Adult Animal Stem Cell-Derived Organoids in Biomedical Research and the One Health Paradigm
Abstract
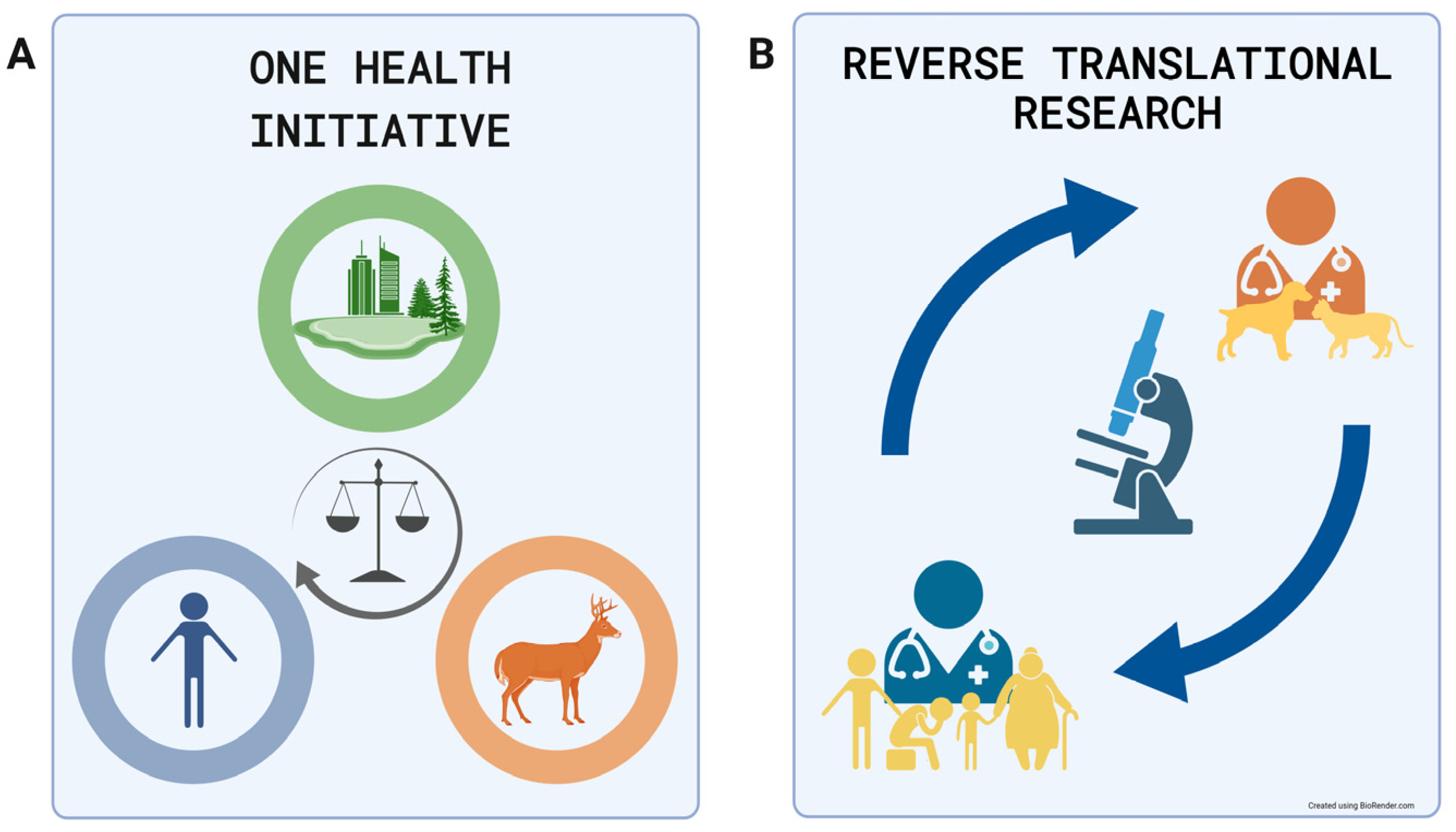
:1. Introduction
2. Brief Overview of Most Common In Vitro Cell Culture Models
2.1. Primary Cell Lines
2.2. 2D Immortalized Cell Lines
2.3. Stem Cell-Derived Organoids
3. Reproducibility and Data Deposition
3.1. Human Organoids
3.2. Murine Organoids
3.3. Other Animal-Derived Organoids
4. Main Research Applications
4.1. Intestinal Organoids
4.2. Modeling Host-Pathogen Interactions
4.3. Liver Organoids
4.4. Renal Organoids
4.5. Pancreatic Organoids
4.6. SARS-COVID-2
4.7. Precision/Personalized Medicine
4.8. Other Applications
4.8.1. Reproductive Physiology and Pathology
4.8.2. Skin Diseases
4.8.3. Ophthalmology
4.8.4. Antivenom Production
5. Summary
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Curry, S.H. Translational Science: Past, Present, and Future. BioTechniques 2008, 44, ii–viii. [Google Scholar] [CrossRef] [PubMed]
- Carbone, L. Pain in Laboratory Animals: The Ethical and Regulatory Imperatives. PLoS ONE 2011, 6, e21578. [Google Scholar] [CrossRef] [PubMed]
- Pritt, S.L.; Hammer, R.E. The Interplay of Ethics, Animal Welfare, and IACUC Oversight on the Reproducibility of Animal Studies. Comp. Med. 2017, 67, 101–105. [Google Scholar] [PubMed]
- Frasch, P.D. Gaps in US Animal Welfare Law for Laboratory Animals: Perspectives From an Animal Law Attorney. ILAR J. 2016, 57, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Hubrecht, R.C.; Carter, E. The 3Rs and Humane Experimental Technique: Implementing Change. Animals 2019, 9, 754. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A. Lost in Translation: The Valley of Death across Preclinical and Clinical Divide–Identification of Problems and Overcoming Obstacles. Transl. Med. Commun. 2019, 4, 18. [Google Scholar] [CrossRef]
- Hickman, D.L.; Johnson, J.; Vemulapalli, T.H.; Crisler, J.R.; Shepherd, R. Commonly Used Animal Models. In Principles of Animal Research for Graduate and Undergraduate Students; Academic Press: Cambridge, MA, USA, 2017; pp. 117–175. [Google Scholar] [CrossRef]
- Perlman, R.L. Mouse Models of Human Disease: An Evolutionary Perspective. Evol. Med. Public Health 2016, 2016, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Farrell, G.; Schattenberg, J.M.; Leclercq, I.; Yeh, M.M.; Goldin, R.; Teoh, N.; Schuppan, D. Mouse Models of Nonalcoholic Steatohepatitis: Toward Optimization of Their Relevance to Human Nonalcoholic Steatohepatitis. Hepatology 2019, 69, 2241–2257. [Google Scholar] [CrossRef]
- Oligschlaeger, Y.; Shiri-Sverdlov, R. NAFLD Preclinical Models: More than a Handful, Less of a Concern? Biomedicines 2020, 8, 28. [Google Scholar] [CrossRef]
- Cook, R.; Karesh, W.; Osofsky, S. One World, One Health: Building Interdisciplinary Bridges to Health in a Globalized World; Wildlife Conservation Society: Bronx, NY, USA, 2004; Available online: https://www.oneworldonehealth.org/sept2004/owohsept04.html (accessed on 18 April 2016).
