Differential microRNA Expression Analysis in Patients with HPV-Infected Ovarian Neoplasms
Abstract
:1. Introduction
2. Results
2.1. HPV16/18 Copy Number Analysis
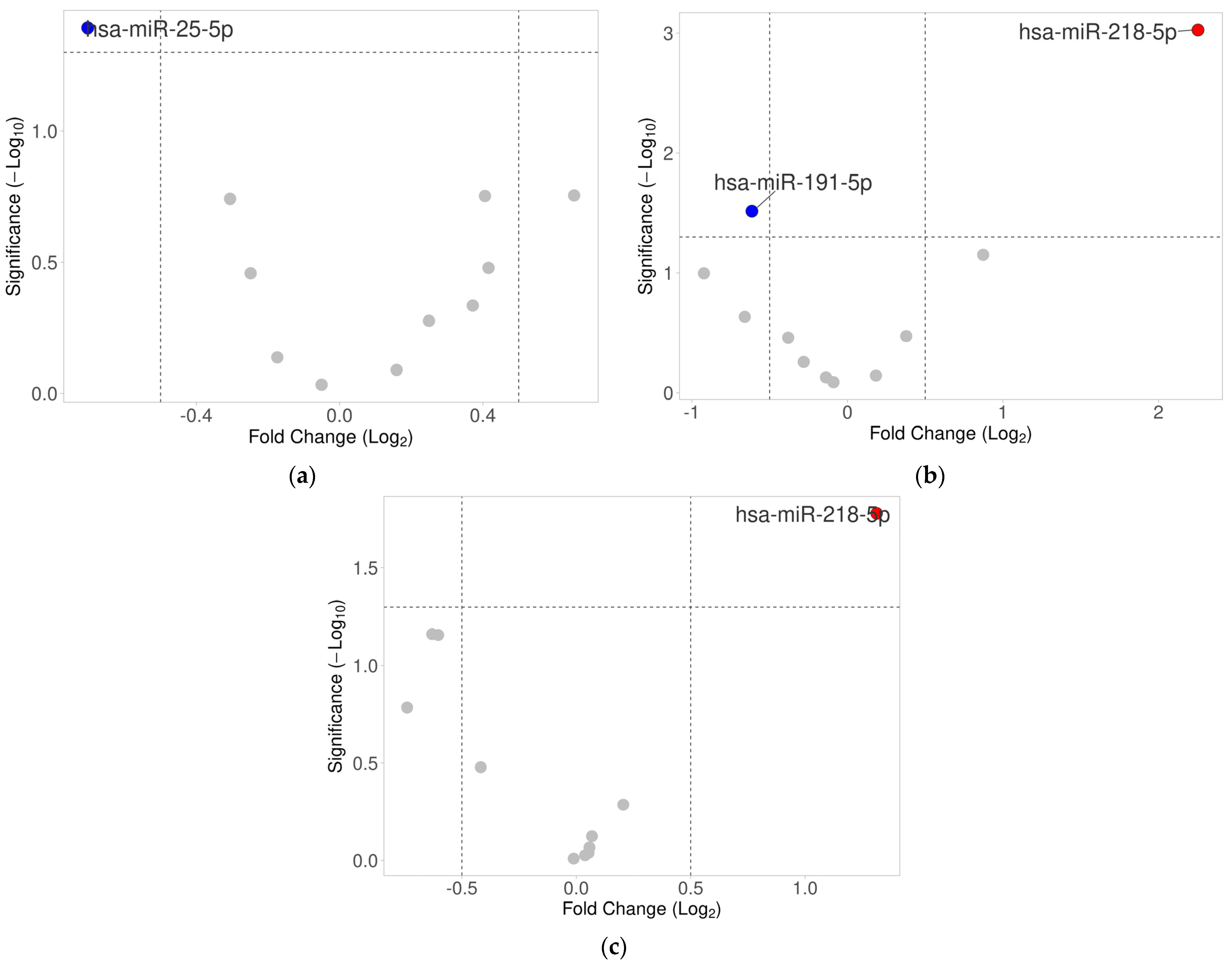
2.2. Differential Expression Analysis of Selected miRNAs in Ovarian Tumor Tissues
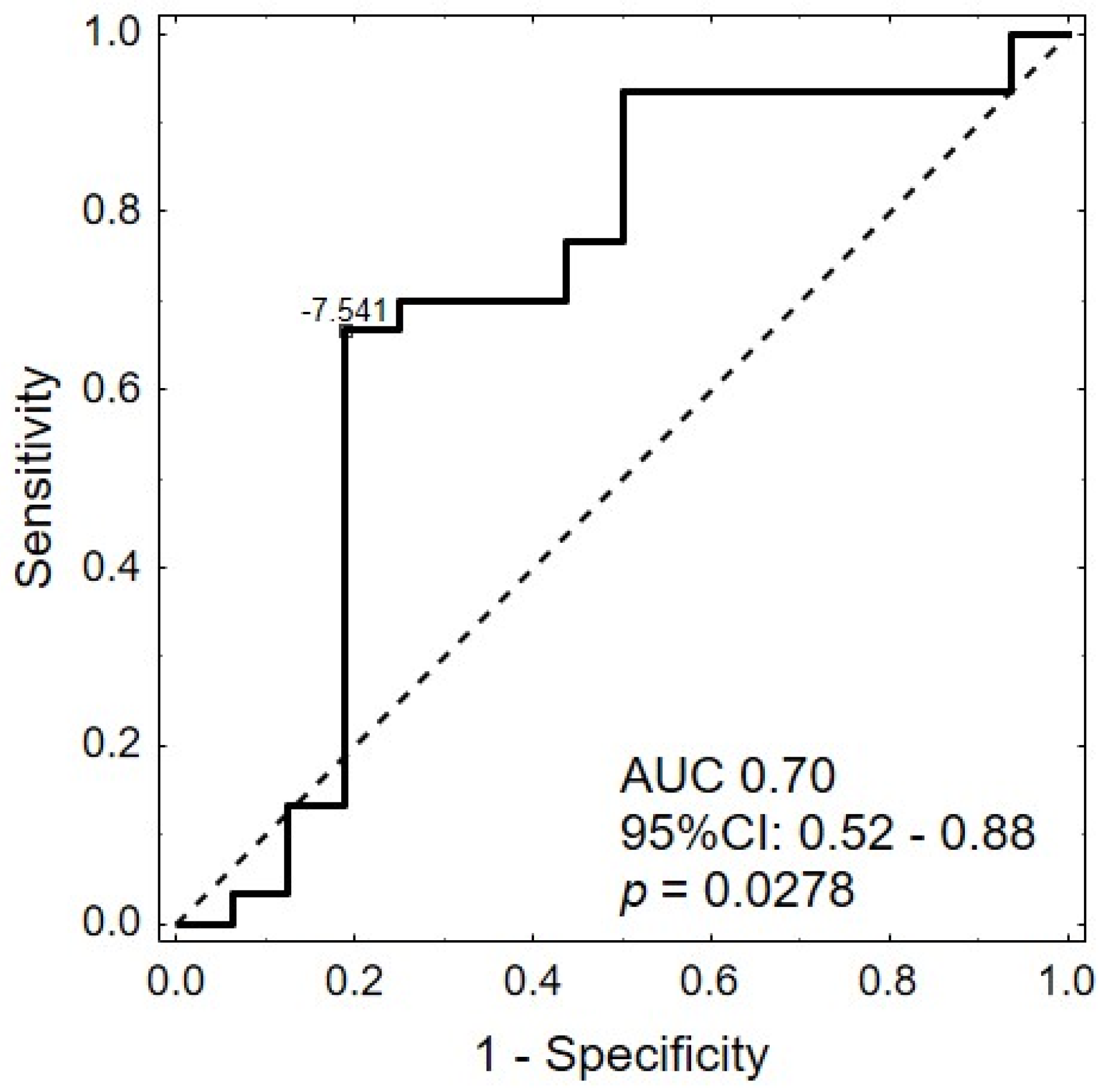
2.3. Expression of miRNAs in Serum
2.4. Association between miRNA Expression Levels and Clinical Features
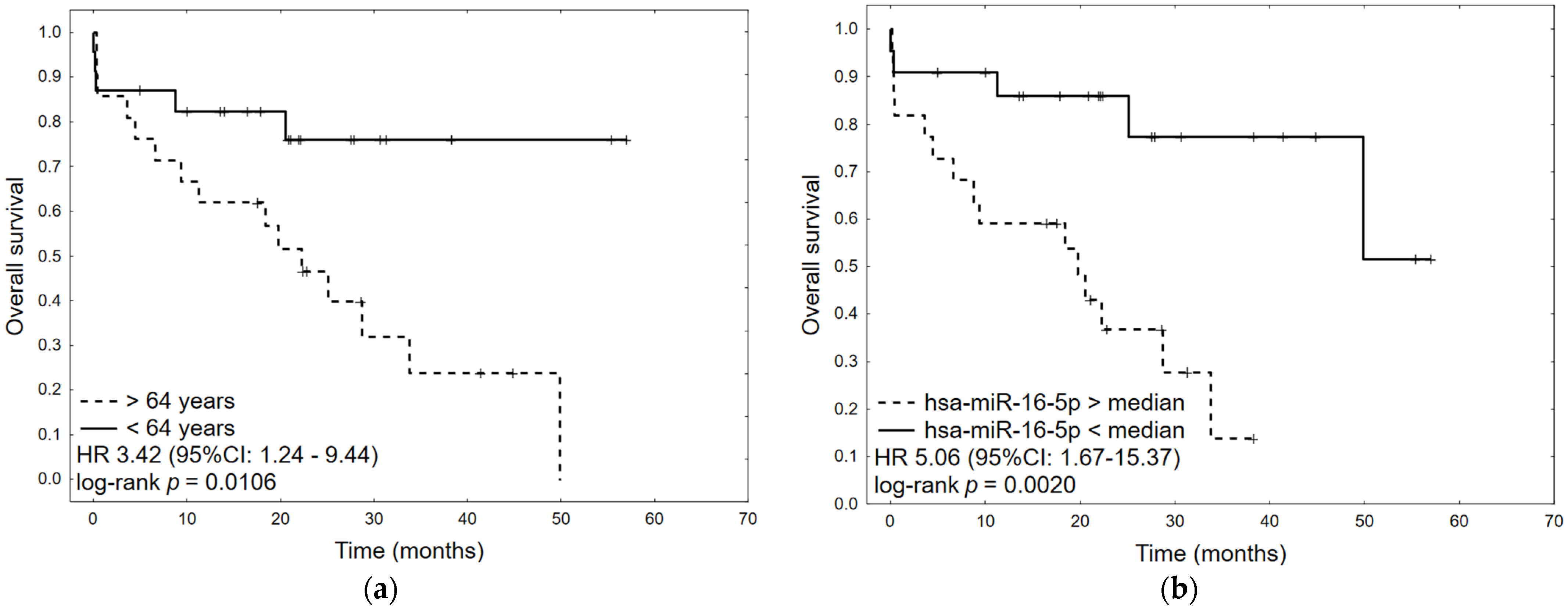
2.5. Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Sample Collection
4.3. DNA Isolation
4.4. MiRNA Isolation
4.5. Reverse Transcription
4.6. Real-Time PCR to Detect microRNA Expression
4.7. HR-HPV Type 16/18 Detection
4.7.1. Nested PCR
4.7.2. Real-Time PCR
4.7.3. Digital Droplet PCR
4.8. Statistical Analysis
4.9. MiRNA Expression Analysis
4.10. Survival Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Global Cancer Observatory. 2022. Available online: https://gco.iarc.fr/ (accessed on 24 January 2023).
