Inconsistencies in the Classification of the Family Cydnidae (Hemiptera: Heteroptera: Pentatomoidea) Revealed by Molecular Apomorphies in the Secondary and Tertiary Structures of 18S rRNA Length-Variable Region L (LVR L)
Abstract
:1. Introduction
2. Results
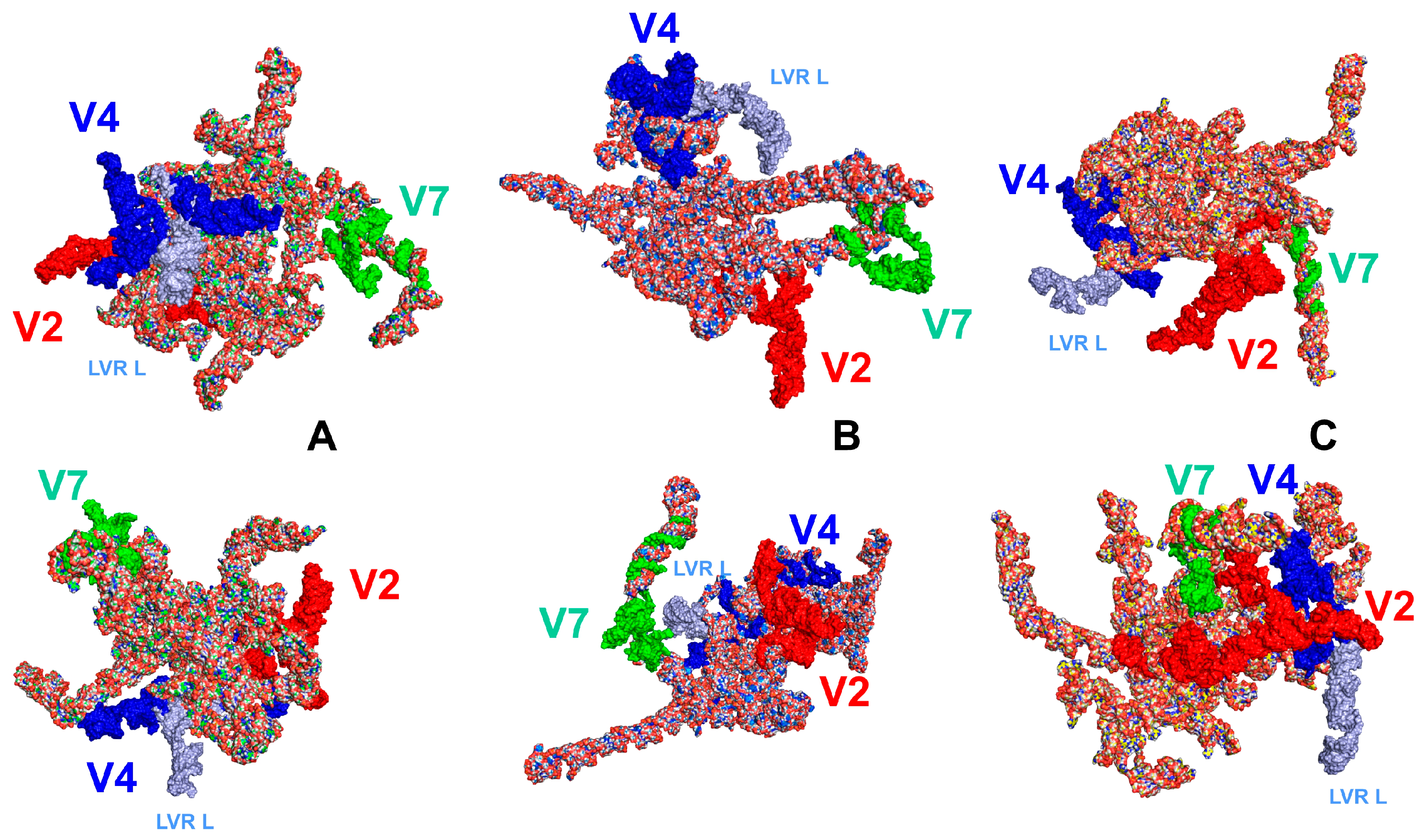
2.1. Hypervariable Region V4 and Length-Variable Region L (LVR L) Sequence Analyses
2.2. 18S rRNA Secondary and Tertiary Structure Models
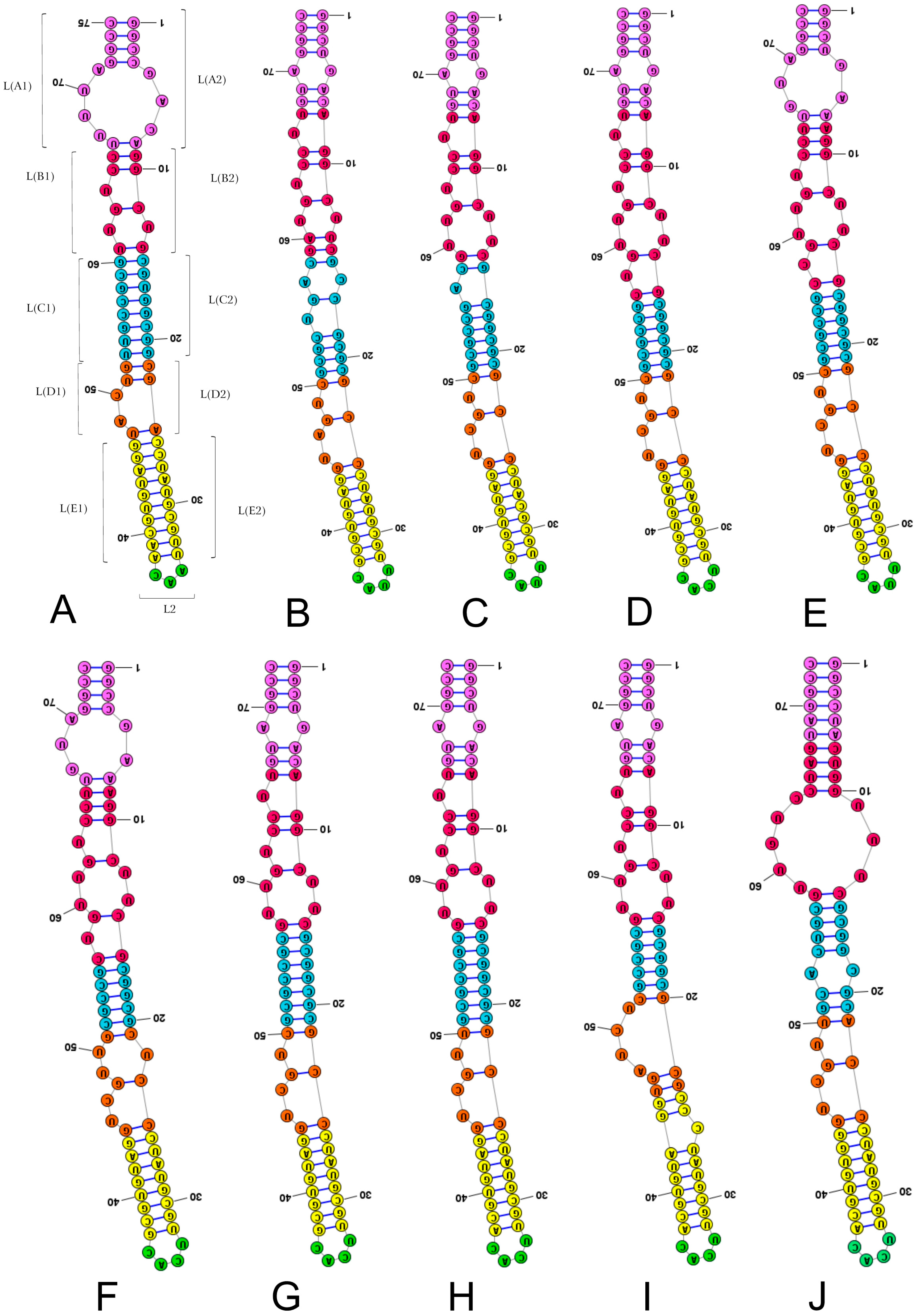
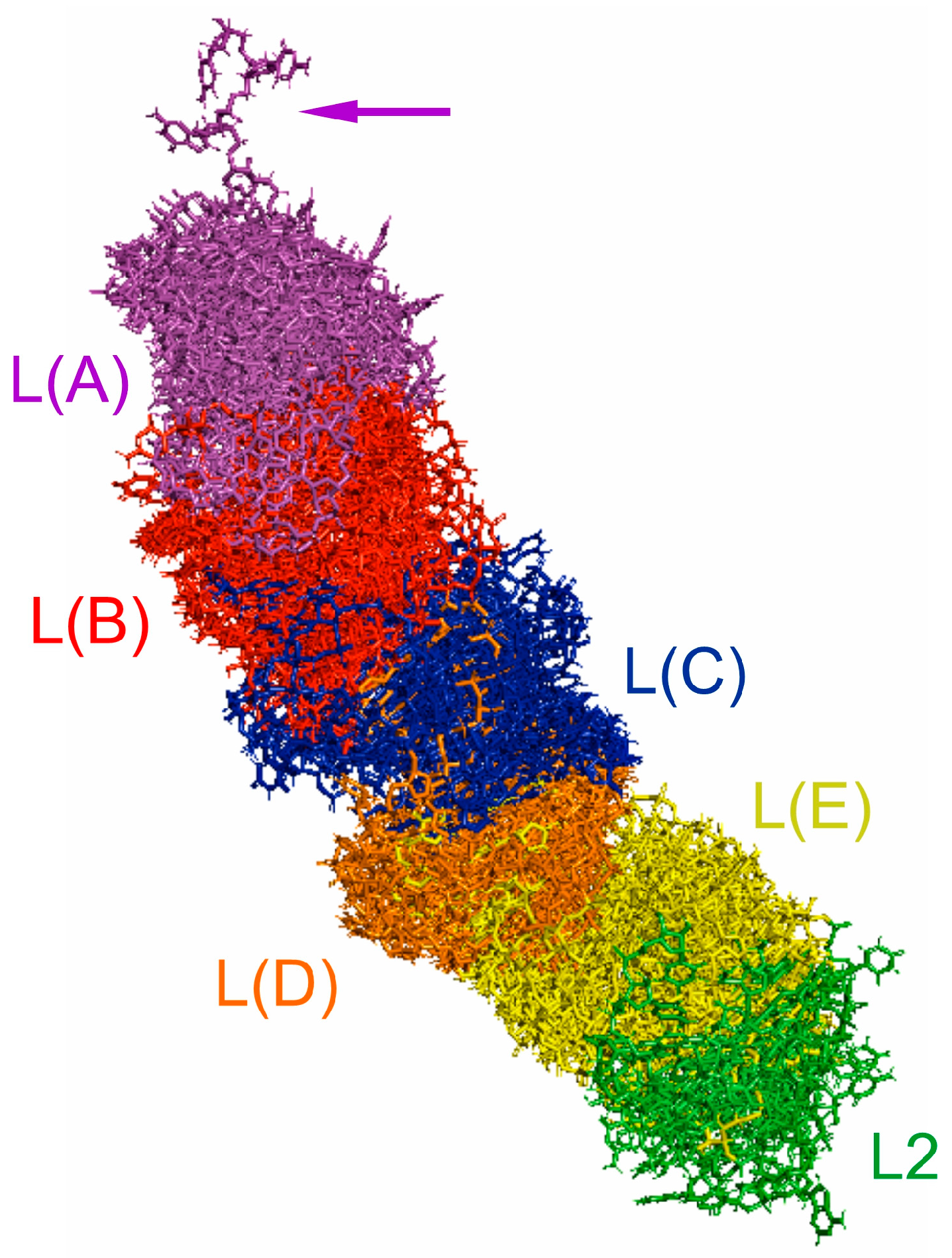
2.3. Length-Variable Region L Secondary Structure
2.4. Length-Variable Region L Tertiary Structure
| Model | Family, Subfamily, Tribe | RMSD Value | |
|---|---|---|---|
| RNAssess Web Server | PyMol | ||
| Rhytidoporus indentatus | Cydnidae: Cydninae: Geotomini | 5.47 | 5.477 |
| Cyrtomenus emarginatus | Cydnidae: Cydninae: Geotomini | 5.88 | 5.881 |
| Thyreocoris scarabaeoides | Thyreocoridae: Thyreocorinae | 6.28 | 6.277 |