- Evans, B.R.; Leighton, F.A. A History of One Health. Rev. Sci. Tech. 2014, 33, 413–420. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; McKinnon, M.; Jeggo, M. One Health: From Concept to Practice. In Confronting Emerging Zoonoses; Springer: Tokyo, Japan, 2014; pp. 163–189. [Google Scholar] [CrossRef]
- Schneider, B.; Balbas-Martinez, V.; Jergens, A.E.; Troconiz, I.F.; Allenspach, K.; Mochel, J.P. Model-Based Reverse Translation between Veterinary and Human Medicine: The One Health Initiative. CPT Pharmacomet. Syst. Pharmacol. 2018, 7, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Bontempo, V. Nutrition and Health of Dogs and Cats: Evolution of Petfood. Vet. Res. Commun. 2005, 29, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.K.; Michel, K.E. Confronting the Problem of Obesity in Dogs and Cats. Vet. Clin. N. Am.—Small Anim. Pract. 2016, 46, xi–xii. [Google Scholar] [CrossRef] [PubMed]
- Bartges, J.; Kushner, R.F.; Michel, K.E.; Sallis, R.; Day, M.J. One Health Solutions to Obesity in People and Their Pets. J. Comp. Pathol. 2017, 156, 326–333. [Google Scholar] [CrossRef]
- Wakshlag, J.; Loftus, J. Canine and Feline Obesity: A Review of Pathophysiology, Epidemiology, and Clinical Management. Vet. Med. Res. Rep. 2014, 2015, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D Cell Cultures—A Comparison of Different Types of Cancer Cell Cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Delie, F.; Rubas, W. A Human Colonic Cell Line Sharing Similarities with Enterocytes as a Model to Examine Oral Absorption: Advantages and Limitations of the Caco-2 Model. Crit. Rev. Ther. Drug Carr. Syst. 1997, 14, 221–286. [Google Scholar] [CrossRef]
- Lea, T. Caco-2 Cell Line. In The Impact of Food Bio-Actives on Gut Health; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 103–111. ISBN 978-3-319-15791-7. [Google Scholar]
- Chandra, L.; Borcherding, D.C.; Kingsbury, D.; Atherly, T.; Ambrosini, Y.M.; Bourgois-Mochel, A.; Yuan, W.; Kimber, M.; Qi, Y.; Wang, Q.; et al. Derivation of Adult Canine Intestinal Organoids for Translational Research in Gastroenterology. BMC Biol. 2019, 17, 33. [Google Scholar] [CrossRef]
- Schmiedlin-Ren, P.; Thummel, K.E.; Fisher, J.M.; Paine, M.F.; Lown, K.S.; Watkins, P.B. Expression of Enzymatically Active CYP3A4 by Caco-2 Cells Grown on Extracellular Matrix-Coated Permeable Supports in the Presence of 1α,25-Dihydroxyvitamin D3. Mol. Pharmacol. 1997, 51, 741–754. [Google Scholar] [CrossRef]
- Anderle, P.; Niederer, E.; Rubas, W.; Hilgendorf, C.; Spahn-Langguth, H.; Wunderli-Allenspach, H.; Merkle, H.P.; Langguth, P. P-Glycoprotein (P-Gp) Mediated Efflux in Caco-2 Cell Monolayers: The Influence of Culturing Conditions and Drug Exposure on P-Gp Expression Levels. J. Pharm. Sci. 1998, 87, 757–762. [Google Scholar] [CrossRef]
- Sun, H.; Chow, E.C.; Liu, S.; Du, Y.; Pang, K.S. The Caco-2 Cell Monolayer: Usefulness and Limitations. Expert. Opin. Drug Metab. Toxicol. 2008, 4, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Korjamo, T.; Honkakoski, P.; Toppinen, M.-R.; Niva, S.; Reinisalo, M.; Palmgrén, J.J.; Mönkkönen, J. Absorption Properties and P-Glycoprotein Activity of Modified Caco-2 Cell Lines. Eur. J. Pharm. Sci. 2005, 26, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, R.; Lee, C.M.; Shugart, E.C.; Benedetti, M.; Charo, R.A.; Gartner, Z.; Hogan, B.; Knoblich, J.; Nelson, C.M.; Wilson, K.M. Human Organoids: A New Dimension in Cell Biology. Mol. Biol. Cell 2019, 30, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures In Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Gabriel, V.; Zdyrski, C.; Sahoo, D.K.; Dao, K.; Bourgois-Mochel, A.; Kopper, J.; Zeng, X.-L.; Estes, M.K.; Mochel, J.P.; Allenspach, K. Standardization and Maintenance of 3D Canine Hepatic and Intestinal Organoid Cultures for Use in Biomedical Research. JoVE (J. Vis. Exp.) 2022, 179, e63515. [Google Scholar] [CrossRef]
- Watanabe, N.; Santostefano, K.E.; Yachnis, A.T.; Terada, N. A Pathologist’s Perspective on Induced Pluripotent Stem Cells. Lab. Investig. 2017, 97, 1126–1132. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-Term Expansion of Epithelial Organoids from Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of Stem Cells in Small Intestine and Colon by Marker Gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Dollé, L.; Theise, N.D.; Schmelzer, E.; Boulter, L.; Gires, O.; van Grunsven, L.A. EpCAM and the Biology of Hepatic Stem/Progenitor Cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G233–G250. [Google Scholar] [CrossRef] [PubMed]
- Zdyrski, C.; Gabriel, V.; Ospina, O.; Wickham, H.; Sahoo, D.K.; Dao, K.; Meza, L.S.A.; Bedos, L.; Honold, S.; Piñeyro, P.; et al. Establishment and Characterization of Novel Canine Organoids with Organ-Specific Physiological Similarity. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zhao, D.; Farnell, M.B.; Kogut, M.H.; Genovese, K.J.; Chapkin, R.S.; Davidson, L.A.; Berghman, L.R.; Farnell, Y.Z. From Crypts to Enteroids: Establishment and Characterization of Avian Intestinal Organoids. Poult. Sci. 2022, 101, 101642. [Google Scholar] [CrossRef] [PubMed]