- Key Statistics for Ovarian Cancer. Available online: https://www.cancer.org/ (accessed on 24 August 2023).
- Köbel, M.; Kalloger, S.E.; Huntsman, D.G.; Santos, J.L.; Swenerton, K.D.; Seidman, J.D.; Gilks, C.B.; on behalf of the Cheryl Brown Ovarian Cancer Outcomes Unit of the British Columbia Cancer Agency, Vancouver BC. Differences in tumor type in low-stage versus high-stage ovarian carcinomas. Int. J. Gynecol. Pathol. 2010, 29, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.M. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Piek, J.M.J.; van Diest, P.J.; Zweemer, R.P.; Jansen, J.W.; Poort-Keesom, R.J.; Menko, F.H.; Gille, J.J.; Jongsma, A.P.; Pals, G.; Kenemans, P.; et al. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. J. Pathol. 2001, 195, 451–456. [Google Scholar] [CrossRef]
- Hebner, C.M.; Laimins, L.A. Human papillomaviruses: Basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 2006, 16, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Paradowska, E.; Jabłońska, A.; Studzińska, M.; Wilczyński, M.; Wilczyński, J.R. Detection and genotyping of CMV and HPV in tumors and fallopian tubes from epithelial ovarian cancer patients. Sci. Rep. 2019, 9, 19935. [Google Scholar] [CrossRef] [PubMed]
- Pathak, S.; Wilczyński, J.R.; Paradowska, E. Factors in Oncogenesis: Viral Infections in Ovarian Cancer. Cancers 2020, 12, 561. [Google Scholar] [CrossRef] [PubMed]
- Al-Shabanah, O.A.; Hafez, M.M.; Hassan, Z.K.; Sayed-Ahmed, M.M.; Abozeed, W.N.; Al-Rejaie, S.S.; Alsheikh, A.A. Human papillomavirus genotyping and integration in ovarian cancer Saudi patients. Virol. J. 2013, 10, 343. [Google Scholar] [CrossRef]
- Wu, Q.J.; Guo, M.; Lu, Z.M.; Li, T.; Qiao, H.Z.; Ke, Y. Detection of human papillomavirus-16 in ovarian malignancy. Br. J. Cancer 2003, 89, 672–675. [Google Scholar] [CrossRef]
- Bilyk, O.O.; Pande, N.T.; Pejovic, T.; Buchinska, L.G. The frequency of human papilloma virus types 16, 18 in upper genital tract of women at high risk of developing ovarian cancer. Exp. Oncol. 2014, 36, 121–124. [Google Scholar]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.I.; Sonoda, T.; et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef]
- Bignotti, E.; Calza, S.; Tassi, R.A.; Zanotti, L.; Bandiera, E.; Sartori, E.; Odicino, F.E.; Ravaggi, A.; Todeschini, P.; Romani, C. Identification of stably expressed reference small non-coding RNA s for micro RNA quantification in high-grade serous ovarian carcinoma tissues. J. Cell. Mol. Med. 2016, 20, 2341–2348. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Yokoi, A.; Kato, T.; Ochiya, T.; Yamamoto, Y. The clinical impact of intra- and extracellular miRNAs in ovarian cancer. Cancer Sci. 2020, 111, 3435–3444. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.H.; Jensen, N.F.; Tarpgaard, L.S.; Qvortrup, C.; Rømer, M.U.; Stenvang, J.; Hansen, T.P.; Christensen, L.L.; Lindebjerg, J.; Hansen, F.; et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol. Oncol. 2013, 7, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Shapira, I.; Oswald, M.; Lovecchio, J.; Khalili, H.; Menzin, A.; Whyte, J.; Dos Santos, L.; Liang, S.; Bhuiya, T.; Keogh, M.; et al. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br. J. Cancer 2014, 110, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.; Dwivedi, S.K.D.; Zhang, Y.; Dey, A.; Shameer, K.; Karthik, R.; Srikantan, S.; Hossen, M.N.; Wren, J.D.; Madesh, M.; et al. MicroRNA-195 controls MICU 1 expression and tumor growth in ovarian cancer. EMBO Rep. 2020, 21, e48483. [Google Scholar] [CrossRef] [PubMed]
- Elias, K.M.; Fendler, W.; Stawiski, K.; Fiascone, S.J.; Vitonis, A.F.; Berkowitz, R.S.; Frendl, G.; Konstantinopoulos, P.; Crum, C.P.; Kedzierska, M.; et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. eLife 2017, 6, e28932. [Google Scholar] [CrossRef]
- Huang, G.L.; Sun, J.; Lu, Y.; Liu, Y.; Cao, H.; Zhang, H.; Calin, G.A. MiR-200 family and cancer: From a meta-analysis view. Mol. Aspects Med. 2019, 70, 57–71. [Google Scholar] [CrossRef]
- Parikh, A.; Lee, C.; Joseph, P.; Marchini, S.; Baccarini, A.; Kolev, V.; Romualdi, C.; Fruscio, R.; Shah, H.; Wang, F.; et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial–mesenchymal transition. Nat. Commun. 2014, 5, 2977. [Google Scholar] [CrossRef]
- Wang, X.; Meng, X.; Li, H.; Liu, W.; Shen, S.; Gao, Z. MicroRNA-25 expression level is an independent prognostic factor in epithelial ovarian cancer. Clin. Transl. Oncol. 2014, 16, 954–958. [Google Scholar] [CrossRef]
