosheva, K.S.; Sorokina, S.A.; Efimova, A.A.; Semenyuk, P.I.; Berkovich, A.K.; Yaroslavov, A.A.; Shifrina, Z.B. Interaction of multicomponent anionic liposomes with cationic pyridylphenylene dendrimer: Does the complex behavior depend on the liposome composition? BBA Biomembr. 2021, 1863, 183761. [Google Scholar] [CrossRef]
- Chawla, P.; Chawla, V.; Maheshwari, R.; Saraf, S.A.; Saraf, S.K. Fullerenes: From Carbon to Nanomedicine. Mini-Rev. Med. Chem. 2010, 10, 662–677. [Google Scholar] [CrossRef]
- Song, M.; Liu, S.; Yin, J.; Wang, H. Interaction of Human Serum Album and C60 Aggregates in Solution. Int. J. Mol. Sci. 2011, 12, 4964–4974. [Google Scholar] [CrossRef]
- Kroto, H.W.; Heath, J.R.; O’Brien, S.C.; Curl, R.F.; Smalley, R.E. C60: Buckminsterfullerene. Nature 1985, 318, 162–163. [Google Scholar] [CrossRef]
- Torres, V.M.; Srdjenovic, B. Biomedical Application of Fullerenes. In Handbook on Fullerene: Synthesis, Properties and Applications; Verner, R.F., Benvegnu, C., Eds.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2011; pp. 199–239. [Google Scholar]
- Moussa, F. [60]Fullerene and Derivatives for Biomedical Applications. In Nanobiomaterials; Elsevier: Amsterdam, The Netherlands, 2018; pp. 113–136. [Google Scholar]
- Kazemzadeh, H.; Mozafari, M. Fullerene-Based Delivery Systems. Drug Discov. Today 2019, 24, 898–905. [Google Scholar] [CrossRef]
- Gharbi, N.; Pressac, M.; Hadchouel, M.; Szwarc, H.; Wilson, S.R.; Moussa, F. [60]Fullerene Is a Powerful Antioxidant in Vivo with No Acute or Subacute Toxicity. Nano Lett. 2005, 5, 2578–2585. [Google Scholar] [CrossRef]
- Roy, P.; Bag, S.; Chakraborty, D.; Dasgupta, S. Exploring the Inhibitory and Antioxidant Effects of Fullerene and Fullerenol on Ribonuclease A. ACS Omega 2018, 3, 12270–12283. [Google Scholar] [CrossRef]
- Zhao, P.-F.; Liu, Z.-Q. Attaching a Dipeptide to Fullerene as an Antioxidant Hybrid against DNA Oxidation. Chem. Res. Toxicol. 2021, 34, 2366–2374. [Google Scholar] [CrossRef] [PubMed]
- Lyon, D.Y.; Adams, L.K.; Falkner, J.C.; Alvarez, P.J.J. Antibacterial Activity of Fullerene Water Suspensions: Effects of Preparation Method and Particle Size. Environ. Sci. Technol. 2006, 40, 4360–4366. [Google Scholar] [CrossRef] [PubMed]