| Macroscytus badius | Cydnidae: Cydninae: Geotomini | 6.46 | 6.459 |
| Stibaropus indonesicus | Cydnidae: Cephalocteinae: Scaptocorini | 6.49 | 6.483 |
| Fromundus pygmaeus | Cydnidae: Cydninae: Geotomini | 6.58 | 6.584 |
| Pseudoscoparipes fraterculus | Cydnidae: Cydninae: Geotomini | 6.70 | 6.693 |
| Adomerus biguttatu | Cydnidae: Sehirinae: Sehirini | 6.82 | 6.824 |
| Garsauria aradoides | Cydnidae: Garsauriinae | 6.85 | 6.851 |
| Parastrachia japonensis | Parastrachiidae | 7.17 | 7.170 |
| Adrisa romani | Cydnidae: Cydninae: Geotomini | 7.73 | 7.725 |
| Cydnus aterrimus | Cydnidae: Cydninae: Cydnini | 8.47 | 8.468 |
| Ochetostethomorpha secunda | Cydnidae: Sehirinae: Sehirini | 9.81 | 9.811 |
| Amaurocoris curtus | Cydnidae: Amaurocorinae | 11.33 | 11.332 |
| Chilocoris piceus | Cydnidae: Cydninae: Cydnini | 11.73 | 11.733 |
| Amnestus zacki | Cydnidae: Amnestinae | 15.12 | 15.116 |