- Kan, C.W.; Hahn, M.A.; Gard, G.B.; Maidens, J.; Huh, J.Y.; Marsh, D.J.; Howell, V.M. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer 2012, 12, 627. [Google Scholar] [CrossRef]
- Pan, C.; Stevic, I.; Müller, V.; Ni, Q.; Oliveira-Ferrer, L.; Pantel, K.; Schwarzenbach, H. Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol. Oncol. 2018, 12, 1935–1948. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Pan, W.; Jin, Y.; Zheng, J. MiR-25 promotes ovarian cancer proliferation and motility by targeting LATS2. Tumor Biol. 2014, 35, 12339–12344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zuo, Z.; Lu, X.; Wang, L.; Wang, H.; Zhu, Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol. Rep. 2012, 27, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Chiantore, M.V.; Mangino, G.; Iuliano, M.; Zangrillo, M.S.; De Lillis, I.; Vaccari, G.; Accardi, R.; Tommasino, M.; Columba Cabezas, S.; Federico, M.; et al. Human papillomavirus E6 and E7 oncoproteins affect the expression of cancer-related microRNAs: Additional evidence in HPV-induced tumorigenesis. J. Cancer Res. Clin. Oncol. 2016, 142, 1751–1763. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, X.; Meng, L.; Li, W.; Li, C.; Li, P.; Xu, S. Changes of miRNA Expression Profiles from Cervical-Vaginal Fluid-Derived Exosomes in Response to HPV16 Infection. BioMed Res. Int. 2020, 2020, 7046894. [Google Scholar] [CrossRef]
- Sadri Nahand, J.; Moghoofei, M.; Salmaninejad, A.; Bahmanpour, Z.; Karimzadeh, M.; Nasiri, M.; Mirzaei, H.R.; Pourhanifeh, M.H.; Bokharaei-Salim, F.; Mirzaei, H.; et al. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: A review. Int. J. Cancer 2020, 146, 305–320. [Google Scholar] [CrossRef]
- Gao, D.; Zhang, Y.; Zhu, M.; Liu, S.; Wang, X. miRNA Expression Profiles of HPV-Infected Patients with Cervical Cancer in the Uyghur Population in China. PLoS ONE 2016, 11, e0164701. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Hussen, B.M.; Shaterabadi, D.; Abak, A.; Shoorei, H.; Taheri, M.; Rakhshan, A. The interaction between human papilloma viruses related cancers and non-coding RNAs. Pathol. Res. Pract. 2022, 234, 153939. [Google Scholar] [CrossRef]
- Wilting, S.M.; Verlaat, W.; Jaspers, A.; Makazaji, N.A.; Agami, R.; Meijer, C.J.; Snijders, P.J.; Steenbergen, R.D. Methylation-mediated transcriptional repression of microRNAs during cervical carcinogenesis. Epigenetics 2013, 8, 220–228. [Google Scholar] [CrossRef]
- Zheng, Z.M.; Wang, X. Regulation of cellular miRNA expression by human papillomaviruses. Biochim. Biophys. Acta 2011, 1809, 668–677. [Google Scholar] [CrossRef]
- Hussen, B.M.; Ahmadi, G.; Marzban, H.; Fard Azar, M.E.; Sorayyayi, S.; Karampour, R.; Nahand, J.S.; Hidayat, H.J.; Moghoofei, M. The role of HPV gene expression and selected cellular MiRNAs in lung cancer development. Microb. Pathog. 2021, 150, 104692. [Google Scholar] [CrossRef] [PubMed]
- Harden, M.E.; Prasad, N.; Griffiths, A.; Munger, K. Modulation of microRNA-mRNA Target Pairs by Human Papillomavirus 16 Oncoproteins. mBio 2017, 8, e02170-16. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.; Saha, S.S.; Sen, S.; Bhattacharya, A.; Bhattacharya, N.P.; Bucha, S.; Sinha, M.; Chowdhury, R.R.; Mondal, N.R.; Chakravarty, B.; et al. Cervical cancer subtypes harbouring integrated and/or episomal HPV16 portray distinct molecular phenotypes based on transcriptome profiling of mRNAs and miRNAs. Cell Death Discov. 2019, 5, 81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jia, L.G.; Wang, P.; Li, J.; Tian, F.; Chu, Z.P.; Kang, S. The expression and significance of lncRNA HOST2 and microRNA let-7b in HPV-positive cervical cancer tissues and cell lines. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2380–2390. [Google Scholar] [CrossRef] [PubMed]
- Lajer, C.B.; Garnæs, E.; Friis-Hansen, L.; Norrild, B.; Therkildsen, M.H.; Glud, M.; Rossing, M.; Lajer, H.; Svane, D.; Skotte, L.; et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: Bridging between HPV-related head and neck cancer and cervical cancer. Br. J. Cancer 2012, 106, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Flemington, E.K. miRNAs in the pathogenesis of oncogenic human viruses. Cancer Lett. 2011, 305, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Gocze, K.; Gombos, K.; Kovacs, K.; Juhasz, K.; Gocze, P.; Kiss, I. MicroRNA expressions in HPV-induced cervical dysplasia and cancer. Anticancer Res. 2015, 35, 523–530. [Google Scholar] [PubMed]