- Piotrovsky, L.B.; Kiselev, O.I. Fullerenes and Viruses. Fuller. Nanotub. Carbon Nanostruct. 2005, 12, 397–403. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, C.; Feng, L.; Yang, K.; Liu, Z. Functional Nanomaterials for Phototherapies of Cancer. Chem. Rev. 2014, 114, 10869–10939. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Mao, R.; Liu, Y. Fullerenes for Cancer Diagnosis and Therapy: Preparation, Biological and Clinical Perspectives. Curr. Drug Metab. 2012, 13, 1035–1045. [Google Scholar] [CrossRef]
- Fernandes, N.B.; Shenoy, R.U.K.; Kajampady, M.K.; DCruz, C.E.M.; Shirodkar, R.K.; Kumar, L.; Verma, R. Fullerenes for the Treatment of Cancer: An Emerging Tool. Environ. Sci. Pollut. Res. 2022, 29, 58607–58627. [Google Scholar] [CrossRef]
- Kokubo, K.; Matsubayashi, K.; Tategaki, H.; Takada, H.; Oshima, T. Facile Synthesis of Highly Water-Soluble Fullerenes More Than Half-Covered by Hydroxyl Groups. ACS Nano 2008, 2, 327–333. [Google Scholar] [CrossRef]
- Anilkumar, P.; Lu, F.; Cao, L.; Luo, P.G.; Liu, J.-H.; Sahu, S.; Tackett, K.N., II; Wang, Y.; Sun, Y.-P. Fullerenes for Applications in Biology and Medicine. Curr. Med. Chem. 2011, 18, 2045–2059. [Google Scholar] [CrossRef]
- Giacalone, F.; D’Anna, F.; Giacalone, R.; Gruttadauria, M.; Riela, S.; Noto, R. Cyclodextrin-[60]Fullerene Conjugates: Synthesis, Characterization, and Electrochemical Behavior. Tetrahedron Lett. 2006, 47, 8105–8108. [Google Scholar] [CrossRef]
- Zhang, L.W.; Yang, J.; Barron, A.R.; Monteiro-Riviere, N.A. Endocytic Mechanisms and Toxicity of a Functionalized Fullerene in Human Cells. Toxicol. Lett. 2009, 191, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Haque, S.A.; Yang, S.-T.; Luo, P.G.; Gu, L.; Kitaygorodskiy, A.; Li, H.; Lacher, S.; Sun, Y.-P. Aqueous Compatible Fullerene−Doxorubicin Conjugates. J. Phys. Chem. C 2009, 113, 17768–17773. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, X.; Ebrahimi, A.; Li, J.; Cui, Q. Fullerene-Biomolecule Conjugates and Their Biomedicinal Applications. Int. J. Nanomed. 2014, 9, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Biswas, R.; Batista Da Rocha, C.; Bennick, R.A.; Zhang, J. Water-Soluble Fullerene Monoderivatives for Biomedical Applications. ChemMedChem 2023, 18, e202300296. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.W.; Monthioux, M.; Luzzi, D.E. Encapsulated C60 in Carbon Nanotubes. Nature 1998, 396, 323–324. [Google Scholar] [CrossRef]