- Puschhof, J.; Post, Y.; Beumer, J.; Kerkkamp, H.M.; Bittenbinder, M.; Vonk, F.J.; Casewell, N.R.; Richardson, M.K.; Clevers, H. Derivation of Snake Venom Gland Organoids for In Vitro Venom Production. Nat. Protoc. 2021, 16, 1494–1510. [Google Scholar] [CrossRef] [PubMed]
- Brandenberg, N.; Hoehnel, S.; Kuttler, F.; Homicsko, K.; Ceroni, C.; Ringel, T.; Gjorevski, N.; Schwank, G.; Coukos, G.; Turcatti, G.; et al. High-Throughput Automated Organoid Culture via Stem-Cell Aggregation in Microcavity Arrays. Nat. Biomed. Eng. 2020, 4, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, H.; Zhang, T.; Cao, L.; Du, Y.; Xie, Y.; Ji, J.; Wu, J. In-Depth Comparison of Matrigel Dissolving Methods on Proteomic Profiling of Organoids. Mol. Cell. Proteom. 2022, 21, 100181. [Google Scholar] [CrossRef] [PubMed]
- Post, Y.; Puschhof, J.; Beumer, J.; Kerkkamp, H.M.; de Bakker, M.A.G.; Slagboom, J.; de Barbanson, B.; Wevers, N.R.; Spijkers, X.M.; Olivier, T.; et al. Snake Venom Gland Organoids. Cell 2020, 180, 233–247.e21. [Google Scholar] [CrossRef] [PubMed]
- Elbadawy, M.; Usui, T.; Mori, T.; Tsunedomi, R.; Hazama, S.; Nabeta, R.; Uchide, T.; Fukushima, R.; Yoshida, T.; Shibutani, M.; et al. Establishment of a Novel Experimental Model for Muscle-Invasive Bladder Cancer Using a Dog Bladder Cancer Organoid Culture. Cancer Sci. 2019, 110, 2806–2821. [Google Scholar] [CrossRef]
- Bartlett, A.P.; Harman, R.M.; Weiss, J.R.; Van de Walle, G.R. Establishment and Characterization of Equine Mammary Organoids Using a Method Translatable to Other Non-Traditional Model Species. Development 2022, 149, dev200412. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Zhang, M.; Wang, H.; Cui, A.; Zhao, J.; Ji, W.; Chen, Y.-G. Establishment of Porcine and Monkey Colonic Organoids for Drug Toxicity Study. Cell Regen 2021, 10, 32. [Google Scholar] [CrossRef]
- Chen, T.-C.; Neupane, M.; Chien, S.-J.; Chuang, F.-R.; Crawford, R.B.; Kaminski, N.E.; Chang, C.-C. Characterization of Adult Canine Kidney Epithelial Stem Cells That Give Rise to Dome-Forming Tubular Cells. Stem Cells Dev. 2019, 28, 1424–1433. [Google Scholar] [CrossRef]
- Liu, M.; Yu, W.; Jin, J.; Ma, M.; An, T.; Nie, Y.; Teng, C.-B. Copper Promotes Sheep Pancreatic Duct Organoid Growth by Activation of an Antioxidant Protein 1-Dependent MEK-ERK Pathway. Am. J. Physiol.—Cell Physiol. 2020, 318, C806–C816. [Google Scholar] [CrossRef] [PubMed]
- Inaba, A.; Kumaki, S.; Arinaga, A.; Tanaka, K.; Aihara, E.; Yamane, T.; Oishi, Y.; Imai, H.; Iwatsuki, K. Generation of Intestinal Chemosensory Cells from Nonhuman Primate Organoids. Biochem. Biophys. Res. Commun. 2021, 536, 20–25. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Zhang, S.Y.; Li, R.X.; Lin, X.; Mi, Y.L.; Zhang, C.Q. Culture and Characterization of Chicken Small Intestinal Crypts. Poult. Sci. 2018, 97, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Töpfer, E.; Pasotti, A.; Telopoulou, A.; Italiani, P.; Boraschi, D.; Ewart, M.-A.; Wilde, C. Bovine Colon Organoids: From 3D Bioprinting to Cryopreserved Multi-Well Screening Platforms. Toxicol. Vitr. 2019, 61, 104606. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.S.; Freund, J.M.; Gonzalez, L.M. Advanced Three-Dimensional Culture of Equine Intestinal Epithelial Stem Cells. Equine Vet. J. 2018, 50, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Mussard, E.; Pouzet, C.; Helies, V.; Pascal, G.; Fourre, S.; Cherbuy, C.; Rubio, A.; Vergnolle, N.; Combes, S.; Beaumont, M. Culture of Rabbit Caecum Organoids by Reconstituting the Intestinal Stem Cell Niche In Vitro with Pharmacological Inhibitors or L-WRN Conditioned Medium. Stem Cell Res. 2020, 48, 101980. [Google Scholar] [CrossRef]
- Gonzalez, L.M.; Williamson, I.; Piedrahita, J.A.; Blikslager, A.T.; Magness, S.T. Cell Lineage Identification and Stem Cell Culture in a Porcine Model for the Study of Intestinal Epithelial Regeneration. PLoS ONE 2013, 8, e66465. [Google Scholar] [CrossRef]
- Adegbola, S.; Moore, J.; Sahnan, K.; Tozer, P.; Phillips, R.; Warusavitarne, J.; Faiz, O.; Hart, A. P005 Establishing a Porcine Model to Translate Anorectal Stem Cell Organoid Models to Elucidate the Aetiology of Perianal Crohn’s Fistulae. J. Crohn’s Colitis 2017, 11, S81–S82. [Google Scholar] [CrossRef]
- Von Furstenberg, R.J.; Gulati, A.S.; Baxi, A.; Doherty, J.M.; Stappenbeck, T.S.; Gracz, A.D.; Magness, S.T.; Henning, S.J. Sorting Mouse Jejunal Epithelial Cells with CD24 Yields a Population with Characteristics of Intestinal Stem Cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G409–G417. [Google Scholar] [CrossRef]
- Koltes, D.A.; Gabler, N.K. Characterization of Porcine Intestinal Enteroid Cultures under a Lipopolysaccharide Challenge1. J. Anim. Sci. 2016, 94, 335–339. [Google Scholar] [CrossRef]
- Powell, R.H.; Behnke, M.S. WRN Conditioned Media Is Sufficient for In Vitro Propagation of Intestinal Organoids from Large Farm and Small Companion Animals. Biol. Open 2017, 6, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fu, F.; Guo, S.; Wang, H.; He, X.; Xue, M.; Yin, L.; Feng, L.; Liu, P. Porcine Intestinal Enteroids: A New Model for Studying Enteric Coronavirus Porcine Epidemic Diarrhea Virus Infection and the Host Innate Response. J. Virol. 2019, 93, e01682-18. [Google Scholar] [CrossRef] [PubMed]