- Park, S.; Eom, K.; Kim, J.; Bang, H.; Wang, H.Y.; Ahn, S.; Kim, G.; Jang, H.; Kim, S.; Lee, D.; et al. MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer 2017, 17, 658. [Google Scholar] [CrossRef]
- Sethi, N.; Wright, A.; Wood, H.; Rabbitts, P. MicroRNAs and head and neck cancer: Reviewing the first decade of research. Eur. J. Cancer 2014, 50, 2619–2635. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, F.; Sun, P. MiR-140-3p impedes the proliferation of human cervical cancer cells by targeting RRM2 to induce cell-cycle arrest and early apoptosis. Bioorg. Med. Chem. 2020, 28, 115283. [Google Scholar] [CrossRef]
- Prat, J.; FIGO Committee on Gynecologic Oncology. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int. J. Gynaecol. Obstet. 2014, 124, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Caiazza, C.; Poltronieri, P.; Mallardo, M. The Roles of miR-25 and Its Targeted Genes in Human Cancer. In Recent Trends in Cancer Biology: Spotlight on Signaling Cascades and MicroRNAs. Cell Signaling Pathways and microRNAs in Cancer Biology; Fayyaz, S., Farooqi, A.A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 129–139. [Google Scholar] [CrossRef]
- Jia, W.; Wu, Y.; Zhang, Q.; Gao, G.; Zhang, C.; Xiang, Y. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol. Clin. Oncol. 2015, 3, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Iwanaga, R.; Drasin, D.J.; Micalizzi, D.S.; Vartuli, R.L.; Tan, A.C.; Ford, H.L. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene 2012, 31, 5162–5171. [Google Scholar] [CrossRef] [PubMed]
- Sárközy, M.; Kahán, Z.; Csont, T. A myriad of roles of miR-25 in health and disease. Oncotarget 2018, 9, 21580–21612. [Google Scholar] [CrossRef]
- Meng, X.; Joosse, S.A.; Müller, V.; Trillsch, F.; Milde-Langosch, K.; Mahner, S.; Geffken, M.; Pantel, K.; Schwarzenbach, H. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br. J. Cancer 2015, 113, 1358–1366. [Google Scholar] [CrossRef]
- Langhe, R.; Norris, L.; Saadeh, F.A.; Blackshields, G.; Varley, R.; Harrison, A.; Gleeson, N.; Spillane, C.; Martin, C.; O’Donnell, D.M.; et al. A novel serum microRNA panel to discriminate benign from malignant ovarian disease. Cancer Lett. 2015, 356, 628–636. [Google Scholar] [CrossRef]
- Li, J.; Yue, H.; Li, W.; Zhu, G.; Zhu, T.; Chen, R.; Lu, X. Bevacizumab confers significant improvements in survival for ovarian cancer patients with low miR-25 expression and high miR-142 expression. J. Ovarian Res. 2021, 14, 166. [Google Scholar] [CrossRef]
- Kumar, M.; Lu, Z.; Takwi, A.A.L.; Chen, W.; Callander, N.S.; Ramos, K.S.; Young, K.H.; Li, Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 2011, 30, 843–853. [Google Scholar] [CrossRef]
- Emmrich, S.; Pützer, B.M. Checks and balances: E2F—microRNA crosstalk in cancer control. Cell Cycle 2010, 9, 2555–2567. [Google Scholar] [CrossRef]
- Miller, D.L.; Davis, J.W.; Taylor, K.H.; Johnson, J.; Shi, Z.; Williams, R.; Atasoy, U.; Lewis, J.S.; Stack, M.S. Identification of a human papillomavirus–associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the cancer genome atlas. Am. J. Pathol. 2015, 185, 679–692. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.-K.; Li, Y.; Hafner, M.; Banerjee, N.S.; Tang, S.; Briskin, D.; Meyers, C.; Chow, L.T.; Xie, X.; et al. MicroRNAs Are Biomarkers of oncogenic human papillomavirus infections. Proc. Natl. Acad. Sci. USA 2014, 111, 4262–4267. [Google Scholar] [CrossRef]
- Narisawa-Saito, M.; Kiyono, T. Basic Mechanisms of High-risk Human Papillomavirus-induced Carcinogenesis: Roles of E6 and E7 Proteins. Cancer Sci. 2007, 98, 1505–1511. [Google Scholar] [CrossRef] [PubMed]
- Shai, A.; Brake, T.; Somoza, C.; Lambert, P.F. The Human Papillomavirus E6 Oncogene Dysregulates the Cell Cycle and Contributes to Cervical Carcinogenesis through Two Independent Activities. Cancer Res. 2007, 67, 1626–1635. [Google Scholar] [CrossRef] [PubMed]
- Mitxelena, J.; Apraiz, A.; Vallejo-Rodríguez, J.; Malumbres, M.; Zubiaga, A.M. E2F7 Regulates Transcription and Maturation of Multiple microRNAs to Restrain Cell Proliferation. Nucleic Acids Res. 2016, 44, 5557–5570. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, F.; He, Z.; Zuo, M.-Z. E2F2/5/8 Serve as Potential Prognostic Biomarkers and Targets for Human Ovarian Cancer. Front. Oncol. 2019, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.; Shalgi, R.; Liran, A.; Landan, G.; Korotayev, K.; Nguyen, G.H.; Enerly, E.; Johnsen, H.; Buganim, Y.; Solomon, H.; et al. P53-repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol. Syst. Biol. 2008, 4, 229. [Google Scholar] [CrossRef]