| Subregion Compared [Synapomorphy Numbering as in Figure 6 and Figure 7] | Target Model | Compared Model | RMSD Value | |
|---|---|---|---|---|
| RNAssess Web Server (Number of Atoms Compared) | PyMol (Number of Atoms Compared) | |||
| L(A) [s1] L(A) [s2] | Thaumastella elizabethae | Garsauria aradoides | 3.98 (413) | 0.194 (543) |
| Adomerus biguttatus | Parastrachia japonensis | 0.20 (352) | 0.201 (352) | |
| L(B) [s3] | Cyrtomenus emarginatus | Rhytidoporus indentatus | 0.21 (538) | 0.205 (556) |
| L(C) [s4] | Adomerus biguttatus | Parastrachia japonensis | 0.25 (422) | 0.252 (423) |
| L(C) [s5] | Macroscytus badius | Cyrtomenus emarginatus | 0.10 (390) | 0.104 (390) |
| Subregion Compared [Synapomorphy Numbering as in Figure 6 and Figure 7] | Target Model | Compared Models | RMSD Value | |
|---|---|---|---|---|
| RNAssess Web Server (Number of Atoms Compared) | PyMol (Number of Atoms Compared) | |||
| L(A) [s6] | Fromundus pygmaeus | Thyreocoris scarabaeoides Stibaropus indonesicus Macroscytus badius Adrisa romani Pseudoscoparipes fraterculus Cydnus aterrimus Chilocoris piceus | 0.20 (453) 0.10 (453) 0.19 (453) 2.11 (288) 2.14 (288) 2.11 (288) 0.88 (194) | 0.196 (453) 0.098 (453) 0.186 (453) 0.173 (453) 0.186 (453) 0.156 (453) 0.906 (424) |
| L(C) [s7] | Thyreocoris scarabaeoides | Adrisa romani Pseudoscoparipes fraterculus | 0.66 (455) 0.20 (455) | 0.660 (455) 0.198 (455) |
| L(D) [s8] | Fromundus pygmaeus | Cyrtomenus emarginatus Macroscytus badius | 0.76 (389) 0.79 (389) | 0.760 (390) 0.785 (390) |
| L(E) [s9] | Fromundus pygmaeus | Stibaropus indonesicus Macroscytus badius Cyrtomenus emarginatus Adrisa romani Pseudoscoparipes fraterculus Ochetostethomorpha secunda | 0.73 (483) 0.32 (483) 0.25 (483) 0.33 (483) 0.37 (449) 0.70 (416) | 0.742 (511) 0.388 (511) 0.330 (511) 0.401 (511) 0.445 (506) 0.746 (502) |
| L(E) [s10] | Adomerus biguttatus | Parastrachia japonensis Thyreocoris scarabaeoides Amaurocoris curtus | 0.83 (416) 0.72 (449) 0.21 (449) | 0.848 (445) 0.718 (449) 0.209 (449) |