- Acharya, M.; Arsi, K.; Donoghue, A.M.; Liyanage, R.; Rath, N.C. Production and Characterization of Avian Crypt-Villus Enteroids and the Effect of Chemicals. BMC Vet. Res. 2020, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Derricott, H.; Luu, L.; Fong, W.Y.; Hartley, C.S.; Johnston, L.J.; Armstrong, S.D.; Randle, N.; Duckworth, C.A.; Campbell, B.J.; Wastling, J.M.; et al. Developing a 3D Intestinal Epithelium Model for Livestock Species. Cell Tissue Res. 2019, 375, 409–424. [Google Scholar] [CrossRef]
- Smith, D.; Price, D.R.G.; Burrells, A.; Faber, M.N.; Hildersley, K.A.; Chintoan-Uta, C.; Chapuis, A.F.; Stevens, M.; Stevenson, K.; Burgess, S.T.G.; et al. The Development of Ovine Gastric and Intestinal Organoids for Studying Ruminant Host-Pathogen Interactions. Front. Cell Infect. Microbiol. 2021, 11, 733811. [Google Scholar] [CrossRef] [PubMed]
- Kardia, E.; Frese, M.; Smertina, E.; Strive, T.; Zeng, X.-L.; Estes, M.; Hall, R.N. Culture and Differentiation of Rabbit Intestinal Organoids and Organoid-Derived Cell Monolayers. Sci. Rep. 2021, 11, 5401. [Google Scholar] [CrossRef] [PubMed]
- Tekes, G.; Ehmann, R.; Boulant, S.; Stanifer, M.L. Development of Feline Ileum- and Colon-Derived Organoids and Their Potential Use to Support Feline Coronavirus Infection. Cells 2020, 9, 2085. [Google Scholar] [CrossRef]
- Thompson, R.E.; Johnson, A.K.; Dini, P.; Turco, M.Y.; Prado, T.M.; Premanandan, C.; Burton, G.J.; Ball, B.A.; Whitlock, B.K.; Pukazhenthi, B.S. Hormone-Responsive Organoids from Domestic Mare and Endangered Przewalski’s Horse Endometrium. Reproduction 2020, 160, 819–831. [Google Scholar] [CrossRef]
- Zhou, J.; Li, C.; Liu, X.; Chiu, M.C.; Zhao, X.; Wang, D.; Wei, Y.; Lee, A.; Zhang, A.J.; Chu, H.; et al. Infection of Bat and Human Intestinal Organoids by SARS-CoV-2. Nat. Med. 2020, 26, 1077–1083. [Google Scholar] [CrossRef]
- Elbadawy, M.; Kato, Y.; Saito, N.; Hayashi, K.; Abugomaa, A.; Kobayashi, M.; Yoshida, T.; Shibutani, M.; Kaneda, M.; Yamawaki, H.; et al. Establishment of Intestinal Organoid from Rousettus Leschenaultii and the Susceptibility to Bat-Associated Viruses, SARS-CoV-2 and Pteropine Orthoreovirus. Int. J. Mol. Sci. 2021, 22, 10763. [Google Scholar] [CrossRef] [PubMed]
- Nantasanti, S.; Spee, B.; Kruitwagen, H.S.; Chen, C.; Geijsen, N.; Oosterhoff, L.A.; van Wolferen, M.E.; Pelaez, N.; Fieten, H.; Wubbolts, R.W.; et al. Disease Modeling and Gene Therapy of Copper Storage Disease in Canine Hepatic Organoids. Stem Cell Rep. 2015, 5, 895–907. [Google Scholar] [CrossRef]
- Kruitwagen, H.S.; Oosterhoff, L.A.; Vernooij, I.G.W.H.; Schrall, I.M.; van Wolferen, M.E.; Bannink, F.; Roesch, C.; van Uden, L.; Molenaar, M.R.; Helms, J.B.; et al. Long-Term Adult Feline Liver Organoid Cultures for Disease Modeling of Hepatic Steatosis. Stem Cell Rep. 2017, 8, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Wiener, D.J.; Basak, O.; Asra, P.; Boonekamp, K.E.; Kretzschmar, K.; Papaspyropoulos, A.; Clevers, H. Establishment and Characterization of a Canine Keratinocyte Organoid Culture System. Vet. Dermatol. 2018, 29, 375-e126. [Google Scholar] [CrossRef] [PubMed]
- Bedos, L.; Wickham, H.; Gabriel, V.; Zdyrski, C.; Allbaugh, R.A.; Sahoo, D.K.; Sebbag, L.; Mochel, J.P.; Allenspach, K. Culture and Characterization of Canine and Feline Corneal Epithelial Organoids: A New Tool for the Study and Treatment of Corneal Diseases. Front. Vet. Sci. 2022, 9, 1050467. [Google Scholar] [CrossRef] [PubMed]
- Jung, P.; Sato, T.; Merlos-Suárez, A.; Barriga, F.M.; Iglesias, M.; Rossell, D.; Auer, H.; Gallardo, M.; Blasco, M.A.; Sancho, E.; et al. Isolation and In Vitro Expansion of Human Colonic Stem Cells. Nat. Med. 2011, 17, 1225–1227. [Google Scholar] [CrossRef] [PubMed]
- Chua, C.W.; Shibata, M.; Lei, M.; Toivanen, R.; Barlow, L.J.; Bergren, S.K.; Badani, K.K.; McKiernan, J.M.; Benson, M.C.; Hibshoosh, H.; et al. Single Luminal Epithelial Progenitors Can Generate Prostate Organoids in Culture. Nat. Cell Biol. 2014, 16, 951–961. [Google Scholar] [CrossRef]
- Tadokoro, T.; Wang, Y.; Barak, L.S.; Bai, Y.; Randell, S.H.; Hogan, B.L.M. IL-6/STAT3 Promotes Regeneration of Airway Ciliated Cells from Basal Stem Cells. Proc. Natl. Acad. Sci. USA 2014, 111, E3641–E3649. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.A.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- Boj, S.F.; Hwang, C.-I.; Baker, L.A.; Chio, I.I.C.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Barkauskas, C.E.; Takeda, N.; Bowie, E.J.; Aghajanian, H.; Wang, Q.; Padmanabhan, A.; Manderfield, L.J.; Gupta, M.; Li, D.; et al. Plasticity of Hopx(+) Type I Alveolar Cells to Regenerate Type II Cells in the Lung. Nat. Commun. 2015, 6, 6727. [Google Scholar] [CrossRef] [PubMed]
- Kessler, M.; Hoffmann, K.; Brinkmann, V.; Thieck, O.; Jackisch, S.; Toelle, B.; Berger, H.; Mollenkopf, H.-J.; Mangler, M.; Sehouli, J.; et al. The Notch and Wnt Pathways Regulate Stemness and Differentiation in Human Fallopian Tube Organoids. Nat. Commun. 2015, 6, 8989. [Google Scholar] [CrossRef] [PubMed]