- Thangavel, C.; Boopathi, E.; Ertel, A.; Lim, M.; Addya, S.; Fortina, P.; Witkiewicz, A.K.; Knudsen, E.S. Regulation of miR106b Cluster through the RB Pathway: Mechanism and Functional Targets. Cell Cycle 2013, 12, 98–111. [Google Scholar] [CrossRef]
- Al-Khanbashi, M.; Caramuta, S.; Alajmi, A.M.; Al-Haddabi, I.; Al-Riyami, M.; Lui, W.O.; Al-Moundhri, M.S. Tissue and Serum miRNA Profile in Locally Advanced Breast Cancer (LABC) in Response to Neo-Adjuvant Chemotherapy (NAC) Treatment. PLoS ONE 2016, 11, e0152032. [Google Scholar] [CrossRef]
- Schneider, A.; Victoria, B.; Lopez, Y.N.; Suchorska, W.; Barczak, W.; Sobecka, A.; Golusinski, W.; Masternak, M.M.; Golusinski, P. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci. Rep. 2018, 8, 675. [Google Scholar] [CrossRef]
- Ferreira, P.; Roela, R.A.; Lopez, R.V.M.; Del Pilar Estevez-Diz, M. The prognostic role of microRNA in epithelial ovarian cancer: A systematic review of literature with an overall survival meta-analysis. Oncotarget 2020, 11, 1085–1095. [Google Scholar] [CrossRef]
- Raue, R.; Frank, A.C.; Syed, S.N.; Brüne, B. Therapeutic Targeting of MicroRNAs in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 2210. [Google Scholar] [CrossRef] [PubMed]
- St-Cyr, G.; Penarroya, D.; Daniel, L.; Giguère, H.; Alkayyal, A.A.; Tai, L.H. Remodeling the tumor immune microenvironment with oncolytic viruses expressing miRNAs. Front. Immunol. 2023, 13, 1071223. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Jia, J.; Yao, L.; Li, Z. Crosstalk of Exosomal Non-Coding RNAs in The Tumor Microenvironment: Novel Frontiers. Front. Immunol. 2022, 13, 900155. [Google Scholar] [CrossRef] [PubMed]
- Kay, P.; Allan, B.; Denny, L.; Hoffman, M.; Williamson, A.L. Detection of HPV 16 and HPV 18 DNA in the blood of patients with cervical cancer. J. Med. Virol. 2005, 75, 435–439. [Google Scholar] [CrossRef]
- Bønløkke, S.; Stougaard, M.; Sorensen, B.S.; Booth, B.B.; Høgdall, E.; Nyvang, G.B.; Lindegaard, J.C.; Blaakær, J.; Bertelsen, J.; Fuglsang, K.; et al. The Diagnostic Value of Circulating Cell-Free HPV DNA in Plasma from Cervical Cancer Patients. Cells 2022, 11, 2170. [Google Scholar] [CrossRef]
- Pao, C.C.; Hor, J.J.; Yang, F.P.; Lin, C.Y.; Tseng, C.J. Detection of human papillomavirus mRNA and cervical cancer cells in peripheral blood of cervical cancer patients with metastasis. J. Clin. Oncol. 1997, 15, 1008–1012. [Google Scholar] [CrossRef]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef]
- Dong, J.; Xu, M. [Corrigendum] A 19-miRNA support vector machine classifier and a 6-miRNA risk score system designed for ovarian cancer patients. Oncol. Rep. 2019, 42, 2855. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, L.; Wang, L.; Zhang, H.; Hu, X. miR-218 functions as a tumor suppressor gene in cervical cancer. Mol. Med. Rep. 2020, 21, 209–219. [Google Scholar] [CrossRef]
- Jiang, Z.; Song, Q.; Zeng, R.; Li, J.; Li, J.; Lin, X.; Chen, X.; Zhang, J.; Zheng, Y. MicroRNA-218 inhibits EMT, migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical cancer. Oncotarget 2016, 7, 45622–45636. [Google Scholar] [CrossRef]
- Huang, Y.; Liang, S.H.; Xiang, L.B.; Han, X.T.; Zhang, W.; Tang, J.; Wu, X.H.; Zhang, M.Q. MiR-218 promoted the apoptosis of human ovarian carcinoma cells via suppression of the WNT/β-catenin signaling pathway. Mol. Biol. 2017, 51, 555–561. [Google Scholar] [CrossRef]
- McBee, W.C.; Gardiner, A.S.; Edwards, R.P.; Lesnock, J.L.; Bhargava, R.; Austin, R.M.; Guido, R.S.; Khan, S.A. MicroRNA Analysis in Human Papillomavirus (HPV)-Associated Cervical Neoplasia and Cancer. J. Carcinog. Mutagen. 2011, 1, 114. [Google Scholar] [CrossRef]
- Zhu, L.; Tu, H.; Liang, Y.; Tang, D. MiR-218 produces anti-tumor effects on cervical cancer cells in vitro. World J. Surg. Oncol. 2018, 16, 204. [Google Scholar] [CrossRef] [PubMed]