- Simon, F.; Peterlik, H.; Pfeiffer, R.; Bernardi, J.; Kuzmany, H. Fullerene Release from the inside of Carbon Nanotubes: A Possible Route toward Drug Delivery. Chem. Phys. Lett. 2007, 445, 288–292. [Google Scholar] [CrossRef]
- Zhou, Z. Liposome Formulation of Fullerene-Based Molecular Diagnostic and Therapeutic Agents. Pharmaceutics 2013, 5, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Hahn, U.; Vögtle, F.; Nierengarten, J.-F. Synthetic Strategies towards Fullerene-Rich Dendrimer Assemblies. Polymers 2012, 4, 501–538. [Google Scholar] [CrossRef]
- Nishioka, T.; Tashiro, K.; Aida, T.; Zheng, J.-Y.; Kinbara, K.; Saigo, K.; Sakamoto, S.; Yamaguchi, K. Molecular Design of a Novel Dendrimer Porphyrin for Supramolecular Fullerene/Dendrimer Hybridization. Macromolecules 2000, 33, 9182–9184. [Google Scholar] [CrossRef]
- Kay, K.-Y.; Han, K.-J.; Yu, Y.-J.; Park, Y.D. Dendritic fullerenes (C60) with photoresponsive azobenzene groups. Tetrahedron Lett. 2002, 43, 5053–5056. [Google Scholar] [CrossRef]
- Rio, Y.; Accorsi, G.; Nierengarten, H.; Rehspringer, J.-L.; Honerlage, B.; Kopitkovas, G.; Chugreev, A.; Dorsselaer, A.V.; Armaroli, N.; Nierengarten, J.-F. Fullerodendrimers with peripheral triethyleneglycol chains: Synthesis, mass spectrometric characterization, and photophysical properties. New J. Chem. 2002, 26, 1146–1154. [Google Scholar] [CrossRef]
- Takaguchi, Y.; Sako, Y.; Yanagimoto, Y.; Tsuboi, S.; Motoyoshiya, J.; Aoyama, H.; Wakahara, T.; Akasaka, T. Facile and reversible synthesis of an acidic water-soluble poly(amidoamine) fullerodendrimer. Tetrahedron Lett. 2003, 44, 5777–5780. [Google Scholar] [CrossRef]
- Takaguchi, Y.; Yanagimoto, Y.; Fujima, S.; Tsuboi, S. Photooxygenation of Olefins, Phenol, and Sulfide Using Fullerodendrimer as Catalyst. Chem. Lett. 2004, 33, 1142–1143. [Google Scholar] [CrossRef]
- Jensen, A.W.; Maru, B.S.; Zhang, X.; Mohanty, D.K.; Fahlman, B.D.; Swanson, D.R.; Tomalia, D.A. Preparation of Fullerene-Shell Dendrimer-Core Nanoconjugates. Nano Lett. 2005, 5, 1171–1173. [Google Scholar] [CrossRef]
- Hahn, U.; Cardinali, F.; Nierengarten, J.-F. Supramolecular chemistry for the self-assembly of fullerene-rich dendrimers. New J. Chem. 2007, 31, 1128–1138. [Google Scholar] [CrossRef]
- Chai, M.; Jensen, A.W.; Abdelhady, H.G.; Tomalia, D.A. AFM Analysis of C60 and a Poly(amido amine) Dendrimer-C60 Nanoconjugate. J. Nanosci. Nanotechnol. 2007, 7, 1401–1405. [Google Scholar] [CrossRef]