- Pringle, S.; Maimets, M.; van der Zwaag, M.; Stokman, M.A.; van Gosliga, D.; Zwart, E.; Witjes, M.J.H.; de Haan, G.; van Os, R.; Coppes, R.P. Human Salivary Gland Stem Cells Functionally Restore Radiation Damaged Salivary Glands. Stem Cells 2016, 34, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Turco, M.Y.; Gardner, L.; Kay, R.G.; Hamilton, R.S.; Prater, M.; Hollinshead, M.S.; McWhinnie, A.; Esposito, L.; Fernando, R.; Skelton, H.; et al. Trophoblast Organoids as a Model for Maternal–Fetal Interactions during Human Placentation. Nature 2018, 564, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Horsley, H.; Dharmasena, D.; Malone-Lee, J.; Rohn, J.L. A Urine-Dependent Human Urothelial Organoid Offers a Potential Alternative to Rodent Models of Infection. Sci. Rep. 2018, 8, 1238. [Google Scholar] [CrossRef] [PubMed]
- Gee, K.; Isani, M.A.; Fode, A.; Maselli, K.M.; Zuber, S.M.; Fowler, K.L.; Squillaro, A.I.; Nucho, L.-M.A.; Grikscheit, T.C. Spleen Organoid Units Generate Functional Human and Mouse Tissue-Engineered Spleen in a Murine Model. Tissue Eng. Part. A 2020, 26, 411–418. [Google Scholar] [CrossRef]
- Rosenbluth, J.M.; Schackmann, R.C.J.; Gray, G.K.; Selfors, L.M.; Li, C.M.-C.; Boedicker, M.; Kuiken, H.J.; Richardson, A.; Brock, J.; Garber, J.; et al. Organoid Cultures from Normal and Cancer-Prone Human Breast Tissues Preserve Complex Epithelial Lineages. Nat. Commun. 2020, 11, 1711. [Google Scholar] [CrossRef]
- Liu, Z.; Anderson, J.D.; Deng, L.; Mackay, S.; Bailey, J.; Kersh, L.; Rowe, S.M.; Guimbellot, J.S. Human Nasal Epithelial Organoids for Therapeutic Development in Cystic Fibrosis. Genes 2020, 11, 603. [Google Scholar] [CrossRef]
- Ogundipe, V.M.L.; Groen, A.H.; Hosper, N.; Nagle, P.W.K.; Hess, J.; Faber, H.; Jellema, A.L.; Baanstra, M.; Links, T.P.; Unger, K.; et al. Generation and Differentiation of Adult Tissue-Derived Human Thyroid Organoids. Stem Cell Rep. 2021, 16, 913–925. [Google Scholar] [CrossRef]
- Mollaki, V. Ethical Challenges in Organoid Use. BioTech 2021, 10, 12. [Google Scholar] [CrossRef]
- Rock, J.R.; Onaitis, M.W.; Rawlins, E.L.; Lu, Y.; Clark, C.P.; Xue, Y.; Randell, S.H.; Hogan, B.L.M. Basal Cells as Stem Cells of the Mouse Trachea and Human Airway Epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 12771–12775. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Huch, M.; Kujala, P.; van de Wetering, M.; Snippert, H.J.; van Es, J.H.; Sato, T.; Stange, D.E.; Begthel, H.; van den Born, M.; et al. Lgr5+ve Stem Cells Drive Self-Renewal in the Stomach and Build Long-Lived Gastric Units In Vitro. Cell Stem Cell 2010, 6, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Bonfanti, P.; Boj, S.F.; Sato, T.; Loomans, C.J.M.; Van De Wetering, M.; Sojoodi, M.; Li, V.S.W.; Schuijers, J.; Gracanin, A.; et al. Unlimited In Vitro Expansion of Adult Bi-Potent Pancreas Progenitors through the Lgr5/R-Spondin Axis. EMBO J. 2013, 32, 2708–2721. [Google Scholar] [CrossRef]
- Huch, M.; Dorrell, C.; Boj, S.F.; van Es, J.H.; Li, V.S.W.; van de Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In Vitro Expansion of Single Lgr5+ Liver Stem Cells Induced by Wnt-Driven Regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef]
- Barkauskas, C.E.; Cronce, M.J.; Rackley, C.R.; Bowie, E.J.; Keene, D.R.; Stripp, B.R.; Randell, S.H.; Noble, P.W.; Hogan, B.L.M. Type 2 Alveolar Cells Are Stem Cells in Adult Lung. J. Clin. Investig. 2013, 123, 3025–3036. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Lewandowski, B.C.; Watson, J.; Aihara, E.; Iwatsuki, K.; Bachmanov, A.A.; Margolskee, R.F.; Jiang, P. Single Lgr5- or Lgr6-Expressing Taste Stem/Progenitor Cells Generate Taste Bud Cells Ex Vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 16401–16406. [Google Scholar] [CrossRef] [PubMed]
- DeWard, A.D.; Cramer, J.; Lagasse, E. Cellular Heterogeneity in the Mouse Esophagus Implicates the Presence of a Nonquiescent Epithelial Stem Cell Population. Cell Rep. 2014, 9, 701–711. [Google Scholar] [CrossRef]
- Maimets, M.; Rocchi, C.; Bron, R.; Pringle, S.; Kuipers, J.; Giepmans, B.N.G.; Vries, R.G.J.; Clevers, H.; de Haan, G.; van Os, R.; et al. Long-Term In Vitro Expansion of Salivary Gland Stem Cells Driven by Wnt Signals. Stem Cell Rep. 2016, 6, 150–162. [Google Scholar] [CrossRef]
- Jamieson, P.R.; Dekkers, J.F.; Rios, A.C.; Fu, N.Y.; Lindeman, G.J.; Visvader, J.E. Derivation of a Robust Mouse Mammary Organoid System for Studying Tissue Dynamics. Development 2017, 144, 1065–1071. [Google Scholar] [CrossRef]
- Xie, Y.; Park, E.-S.; Xiang, D.; Li, Z. Long-Term Organoid Culture Reveals Enrichment of Organoid-Forming Epithelial Cells in the Fimbrial Portion of Mouse Fallopian Tube. Stem Cell Res. 2018, 32, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Onishi, N.; Takami, H.; Seishima, R.; Inoue, H.; Hirata, Y.; Kameyama, K.; Tsuchihashi, K.; Sugihara, E.; Uchino, S.; et al. Development of a Functional Thyroid Model Based on an Organoid Culture System. Biochem. Biophys. Res. Commun. 2018, 497, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.P.; Lapi, E.; Martínez de Villarreal, J.; Álvaro-Espinosa, L.; Fernández-Barral, A.; Barbáchano, A.; Domínguez, O.; Laughney, A.M.; Megías, D.; Muñoz, A.; et al. Urothelial Organoids Originating from Cd49fhigh Mouse Stem Cells Display Notch-Dependent Differentiation Capacity. Nat. Commun. 2019, 10, 4407. [Google Scholar] [CrossRef] [PubMed]
- Pinel, L.; Cyr, D.G. Self-Renewal and Differentiation of Rat Epididymal Basal Cells Using a Novel In Vitro Organoid Model. Biol. Reprod. 2021, 105, 987–1001. [Google Scholar] [CrossRef]