| Subregion Compared [Autapomorphy Numbering as in Figure 6 and Figure 7] | Target Model against which All Other Species Were Compared | Range of RMSD Values |
|---|---|---|
| L(A) [a5] L(A) [a6] L(B) [a4] L(B) [a7] L(C) [a8] L(E) [a1] | Amaurocoris curtus | 1.1 **–11.5 ** |
| Amnestus zacki | 10.3 **–11.7 ** | |
| Thyreocoris scarabaeoides | 3.7 **–10.8 * | |
| Amnestus zacki | 3.9 **–18.4 ** | |
| Amnestus zacki | 4.4 **–6.9 ** | |
| Thaumastella elizabethae | 2.7 *–11.3 ** | |
| L(E) [a3] | Cydnus aterrimus | 1.1 **–11.9 ** |
| L(E) [a9] | Amnestus zacki | 6.7 **–11.9 ** |
| L(E) [a10] | Chilocoris piceus | 3.9 **–10.7 ** |
| L2 [a2] | Thaumastella elizabethae | 1.1 **–4.8 ** |
| L2 [a11] | Chilocoris piceus | 2.8 **–5.7 ** |
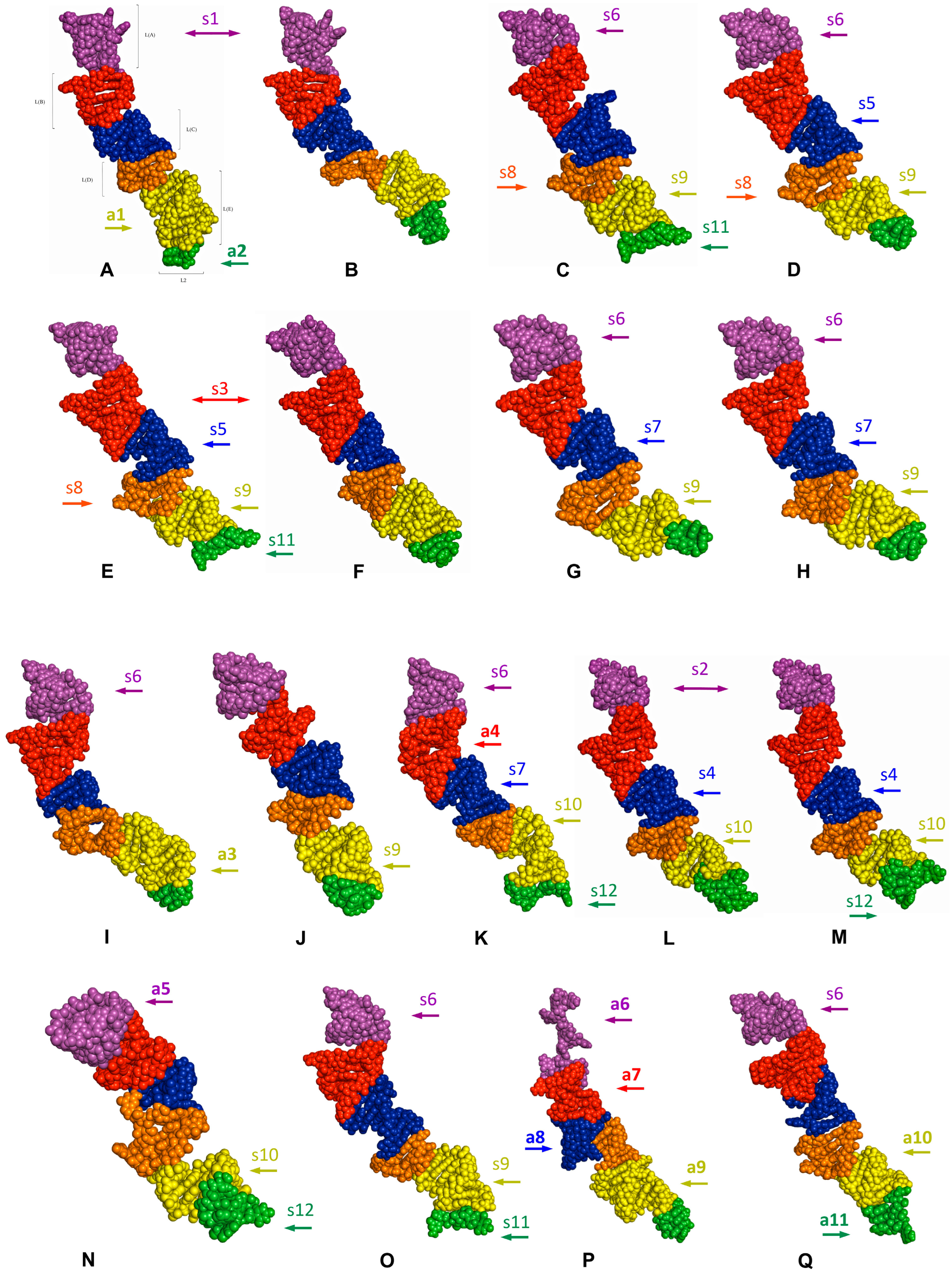
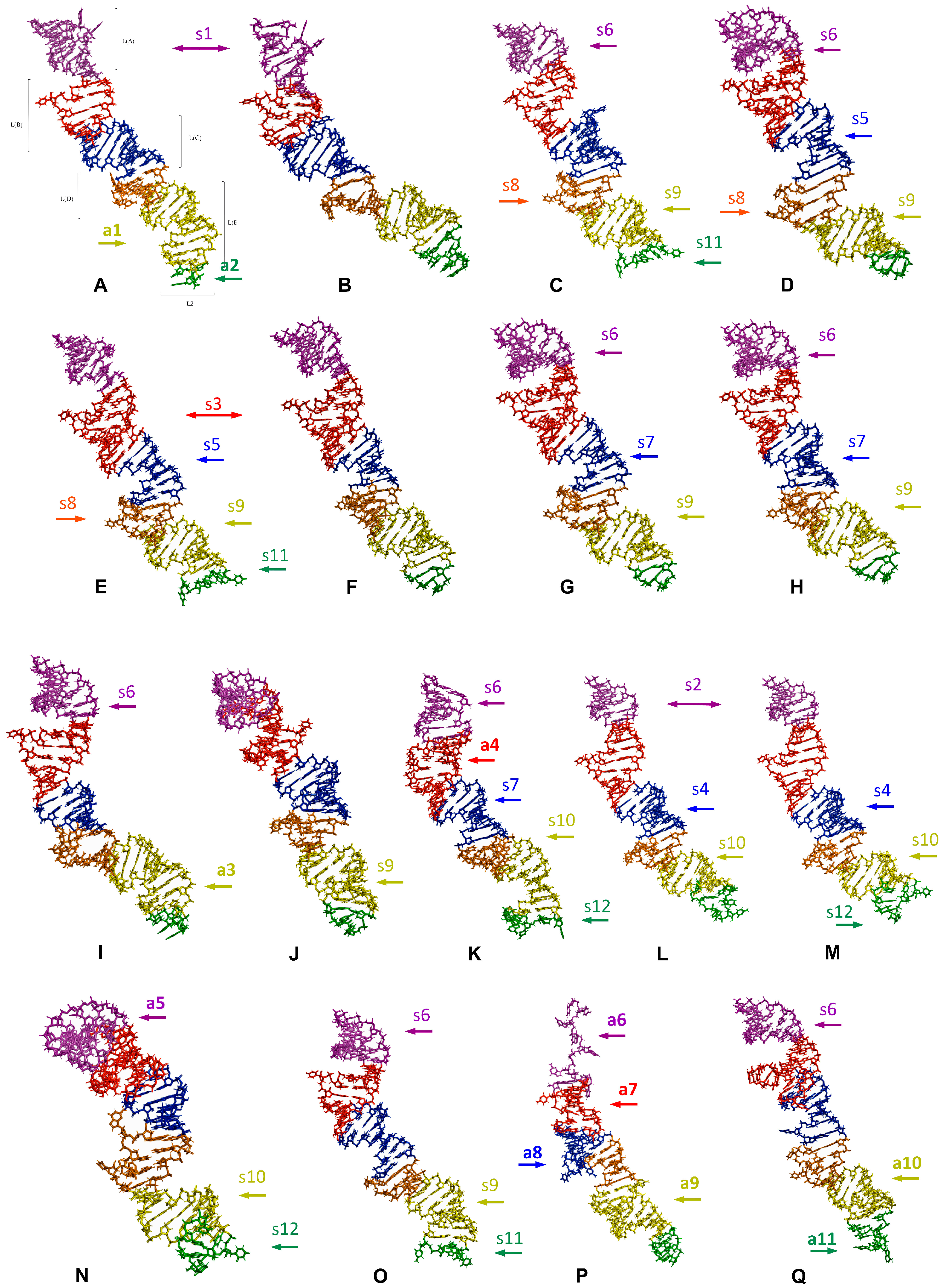
3. Discussion