- Scanu, D.; Yevlampieva, N.P.; Deschenaux, R. Polar and Electrooptical Properties of [60]Fullerene-Containing Poly(benzyl ether) Dendrimers in Solution. Macromolecules 2007, 40, 1133–1139. [Google Scholar] [CrossRef]
- Hahn, U.; Nierengarten, J.-F.; Vögtle, F.; Listorti, A.; Monti, F.; Armaroli, N. Fullerene-rich dendrimers: Divergent synthesis and photophysical properties. New J. Chem. 2009, 33, 337–344. [Google Scholar] [CrossRef]
- Hahn, U.; Nierengarten, J.-F.; Delavaux-Nicot, B.; Monti, F.; Chiorboli, C.; Armaroli, N. Fullerodendrimers with a perylenediimide core. New J. Chem. 2011, 35, 2234–2244. [Google Scholar] [CrossRef]
- Kim, J.; Yun, M.H.; Lee, J.; Kim, J.Y.; Wudl, F.; Yang, C. A synthetic approach to a fullerene-rich dendron and its linear polymer via ring-opening metathesis polymerization. Chem. Commun. 2011, 47, 3078. [Google Scholar] [CrossRef]
- Hung, C.-H.; Chang, W.-W.; Liu, S.-C.; Wu, S.-J.; Chu, C.-C.; Tsai, Y.-J.; Imae, T. Self-aggregation of amphiphilic [60]fullerenyl focal point functionalized PAMAM dendrons into pseudodendrimers: DNA binding involving dendriplex formation. J. Biomed. Mater. Res. Part A 2015, 103, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Vonlanthen, M.; Gonzalez-Ortega, J.; Porcu, P.; Ruiu, A.; Rodríguez-Alba, E.; Cevallos-Vallejo, A.; Rivera, E. Pyrene-labeled dendrimers functionalized with fullerene C60 or porphyrin core as light harvesting antennas. Synth. Met. 2018, 245, 195–201. [Google Scholar] [CrossRef]
- Schonberger, H.; Schwab, C.H.; Hirsch, A.; Gasteiger, J. Molecular Modeling of Fullerene Dendrimers. J. Mol. Model. 2000, 6, 379–395. [Google Scholar] [CrossRef]
- Nierengarten, J.-F. Dendritic encapsulation of active core molecules. C. R. Chim. 2003, 6, 725–733. [Google Scholar] [CrossRef]
- Kujawski, M.; Rakesh, L.; Gala, K.; Jensen, A.; Fahlman, B.; Feng, Z.R.; Mohanty, D.K. Molecular Dynamics Simulation of Polyamidoamine Dendrimer-Fullerene Conjugates:Generations Zero Through Four. J. Nanosci. Nanotechnol. 2007, 7, 1670–1674. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, P.; Kim, S.H.; Chen, P.; Chen, R.; Spuches, A.M.; Brown, J.M.; Lamm, M.H.; Ke, P.C. Dendrimer-fullerenol soft-condensed nanoassembly. J. Phys. Chem. C 2012, 116, 15775–15781. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Albrecht, K.; Kasai, Y.; Kuramoto, Y.; Yamamoto, K. Dynamic control of dendrimer–fullerene association by axial coordination to the core. Chem. Commun. 2013, 49, 6861. [Google Scholar] [CrossRef]