- Kar, S.K.; Wells, J.M.; Ellen, E.D.; te Pas, M.F.W.; Madsen, O.; Groenen, M.A.M.; Woelders, H. Organoids: A Promising New in Vitro Platform in Livestock and Veterinary Research. Vet. Res. 2021, 52, 43. [Google Scholar] [CrossRef]
- Andersson, L. Domestic Animals as Models for Biomedical Research. Ups. J. Med. Sci. 2016, 121, 1–11. [Google Scholar] [CrossRef]
- Axelsson, E.; Ratnakumar, A.; Arendt, M.-L.; Maqbool, K.; Webster, M.T.; Perloski, M.; Liberg, O.; Arnemo, J.M.; Hedhammar, Å.; Lindblad-Toh, K. The Genomic Signature of Dog Domestication Reveals Adaptation to a Starch-Rich Diet. Nature 2013, 495, 360–364. [Google Scholar] [CrossRef]
- Beaumont, M.; Blanc, F.; Cherbuy, C.; Egidy, G.; Giuffra, E.; Lacroix-Lamandé, S.; Wiedemann, A. Intestinal Organoids in Farm Animals. Vet. Res. 2021, 52, 33. [Google Scholar] [CrossRef]
- Linder, D.E.; Santiago, S.; Halbreich, E.D. Is There a Correlation Between Dog Obesity and Human Obesity? Preliminary Findings of Overweight Status Among Dog Owners and Their Dogs. Front. Vet. Sci. 2021, 8, 654617. [Google Scholar] [CrossRef]
- Mahe, M.M.; Aihara, E.; Schumacher, M.A.; Zavros, Y.; Montrose, M.H.; Helmrath, M.A.; Sato, T.; Shroyer, N.F. Establishment of Gastrointestinal Epithelial Organoids. Curr. Protoc. Mouse Biol. 2013, 3, 217–240. [Google Scholar] [CrossRef]
- Inaba, A.; Arinaga, A.; Tanaka, K.; Endo, T.; Hayatsu, N.; Okazaki, Y.; Yamane, T.; Oishi, Y.; Imai, H.; Iwatsuki, K. Interleukin-4 Promotes Tuft Cell Differentiation and Acetylcholine Production in Intestinal Organoids of Non-Human Primate. Int. J. Mol. Sci. 2021, 22, 7921. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell 2018, 23, 787–793.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Scoville, D.; He, X.C.; Mahe, M.M.; Box, A.; Perry, J.M.; Smith, N.R.; Lei, N.Y.; Davies, P.S.; Fuller, M.K.; et al. Isolation and Characterization of Intestinal Stem Cells Based on Surface Marker Combinations and Colony-Formation Assay. Gastroenterology 2013, 145, 383–395.e21. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.A.; Wang, Y.; Gracz, A.D.; Sims, C.E.; Magness, S.T.; Allbritton, N.L. Optimization of 3-D Organotypic Primary Colonic Cultures for Organ-on-Chip Applications. J. Biol. Eng. 2014, 8, 9. [Google Scholar] [CrossRef] [PubMed]
- Khalil, H.A.; Lei, N.Y.; Brinkley, G.; Scott, A.; Wang, J.; Kar, U.K.; Jabaji, Z.B.; Lewis, M.; Martín, M.G.; Dunn, J.C.Y.; et al. A Novel Culture System for Adult Porcine Intestinal Crypts. Cell Tissue Res. 2016, 365, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Gracz, A.D.; Puthoff, B.J.; Magness, S.T. Identification, Isolation, and Culture of Intestinal Epithelial Stem Cells from Murine Intestine. Methods Mol. Biol. 2012, 879, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Von Furstenberg, R.J.; Li, J.; Stolarchuk, C.; Feder, R.; Campbell, A.; Kruger, L.; Gonzalez, L.M.; Blikslager, A.T.; Cardona, D.M.; McCall, S.J.; et al. Porcine Esophageal Submucosal Gland Culture Model Shows Capacity for Proliferation and Differentiation. Cell Mol. Gastroenterol. Hepatol. 2017, 4, 385–404. [Google Scholar] [CrossRef] [PubMed]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.-L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R.; et al. Human Intestinal Enteroids: A New Model to Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J. Virol. 2016, 90, 43–56. [Google Scholar] [CrossRef]
- Formeister, E.J.; Sionas, A.L.; Lorance, D.K.; Barkley, C.L.; Lee, G.H.; Magness, S.T. Distinct SOX9 Levels Differentially Mark Stem/Progenitor Populations and Enteroendocrine Cells of the Small Intestine Epithelium. Am. J. Physiol. Gastrointest. Liver Physiol. 2009, 296, G1108–G1118. [Google Scholar] [CrossRef]
- Kramer, N.; Pratscher, B.; Meneses, A.M.C.; Tschulenk, W.; Walter, I.; Swoboda, A.; Kruitwagen, H.S.; Schneeberger, K.; Penning, L.C.; Spee, B.; et al. Generation of Differentiating and Long-Living Intestinal Organoids Reflecting the Cellular Diversity of Canine Intestine. Cells 2020, 9, 822. [Google Scholar] [CrossRef]
- Gabriel, V.; Zdyrski, C.; Sahoo, D.K.; Dao, K.; Bourgois-Mochel, A.; Atherly, T.; Martinez, M.N.; Volpe, D.A.; Kopper, J.; Allenspach, K.; et al. Canine Intestinal Organoids in a Dual-Chamber Permeable Support System. JoVE (J. Vis. Exp.) 2022, 181, e63612. [Google Scholar] [CrossRef]
- Ambrosini, Y.M.; Park, Y.; Jergens, A.E.; Shin, W.; Min, S.; Atherly, T.; Borcherding, D.C.; Jang, J.; Allenspach, K.; Mochel, J.P.; et al. Recapitulation of the Accessible Interface of Biopsy-Derived Canine Intestinal Organoids to Study Epithelial-Luminal Interactions. PLoS ONE 2020, 15, e0231423. [Google Scholar] [CrossRef]
- Duzagac, F.; Saorin, G.; Memeo, L.; Canzonieri, V.; Rizzolio, F. Microfluidic Organoids-on-a-chip: Quantum Leap in Cancer Research. Cancers 2021, 13, 737. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Stappenbeck, T.S. In Vitro Expansion and Genetic Modification of Gastrointestinal Stem Cells in Spheroid Culture. Nat. Protoc. 2013, 8, 2471–2482. [Google Scholar] [CrossRef]