- Braza-Boïls, A.; Marí-Alexandre, J.; Gilabert, J.; Sánchez-Izquierdo, D.; España, F.; Estellés, A.; Gilabert-Estellés, J. MicroRNA expression profile in endometriosis: Its relation to angiogenesis and fibrinolytic factors. Hum. Reprod. 2014, 29, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Abdullah, S.T.; Taheri, M.; Samadian, M. A review on the role of miR-16-5p in the carcinogenesis. Cancer Cell Int. 2022, 22, 342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lai, X.; Yue, Q.; Cao, F.; Zhang, Y.; Sun, Y.; Tian, J.; Lu, Y.; He, L.; Bai, J.; et al. Bone Marrow Mesenchymal Stem Cells-Derived Exosomal microRNA-16-5p Restrains Epithelial-Mesenchymal Transition in Breast Cancer Cells via EPHA1/NF-κB Signaling Axis. Genomics 2022, 114, 110341. [Google Scholar] [CrossRef] [PubMed]
- Krell, A.; Wolter, M.; Stojcheva, N.; Hertler, C.; Liesenberg, F.; Zapatka, M.; Weller, M.; Malzkorn, B.; Reifenberger, G. MiR-16-5p is frequently down-regulated in astrocytic gliomas and modulates glioma cell proliferation, apoptosis and response to cytotoxic therapy. Neuropath. Appl. Neurobiol. 2019, 45, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hu, S.; Li, X.; Liu, Z.; Han, D.; Wang, Y.; Wei, L.; Zhang, G.; Wang, X. MiR-16-5p Suppresses Breast Cancer Proliferation by Targeting ANLN. BMC Cancer 2021, 21, 1188. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, C.; Cui, H. Long Non-Coding RNA SNHG22 Facilitates Hepatocellular Carcinoma Tumorigenesis and Angiogenesis via DNA Methylation of microRNA miR-16-5p. Bioengineered 2021, 12, 7446–7458. [Google Scholar] [CrossRef]
- Salmasi, S.; Sharifi, M.; Rashidi, B. Ovarian stimulation and exogenous progesterone affect the endometrial miR-16-5p, VEGF protein expression, and angiogenesis. Microvasc. Res. 2021, 133, 104074. [Google Scholar] [CrossRef]
- Yan, Z.; Liu, Y.; Wei, Y.; Zhao, N.; Zhang, Q.; Wu, C.; Chang, Z.; Xu, Y. The functional consequences and prognostic value of dosage sensitivity in ovarian cancer. Mol. Biosyst. 2017, 13, 380–391. [Google Scholar] [CrossRef]
- Wyman, S.K.; Parkin, R.K.; Mitchell, P.S.; Fritz, B.R.; O’Briant, K.; Godwin, A.K.; Urban, N.; Drescher, C.W.; Knudsen, B.S.; Tewari, M. Repertoire of microRNAs in Epithelial Ovarian Cancer as Determined by Next Generation Sequencing of Small RNA cDNA Libraries. PLoS ONE 2009, 4, e5311. [Google Scholar] [CrossRef] [PubMed]
- Nam, E.J.; Yoon, H.; Kim, S.W.; Kim, H.; Kim, Y.T.; Kim, J.H.; Kim, J.W.; Kim, S. MicroRNA Expression Profiles in Serous Ovarian Carcinoma. Clin. Cancer Res. 2008, 14, 2690–2695. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.V.; Fedorov, I.S.; Asaturova, A.V.; Sannikova, M.V.; Tregubova, A.V.; Mayboroda, O.A.; Khabas, G.N.; Frankevich, V.E.; Sukhikh, G.T. Blood Plasma Small Non-Coding RNAs as Diagnostic Molecules for the Progesterone-Receptor-Negative Phenotype of Serous Ovarian Tumors. Int. J. Mol. Sci. 2023, 24, 12214. [Google Scholar] [CrossRef] [PubMed]
- Saral, M.A.; Tuncer, S.B.; Odemis, D.A.; Erdogan, O.S.; Erciyas, S.K.; Saip, P.; Ozel, S.; Yazici, H. New biomarkers in peripheral blood of patients with ovarian cancer: High expression levels of miR-16-5p, miR-17-5p, and miR-638. Arch. Gynecol. Obstet. 2022, 305, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.V.; Asaturova, A.V.; Sannikova, M.V.; Khabas, G.N.; Chagovets, V.V.; Fedorov, I.S.; Frankevich, V.E.; Sukhikh, G.T. Search for new participants in the pathogenesis of high-grade serous ovarian cancer with the potential to be used as diagnostic molecules. Life 2022, 12, 2017. [Google Scholar] [CrossRef] [PubMed]
- Paradowska, E.; Haręża, D.; Kania, K.D.; Jarych, D.; Wilczyński, M.; Malinowski, A.; Kawecka, M.; Wilczyński, J.R. A Prevalence of Human Papillomavirus, Cytomegalovirus, and Epstein-Barr Virus Infections in Ovarian Cancer Patients. 2024; submitted. [Google Scholar]
- Stevenson, A.; Wakeham, K.; Pan, J.; Kavanagh, K.; Millan, D.; Bell, S.; McLellan, D.; Graham, S.V.; Cuschieri, K. Droplet digital PCR quantification suggests that higher viral load correlates with improved survival in HPV-positive oropharyngeal tumours. J. Clin. Virol. 2020, 129, 104505. [Google Scholar] [CrossRef]
- Hanna, G.J.; Supplee, J.G.; Kuang, Y.; Mahmood, U.; Lau, C.J.; Haddad, R.I.; Jänne, P.A.; Paweletz, C.P. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann. Oncol. 2018, 29, 1980–1986. [Google Scholar] [CrossRef]
- Van Heetvelde, M.; Van Loocke, W.; Trypsteen, W.; Baert, A.; Vanderheyden, K.; Crombez, B.; Vandesompele, J.; De Leeneer, K.; Claes, K.B.M. Evaluation of relative quantification of alternatively spliced transcripts using droplet digital PCR. Biomol. Detect. Quantif. 2017, 13, 40–48. [Google Scholar] [CrossRef]
- Grabia, S.; Smyczynska, U.; Pagacz, K.; Fendler, W. NormiRazor: Tool applying GPU-accelerated computing for determination of internal references in microRNA transcription studies. BMC Bioinform. 2020, 21, 425. [Google Scholar] [CrossRef]