- Albrecht, K.; Kasai, Y.; Kuramoto, Y.; Yamamoto, K. A fourth-generation carbazole-phenylazomethine dendrimer as a size-selective host for fullerenes. Chem. Commun. 2013, 49, 865–867. [Google Scholar] [CrossRef]
- Eckert, J.-F.; Byrne, D.; Nicoud, J.-F.; Oswald, L.; Nierengarten, J.-F.; Numata, M.; Ikeda, A.; Shinkai, S.; Armaroli, N. Polybenzyl Ether Dendrimers for the Complexation of [60]Fullerenes. New J. Chem. 2000, 24, 749–758. [Google Scholar] [CrossRef]
- Gorzkiewicz, M.; Konopka, M.; Janaszewska, A.; Tarasenko, I.I.; Sheveleva, N.N.; Gajek, A.; Neelov, I.M.; Klajnert-Maculewicz, B. Application of New Lysine-Based Peptide Dendrimers D3K2 and D3G2 for Gene Delivery: Specific Cytotoxicity to Cancer Cells and Transfection in Vitro. Bioorg. Chem. 2020, 95, 103504. [Google Scholar] [CrossRef]
- Ikeda, A.; Doi, Y.; Nishiguchi, K.; Kitamura, K.; Hashizume, M.; Kikuchi, J.; Yogo, K.; Ogawa, T.; Takeya, T. Induction of Cell Death by Photodynamic Therapy with Water-Soluble Lipid-Membrane-Incorporated [60]Fullerene. Org. Biomol. Chem. 2007, 5, 1158. [Google Scholar] [CrossRef] [PubMed]
- Kulchitsky, V.A.; Alexandrova, R.; Suziedelis, K.; Paschkevich, S.G.; Potkin, V.I. Perspectives of Fullerenes, Dendrimers, and Heterocyclic Compounds Application in Tumor Treatment. Recent Pat. Nanomed. 2015, 4, 82–89. [Google Scholar] [CrossRef]
- Markelov, D.A.; Mazo, M.A.; Balabaev, N.K.; Gotlib, Y.Y. Temperature Dependence of the Structure of a Carbosilane Dendrimer with Terminal Cyanobiphenyl Groups: Molecular-Dynamics Simulation. Polym. Sci. Ser. A 2013, 55, 53–60. [Google Scholar] [CrossRef]
- Markelov, D.A.; Polotsky, A.A.; Birshtein, T.M. Formation of a “Hollow” Interior in the Fourth-Generation Dendrimer with Attached Oligomeric Terminal Segments. J. Phys. Chem. B 2014, 118, 14961–14971. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tang, P.; Qiu, F.; Shi, A.C. Density Functional Study for Homodendrimers and Amphiphilic Dendrimers. J. Phys. Chem. B 2016, 120, 5553–5563. [Google Scholar] [CrossRef] [PubMed]
- Markelov, D.A.; Semisalova, A.S.; Mazo, M.A. Formation of a Hollow Core in Dendrimers in Solvents. Macromol. Chem. Phys. 2021, 222, 2100085. [Google Scholar] [CrossRef]