- Jung, K.; Eyerly, B.; Annamalai, T.; Lu, Z.; Saif, L.J. Structural Alteration of Tight and Adherens Junctions in Villous and Crypt Epithelium of the Small and Large Intestine of Conventional Nursing Piglets Infected with Porcine Epidemic Diarrhea Virus. Vet. Microbiol. 2015, 177, 373–378. [Google Scholar] [CrossRef]
- Hamilton, C.A.; Young, R.; Jayaraman, S.; Sehgal, A.; Paxton, E.; Thomson, S.; Katzer, F.; Hope, J.; Innes, E.; Morrison, L.J.; et al. Development of in Vitro Enteroids Derived from Bovine Small Intestinal Crypts. Vet. Res. 2018, 49, 54. [Google Scholar] [CrossRef]
- Co, J.Y.; Margalef-Català, M.; Li, X.; Mah, A.T.; Kuo, C.J.; Monack, D.M.; Amieva, M.R. Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions. Cell Rep. 2019, 26, 2509–2520.e4. [Google Scholar] [CrossRef] [PubMed]
- Kruitwagen, H.S.; Oosterhoff, L.A.; van Wolferen, M.E.; Chen, C.; Nantasanti Assawarachan, S.; Schneeberger, K.; Kummeling, A.; van Straten, G.; Akkerdaas, I.C.; Vinke, C.R.; et al. Long-Term Survival of Transplanted Autologous Canine Liver Organoids in a COMMD1-Deficient Dog Model of Metabolic Liver Disease. Cells 2020, 9, 410. [Google Scholar] [CrossRef]
- Van Den Bossche, L.; Schoonenberg, V.A.C.; Burgener, I.A.; Penning, L.C.; Schrall, I.M.; Kruitwagen, H.S.; van Wolferen, M.E.; Grinwis, G.C.M.; Kummeling, A.; Rothuizen, J.; et al. Aberrant Hepatic Lipid Storage and Metabolism in Canine Portosystemic Shunts. PLoS ONE 2017, 12, e0186491. [Google Scholar] [CrossRef]
- Wu, X.; Chien, H.; van Wolferen, M.E.; Kruitwagen, H.S.; Oosterhoff, L.A.; Penning, L.C. Reduced FXR Target Gene Expression in Copper-Laden Livers of COMMD1-Deficient Dogs. Vet. Sci. 2019, 6, 78. [Google Scholar] [CrossRef]
- Zou, W.Y.; Blutt, S.E.; Crawford, S.E.; Ettayebi, K.; Zeng, X.-L.; Saxena, K.; Ramani, S.; Karandikar, U.C.; Zachos, N.C.; Estes, M.K. Human Intestinal Enteroids: New Models to Study Gastrointestinal Virus Infections. Methods Mol. Biol. 2019, 1576, 229–247. [Google Scholar] [CrossRef] [PubMed]
- Haaker, M.W.; Kruitwagen, H.S.; Vaandrager, A.B.; Houweling, M.; Penning, L.C.; Molenaar, M.R.; van Wolferen, M.E.; Oosterhoff, L.A.; Spee, B.; Helms, J.B. Identification of Potential Drugs for Treatment of Hepatic Lipidosis in Cats Using an in Vitro Feline Liver Organoid System. J. Vet. Intern. Med. 2020, 34, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Gaush, C.R.; Hard, W.L.; Smith, T.F. Characterization of an Established Line of Canine Kidney Cells (MDCK). Exp. Biol. Med. 1966, 122, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Humann-Ziehank, E. Selenium, Copper and Iron in Veterinary Medicine—From Clinical Implications to Scientific Models. J. Trace Elem. Med. Biol. 2016, 37, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Agbunag, C.; Lee, K.E.; Buontempo, S.; Bar-Sagi, D. Pancreatic Duct Epithelial Cell Isolation and Cultivation in Two-Dimensional and Three-Dimensional Culture Systems. Methods Enzym. 2006, 407, 703–710. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Mealey, K.L.; Martinez, S.E.; Villarino, N.F.; Court, M.H. Personalized Medicine: Going to the Dogs? Hum. Genet. 2019, 138, 467–481. [Google Scholar] [CrossRef]
- Thompson, R.E.; Meyers, M.A.; Pukazhenthi, B.S.; Hollinshead, F.K. Evaluation of Growth, Viability, and Structural Integrity of Equine Endometrial Organoids Following Cryopreservation. Cryobiology 2022, 104, 56–62. [Google Scholar] [CrossRef]
- Thompson, R.E.; Meyers, M.A.; Veeramachaneni, D.N.R.; Pukazhenthi, B.S.; Hollinshead, F.K. Equine Oviductal Organoid Generation and Cryopreservation. Methods Protoc. 2022, 5, 51. [Google Scholar] [CrossRef]
- Dundon, M.; Madden, O.; Comizzoli, P. Three-Dimensional Culture of Endometrial Cells from Domestic Cats: A New In Vitro Platform for Assessing Plastic Toxicity. PLoS ONE 2019, 14, e0217365. [Google Scholar] [CrossRef]
- Wiener, D.J.; Doherr, M.G.; Müller, E.J.; Suter, M.M.; Welle, M.M. Comparative Assessment of a Canine-Specific Medium to Support Colony Formation from Canine Hair Follicular Keratinocytes. Vet. Dermatol. 2015, 26, 198-e42. [Google Scholar] [CrossRef] [PubMed]
- Wiener, D.J.; Studer, I.C.; Brunner, M.A.T.; Hermann, A.; Vincenti, S.; Zhang, M.; Groch, K.R.; Welle, M.M. Characterization of Canine Epidermal Organoid Cultures by Immunohistochemical Analysis and Quantitative PCR. Vet. Dermatol. 2021, 32, 179-e44. [Google Scholar] [CrossRef] [PubMed]
- Grego, K.F.; Vieira, S.E.M.; Vidueiros, J.P.; Serapicos, E.d.O.; Barbarini, C.C.; da Silveira, G.P.M.; Rodrigues, F.d.S.; Alves, L.d.C.F.; Stuginski, D.R.; Rameh-de-Albuquerque, L.C.; et al. Maintenance of Venomous Snakes in Captivity for Venom Production at Butantan Institute from 1908 to the Present: A Scoping History. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021, 27, e20200068. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.M.; León, G.; Burnouf, T. Antivenoms for the Treatment of Snakebite Envenomings: The Road Ahead. Biologicals 2011, 39, 129–142. [Google Scholar] [CrossRef]
- Habib, A.G.; Musa, B.M.; Iliyasu, G.; Hamza, M.; Kuznik, A.; Chippaux, J.-P. Challenges and Prospects of Snake Antivenom Supply in Sub-Saharan Africa. PLoS Negl. Trop. Dis. 2020, 14, e0008374. [Google Scholar] [CrossRef]
- Chippaux, J.P. Snake-Bites: Appraisal of the Global Situation. Bull. World Health Organ. 1998, 76, 515–524. [Google Scholar]