- Stawiski, K.; Kaszkowiak, M.; Mikulski, D.; Hogendorf, P.; Durczyński, A.; Strzelczyk, J.; Chowdhury, D.; Fendler, W. OmicSelector: Automatic feature selection and deep learning modeling for omic experiments. BioRxiv 2022. [Google Scholar] [CrossRef]

| Variable | N | % |
|---|---|---|
| FIGO | ||
| I | 5 | 10.9 |
| II | 2 | 4.3 |
| III | 29 | 63 |
| IV | 9 | 19.6 |
| ND | 1 | 2.2 |
| EON type | ||
| High-grade serous ovarian cancer | 33 | 71.7 |
| Borderline ovarian tumor | 5 | 10.9 |
| Clear-cell ovarian cancer | 3 | 6.5 |
| Mucinous ovarian cancer | 3 | 6.5 |
| Others | 2 | 4.3 |
| HPV Status | Prevalence; N (%) | |
|---|---|---|
| Tumor | Blood | |
| HPV-infected | 30/46 (65.2) | 29/46 (63.0) |
| HPV16 | 14/30 (46.7) | 10/29 (34.5) |
| HPV18 | 9/30 (30.0) | 9/29 (31.0) |
| HPV16/18 coinfection | 7/30 (23.3) | 10/29 (34.5) |
| Uninfected | 16/46 (34.8) | 17/46 (37.0) |
| miRNA | HPV- Positive Mean | HPV- Positive SD | HPV- Negative Mean | HPV- Negative SD | FC | log2FC | p-Value |
|---|---|---|---|---|---|---|---|
| hsa-miR-25-5p | −7.53 | 0.80 | −6.83 | 1.15 | 0.61 | −0.70 | 0.0405 |
| hsa-miR-203a-3p | −0.97 | 1.34 | −1.62 | 1.61 | 1.57 | 0.65 | 0.1761 |
| hsa-miR-21-5p | 6.23 | 1.08 | 5.82 | 0.88 | 1.32 | 0.41 | 0.1770 |
| hsa-miR-191-5p | 1.01 | 0.67 | 1.32 | 0.75 | 0.81 | −0.31 | 0.1813 |
| hsa-let-7b-5p | 2.41 | 1.12 | 2.00 | 1.47 | 1.33 | 0.42 | 0.3320 |
| hsa-miR-16-5p | 3.70 | 0.88 | 3.95 | 0.82 | 0.84 | −0.25 | 0.3481 |
| hsa-miR-140-3p | −0.84 | 1.21 | −1.21 | 1.78 | 1.29 | 0.37 | 0.4624 |
| hsa-miR-34a-5p | 0.04 | 1.04 | −0.21 | 1.36 | 1.19 | 0.25 | 0.5285 |
| hsa-miR-218-5p | −1.81 | 1.34 | −1.64 | 1.73 | 0.89 | −0.17 | 0.7284 |
| hsa-miR-200a-3p | −0.20 | 1.75 | −0.36 | 2.34 | 1.12 | 0.16 | 0.8134 |
| hsa-miR-9-5p | −4.35 | 1.74 | −4.30 | 1.74 | 0.97 | −0.05 | 0.9260 |
| Variable | OS | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||||||
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |||
| Lower | Upper | Lower | Upper | |||||
| Age | 1.04 | 1.00 | 1.08 | 0.0401 | 1.05 | 1.01 | 1.08 | 0.0166 |
| HPV16 and/or HPV18 in tumor | 1.01 | 0.40 | 2.60 | 0.9767 | ||||
| Stage 4 FIGO | 1.72 | 0.68 | 4.35 | 0.2492 | ||||
| HGSOC | 0.80 | 0.31 | 2.09 | 0.6521 | ||||
| hsa-miR-21-5p > median | 2.12 | 0.85 | 5.23 | 0.1051 | ||||
| hsa-miR-191-5p > median | 0.92 | 0.38 | 2.22 | 0.8492 | ||||
| hsa-miR-9-5p > median | 1.10 | 0.46 | 2.68 | 0.8272 | ||||
| hsa-miR-16-5p > median | 5.06 | 1.67 | 15.37 | 0.0042 | 5.50 | 1.78 | 16.99 | 0.0030 |
| hsa-miR-25-5p > median | 0.47 | 0.18 | 1.19 | 0.1107 | ||||
| hsa-miR-34a-5p > median | 1.03 | 0.43 | 2.50 | 0.9430 | ||||
| hsa-miR-200a-3p > median | 0.69 | 0.28 | 1.67 | 0.4071 | ||||
| hsa-miR-203a-3p > median | 0.60 | 0.24 | 1.48 | 0.2653 | ||||
| hsa-miR-218-5p > median | 2.26 | 0.87 | 5.90 | 0.0953 | ||||
| hsa-let-7b-5p > median | 1.04 | 0.43 | 2.51 | 0.9354 | ||||
| hsa-miR-140-3p > median | 0.89 | 0.36 | 2.19 | 0.7928 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarych, D.; Mikulski, D.; Wilczyński, M.; Wilczyński, J.R.; Kania, K.D.; Haręża, D.; Malinowski, A.; Perdas, E.; Nowak, M.; Paradowska, E. Differential microRNA Expression Analysis in Patients with HPV-Infected Ovarian Neoplasms. Int. J. Mol. Sci. 2024, 25, 762. https://doi.org/10.3390/ijms25020762
Jarych D, Mikulski D, Wilczyński M, Wilczyński JR, Kania KD, Haręża D, Malinowski A, Perdas E, Nowak M, Paradowska E. Differential microRNA Expression Analysis in Patients with HPV-Infected Ovarian Neoplasms. International Journal of Molecular Sciences. 2024; 25(2):762. https://doi.org/10.3390/ijms25020762
Chicago/Turabian StyleJarych, Dariusz, Damian Mikulski, Miłosz Wilczyński, Jacek R. Wilczyński, Katarzyna D. Kania, Daria Haręża, Andrzej Malinowski, Ewelina Perdas, Mateusz Nowak, and Edyta Paradowska. 2024. "Differential microRNA Expression Analysis in Patients with HPV-Infected Ovarian Neoplasms" International Journal of Molecular Sciences 25, no. 2: 762. https://doi.org/10.3390/ijms25020762
APA StyleJarych, D., Mikulski, D., Wilczyński, M., Wilczyński, J. R., Kania, K. D., Haręża, D., Malinowski, A., Perdas, E., Nowak, M., & Paradowska, E. (2024). Differential microRNA Expression Analysis in Patients with HPV-Infected Ovarian Neoplasms. International Journal of Molecular Sciences, 25(2), 762. https://doi.org/10.3390/ijms25020762