- Kłos, J.S.; Sommer, J.-U. Properties of Dendrimers with Flexible Spacer-Chains: A Monte Carlo Study. Macromolecules 2009, 42, 4878–4886. [Google Scholar] [CrossRef]
- Rudnick, J.; Gaspari, G. The Aspherity of Random Walks. J. Phys. A Math. Gen. 1986, 19, L191–L193. [Google Scholar] [CrossRef]
- Theodorou, D.N.; Suter, U.W. Shape of Unperturbed Linear Polymers: Polypropylene. Macromolecules 1985, 18, 1206–1214. [Google Scholar] [CrossRef]
- Roberts, B.P.; Scanlon, M.J.; Krippner, G.Y.; Chalmers, D.K. Molecular Dynamics of Poly(L-lysine) Dendrimers with Naphthalene Disulfonate Caps. Macromolecules 2009, 42, 2775–2783. [Google Scholar] [CrossRef]
- Ohshima, H. Theory of Colloid and Interfacial Electric Phenomena; Interface Science and Technology; Academic Press: Cambridge, MA, USA, 2006; Volume 12, p. 473. ISBN 9780123706423. [Google Scholar]
- Eslami, H.; Khani, M.; Muller-Plathe, F. Gaussian Charge Distributions for Incorporation of Electrostatic Interactions in Dissipative Particle Dynamics: Application to Self-Assembly of Surfactants. J. Chem. Theory Comput. 2019, 15, 4197–4207. [Google Scholar] [CrossRef] [PubMed]
- Delgado, A.V.; Gonzalez-Caballero, F.; Hunter, R.J.; Koopal, L.K.; Lyklema, J. Measurement and interpretation of electrokinetic phenomena. Pure Appl. Chem. 2005, 77, 1753–1805. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-Chain Torsion Potentials for the Amber Ff99SB Protein Force Field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Mikhtaniuk, S.; Bezrodnyi, V.; Shavykin, O.; Neelov, I.; Sheveleva, N.; Penkova, A.; Markelov, D. Comparison of Structure and Local Dynamics of Two Peptide Dendrimers with the Same Backbone but with Different Side Groups in Their Spacers. Polymers 2020, 12, 1657. [Google Scholar] [CrossRef]
- Stroet, M.; Caron, B.; Visscher, K.M.; Geerke, D.P.; Malde, A.K.; Mark, A.E. Automated Topology Builder Version 3.0: Prediction of Solvation Free Enthalpies in Water and Hexane. J. Chem. Theory Comput. 2018, 14, 5834–5845. [Google Scholar] [CrossRef] [PubMed]
- Darinskii, A.; Gotlib, Y.Y.; Lyulin, A.; Neyelov, L.M. Computer Simulation of Local Dynamics of a Polymer Chain in the Orienting Field of the Liquid Crystal Type. Polym. Sci. USSR 1991, 33, 1116–1125. [Google Scholar] [CrossRef]