| Biomedical Model For | Species | Taxonomical Name | Author | Organ/Intestinal Segment |
|---|---|---|---|---|
| Antivenom Production | Red spitting cobra | Naja pallida | Post et al. 2020 [41] | Venom Gland |
| Snouted cobra | Naja annulifera | |||
| Cape cobra | Naja nivea | |||
| Chinese cobra | Naja atra | |||
| Cape coral snake | Aspidelaps lubricus cowlesi | |||
| West African carpet viper | Echis ocellatus | |||
| Chinese moccasin | Deinagkistrodon acutus | |||
| Western diamondback rattlesnake | Crotalus atrox | |||
| Puff adder | Bitis arietans | |||
| Urinary Bladder Cancer Model | Dog | Canis lupus familiaris | Elbadawy et al. 2022 [42] | Urinary Bladder |
| Mammary Gland Cancer | Prarie Vole | Microtus ochrogaster | Barlett et al. 2022 [43] | Mammary Gland |
| Pig | Sus scrofa domesticus | |||
| White-tailed deer | Odocoileus virginianus | |||
| Horse | Equus ferus caballus | |||
| European rabbit | Oryctolagus cunniculus | |||
| Cat | Felis catus domestica | |||
| Drug Permeability and Toxicity Testing | Crab-eating Macaque | Macaca fascicularis | Li et al.2021 [44] | Colon |
| Dog | Canis lupus familiaris | Chen et al. 2019 [45] | Kidney | |
| Inflammatory Bowel Disease | Chandra et al. 2019 [23] | Intestine | ||
| Influence of Copper on Pancreas | Sheep | Ovis Aries | Liu et al. 2021 [46] | Pancreas |
| Intestinal Physiology Investigation | Rhesus Macaque | Macaca mulatta | Inaba et al. 2021 [47] | Full Intestine |
| Japanese Macaque | Macaca fuscata | Full Intestine | ||
| Chicken | Gallus gallus domesticus | Li et al. 2018 [48] | Jejunum | |
| Cattle | Bos taurus | Topfer et al. 2019 [49] | Colon | |
| Horse | Equus ferus caballus | Stewart et al. 2017 [50] | Jejunum | |
| European rabbit | Oryctolagus cunniculus | Mussard et al. 2020 [51] | Cecum | |
| Pig | Sus scrofa domesticus | Gonzalez et al. 2013 [52] | Jejunum | |
| Adegbola et al. 2017 [53] | Anus, Rectum | |||
| von Furstenberg et al. 2017 [54] | Esophageal Glands | |||
| Inflammation Signaling and Edotoxine Tolerance | Pig | Sus scrofa domesticus | Koltes et al. 2016 [55] | Duodenum |
| Pathogen-Host Interaction | Pig | Sus scrofa domesticus | Powell et al. 2017 [56] | Ileum |
| Chicken | Gallus gallus domesticus | |||
| Pig | Sus scrofa domesticus | Li et al. 2019 [57] | Colon | |
| Dog | Canis lupus familiaris | Powell et al. 2017 [56] | Ileum | |
| Acharya et al. 2020 [58] | Small Intestine | |||
| Cattle | Bos taurus | Derricott et al. 2019 [59] | Jejunum | |
| Powell et al. 2017 [56] | Ileum | |||
| Sheep | Ovis Aries | |||
| Smith et al. 2021 [60] | Abomasum | |||
| Horse | Equus ferus caballus | Powell et al. 2017 [56] | Ileum | |
| European rabbit | Oryctolagus cunniculus | Kardia et al. 2021 [61] | Duodenum | |
| Ileum | ||||
| Colon | ||||
| Cat | Felis catus domestica | Powell et al. 2017 [56] | Ileum | |
| Tekes et al. 2020 [62] | Colon | |||
| Reproductive Mechanisms | Horse | Equus ferus caballus | Thompson et al. 2020 [63] | Endometrium |
| Przewalski’s horse | Equus ferus prezewalski | |||
| Horse | Equus ferus caballus | Oviduct | ||
| SARS-COVID-2 | Chinese rufous horseshoe bat | Rhinolophus sinicus | Zhou et al. 2020 [64] | Small Intestine |
| Leschenault’s rousette | Rousettus leschenualtia | Elbadawy et al. 2021 [65] | Intestine | |
| Wilson’s Disease | Dog | Canis lupus familiaris | Nantasanti et al. 2015 [66] | Liver |
| Hepatic Steatosis | Cat | Felis catus domestica | Kruitwagen et al. 2017 [67] | |
| Epidermal Function and Cutaneus Disorders | Dog | Canis lupus familiaris | Wiener et al. 2018 [68] | Skin |
| Ophthalmology | Dog | Canis lupus familiaris | Bedos et al. 2022 [69] | Cornea |
| Cat | Felis catus domestica | Bedos et al. 2022 [69] | Cornea |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabriel, V.; Zdyrski, C.; Sahoo, D.K.; Ralston, A.; Wickham, H.; Bourgois-Mochel, A.; Ahmed, B.; Merodio, M.M.; Paukner, K.; Piñeyro, P.; et al. Adult Animal Stem Cell-Derived Organoids in Biomedical Research and the One Health Paradigm. Int. J. Mol. Sci. 2024, 25, 701. https://doi.org/10.3390/ijms25020701
Gabriel V, Zdyrski C, Sahoo DK, Ralston A, Wickham H, Bourgois-Mochel A, Ahmed B, Merodio MM, Paukner K, Piñeyro P, et al. Adult Animal Stem Cell-Derived Organoids in Biomedical Research and the One Health Paradigm. International Journal of Molecular Sciences. 2024; 25(2):701. https://doi.org/10.3390/ijms25020701
Chicago/Turabian StyleGabriel, Vojtech, Christopher Zdyrski, Dipak K. Sahoo, Abigail Ralston, Hannah Wickham, Agnes Bourgois-Mochel, Basant Ahmed, Maria M. Merodio, Karel Paukner, Pablo Piñeyro, and et al. 2024. "Adult Animal Stem Cell-Derived Organoids in Biomedical Research and the One Health Paradigm" International Journal of Molecular Sciences 25, no. 2: 701. https://doi.org/10.3390/ijms25020701
APA StyleGabriel, V., Zdyrski, C., Sahoo, D. K., Ralston, A., Wickham, H., Bourgois-Mochel, A., Ahmed, B., Merodio, M. M., Paukner, K., Piñeyro, P., Kopper, J., Rowe, E. W., Smith, J. D., Meyerholz, D., Kol, A., Viall, A., Elbadawy, M., Mochel, J. P., & Allenspach, K. (2024). Adult Animal Stem Cell-Derived Organoids in Biomedical Research and the One Health Paradigm. International Journal of Molecular Sciences, 25(2), 701. https://doi.org/10.3390/ijms25020701