- Ennari, J.A.; Elomaa, M.; Neelov, I.; Sundholm, F. Modeling of water-free and water containing solid polyelectrolytes. Polymer 2000, 41, 985–990. [Google Scholar] [CrossRef]
- Ennari, J.; Neelov, I.; Sundholm, F. Molecular dynamics simulation of the PEO sulfonic acid anion in water. Comput. Theor. Polym. Sci. 2000, 10, 403–410. [Google Scholar] [CrossRef]
- Neelov, I.M.; Binder, K. Brownian dynamics of grafted polymer chains: Time dependent properties. Macromol. Theory Simul. 1995, 4, 1063–1084. [Google Scholar] [CrossRef]
- Neelov, I.M.; Adolf, D.B.; McLeish, T.C.B.; Paci, E. Molecular Dynamics Simulation of Dextran Extension by Constant Force in Single Molecule AFM. Biophys. J. 2006, 91, 3579–3588. [Google Scholar] [CrossRef]
- Gowdy, J.; Batchelor, M.; Neelov, I.; Paci, E. Nonexponential Kinetics of Loop Formation in Proteins and Peptides: A Signature of Rugged Free Energy Landscapes? J. Phys. Chem. B 2017, 121, 9518–9525. [Google Scholar] [CrossRef] [PubMed]
- Bezrodnyi, V.V.; Shavykin, O.V.; Mikhtaniuk, S.E.; Neelov, I.M.; Sheveleva, N.N.; Markelov, D.A. Why the Orientational Mobility in Arginine and Lysine Spacers of Peptide Dendrimers Designed for Gene Delivery Is Different? Int. J. Mol. Sci. 2020, 21, 9749. [Google Scholar] [CrossRef] [PubMed]
- Shavykin, O.V.; Neelov, I.M.; Darinskii, A.A. Is the Manifestation of the Local Dynamics in the Spin–Lattice NMR Relaxation in Dendrimers Sensitive to Excluded Volume Interactions? Phys. Chem. Chem. Phys. 2016, 18, 24307–24317. [Google Scholar] [CrossRef] [PubMed]
- Shavykin, O.V.; Mikhailov, I.V.; Darinskii, A.A.; Neelov, I.M.; Leermakers, F.A.M. Effect of an Asymmetry of Branching on Structural Characteristics of Dendrimers Revealed by Brownian Dynamics Simulations. Polymer 2018, 146, 256–266. [Google Scholar] [CrossRef]
- Shavykin, O.V.; Leermakers, F.A.M.; Neelov, I.M.; Darinskii, A.A. Self-Assembly of Lysine-Based Dendritic Surfactants Modeled by the Self-Consistent Field Approach. Langmuir 2018, 34, 1613–1626. [Google Scholar] [CrossRef]
- Mikhailov, I.V.; Darinskii, A.A. Does symmetry of branching affect the properties of dendrimers? Polym. Sci. Ser. A 2014, 56, 534–544. [Google Scholar] [CrossRef]
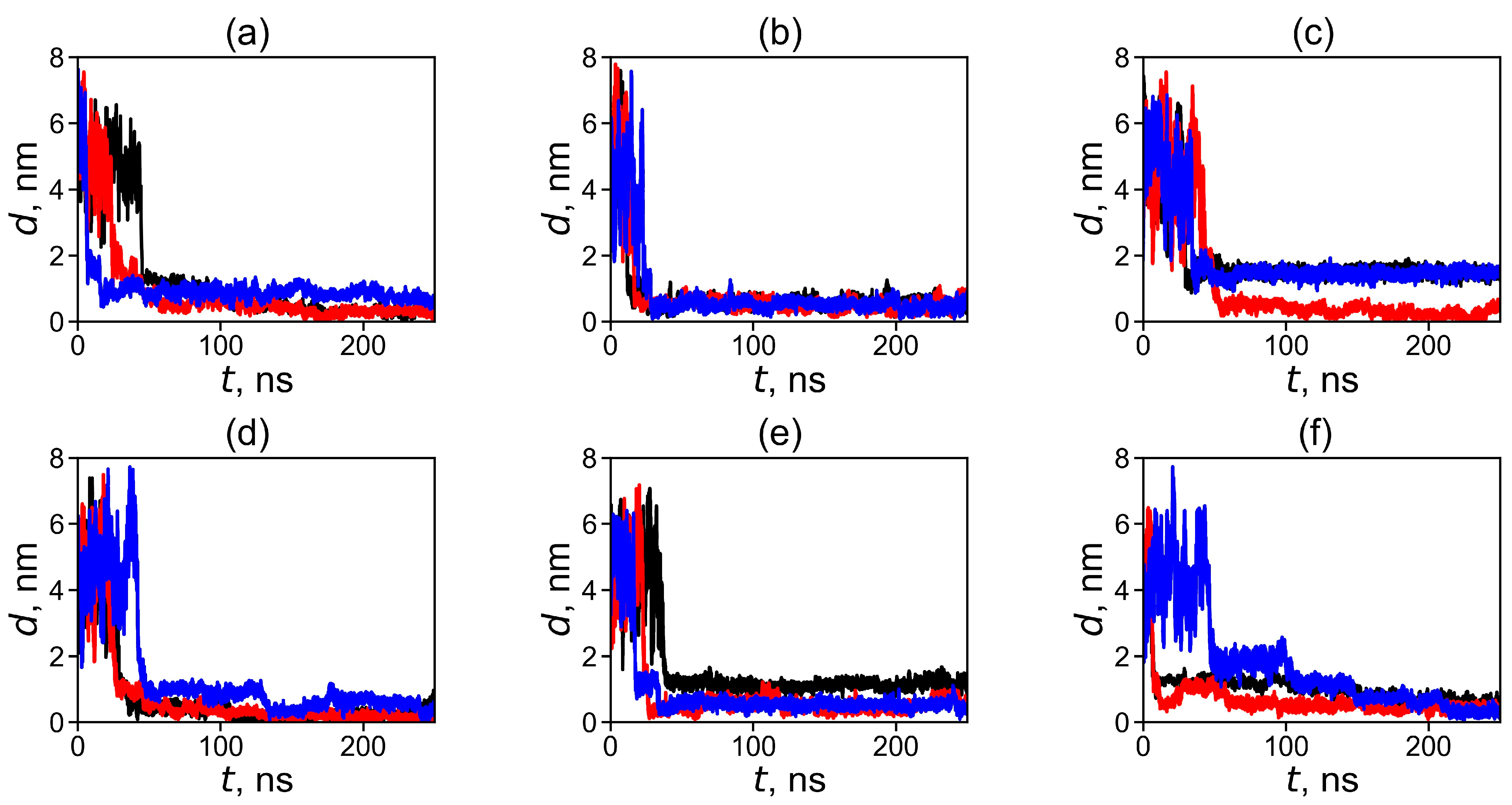
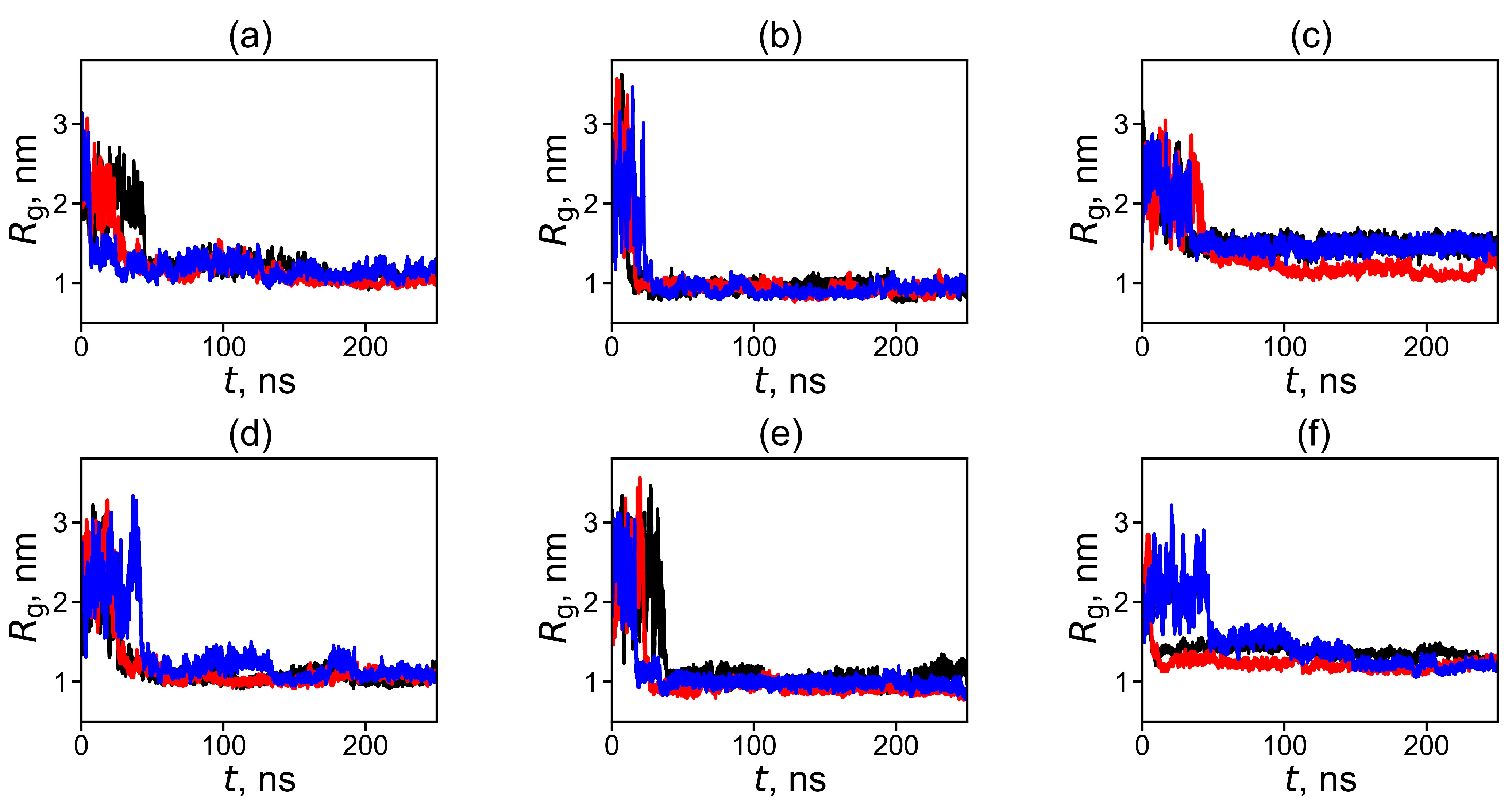

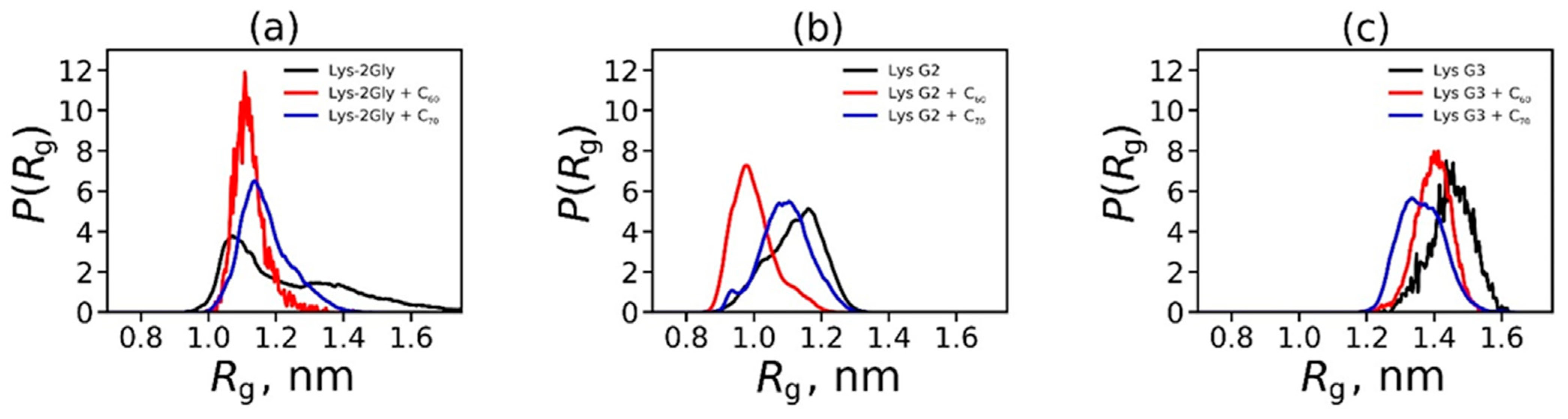

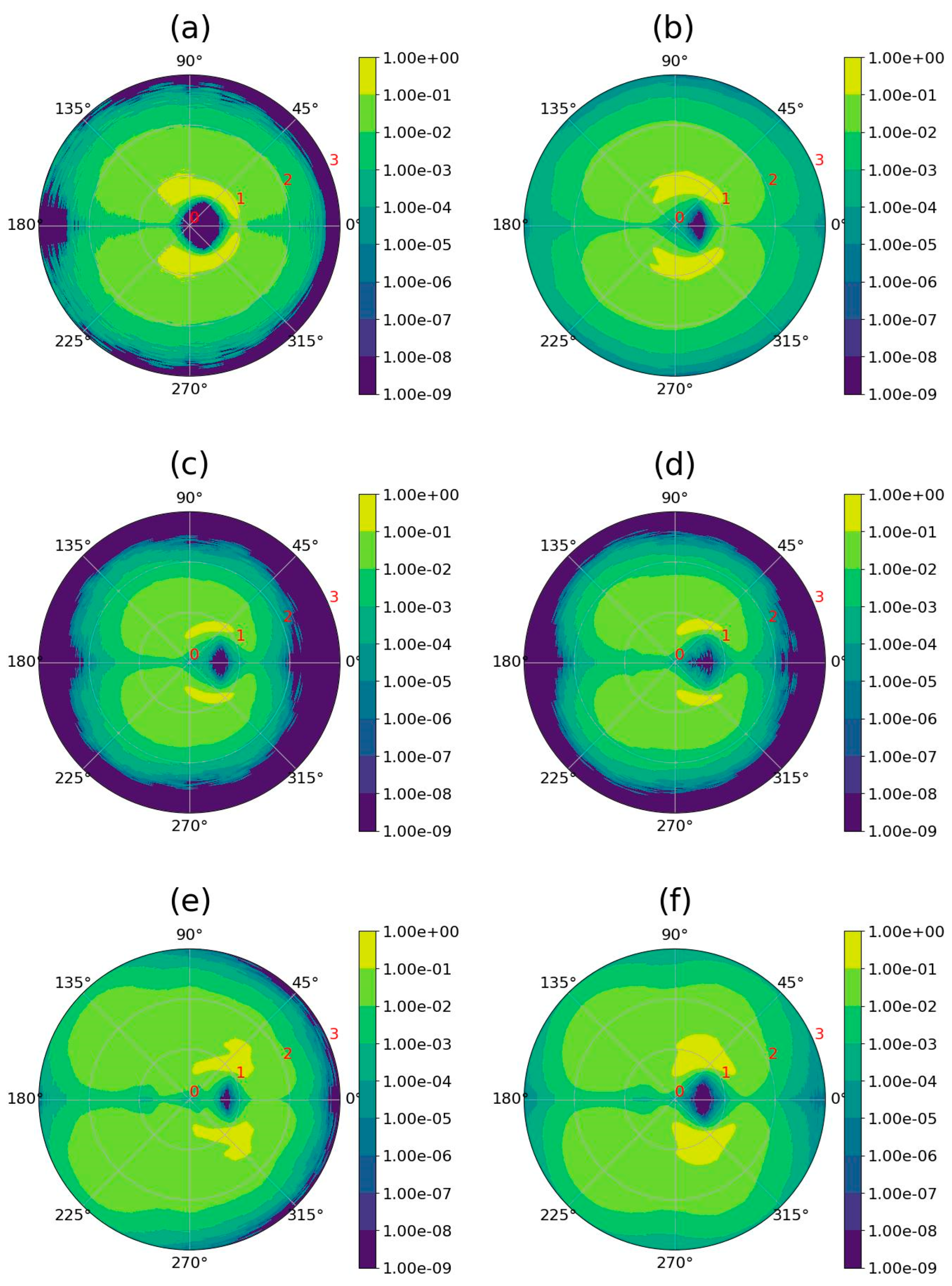
| Dendrimer/Fullertene | α | Rg | d |
|---|---|---|---|
| Lys-2Gly | 0.013 | 1.24 | - |
| Lys-2Gly + C60 | 0.006 | 1.04 | 0.51 |
| Lys-2Gly + C70 | 0.006 | 1.08 | 0.57 |
| Lys G2 | 0.021 | 1.12 | - |
| Lys G2 + C60 | 0.009 | 0.91 | 0.72 |
| Lys G2 + C70 | 0.018 | 0.98 | 0.77 |
| Lys G3 | 0.015 | 1.45 | - |
| Lys G3 + C60 | 0.018 | 1.46 | 0.81 |
| Lys G3 + C70 | 0.012 | 1.27 | 0.78 |
| System | Qmax [e] | ζ [mV] | NHB |
|---|---|---|---|
| Lys-2Gly | 9.3 | 21.2 | 101 |
| Lys-2Gly + C60 | 11.3 | 35.1 | 96 |
| Lys-2Gly + C70 | 11.3 | 35.2 | 100 |
| Lys G2 | 10.1 | 30.9 | 56 |
| Lys G2 + C60 | 11.5 | 52.8 | 55 |
| Lys G2 + C70 | 11.4 | 45.1 | 56 |
| Lys G3 | 15.7 | 37.4 | 107 |
| Lys G3 + C60 | 15.8 | 40.1 | 105 |
| Lys G3 + C70 | 15.5 | 42.3 | 104 |
| Dendrimer | Md, g/mol | Qbare [e] | Nend | Nins | Nres |
|---|---|---|---|---|---|
| Lys-2Gly | 3675.44 | 16 | 16 | 28 | 44 |
| Lys G2 | 1895.63 | 16 | 16 | 0 | 16 |
| Lys G3 | 3962.5 | 32 | 32 | 0 | 32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bezrodnyi, V.V.; Mikhtaniuk, S.E.; Shavykin, O.V.; Sheveleva, N.N.; Markelov, D.A.; Neelov, I.M. A Molecular Dynamics Simulation of Complexes of Fullerenes and Lysine-Based Peptide Dendrimers with and without Glycine Spacers. Int. J. Mol. Sci. 2024, 25, 691. https://doi.org/10.3390/ijms25020691
Bezrodnyi VV, Mikhtaniuk SE, Shavykin OV, Sheveleva NN, Markelov DA, Neelov IM. A Molecular Dynamics Simulation of Complexes of Fullerenes and Lysine-Based Peptide Dendrimers with and without Glycine Spacers. International Journal of Molecular Sciences. 2024; 25(2):691. https://doi.org/10.3390/ijms25020691
Chicago/Turabian StyleBezrodnyi, Valeriy V., Sofia E. Mikhtaniuk, Oleg V. Shavykin, Nadezhda N. Sheveleva, Denis A. Markelov, and Igor M. Neelov. 2024. "A Molecular Dynamics Simulation of Complexes of Fullerenes and Lysine-Based Peptide Dendrimers with and without Glycine Spacers" International Journal of Molecular Sciences 25, no. 2: 691. https://doi.org/10.3390/ijms25020691
APA StyleBezrodnyi, V. V., Mikhtaniuk, S. E., Shavykin, O. V., Sheveleva, N. N., Markelov, D. A., & Neelov, I. M. (2024). A Molecular Dynamics Simulation of Complexes of Fullerenes and Lysine-Based Peptide Dendrimers with and without Glycine Spacers. International Journal of Molecular Sciences, 25(2), 691. https://doi.org/10.3390/ijms25020691