3.1. Potential Plesiomorphies and Apomorphies in LVR L Secondary Structures
3.2. Potential Synapomorphies and Autapomorphies in LVR L Tertiary Structure
3.3. Systematic Position of the Family Thaumastellidae
3.4. Classification of the Family Cydnidae versus Morpho-Molecular Apomorphies in the LVR L
4. Materials and Methods
4.1. Selection of Taxa
4.2. DNA Extraction
4.3. PCR Amplification, Purification and Sequencing
4.4. Reconstruction of 18S rRNA Secondary Structure Models
4.5. Prediction of LVR L Secondary Structure
4.6. Prediction of 18S rRNA Tertiary Structures
4.7. Prediction of LVR L Tertiary Structure
4.8. Concept of the Morpho-Molecular Structures Potentially Serving as Derived Characters
5. Conclusions
- Comparisons of the predicted tertiary structures of the LVR L of the 18S rRNA in species representing all presently recognised and accepted subfamilies and tribes within the family Cydnidae revealed inconsistencies in their classifications.
- The present comparative analyses of the LVR L of the 18S rRNA secondary and tertiary structures support earlier findings that irrespective of its internal classification, Thaumastellidae is not a member of the family Cydnidae and should be recognised as a distinct Pentatomoidea family.
- The analysis did not identify one synapomorphy that was present across all presently acknowledged subfamilies of Cydnidae. This absence was observable in the primary, secondary, and tertiary structures of the studied 18S ribosomal RNA region. Furthermore, no autapomorphy was detected in the examined region to differentiate Cydnidae as a monophyletic group within the ‘cydnoid’ complex. These findings are consistent with previous hypotheses suggesting that the origin of this family is non-monophyletic.
- The predicted secondary and tertiary structures of the LVR L of the 18S rRNA of the family Parastrachiidae and the subfamily Sehirinae (Cydnidae) confirm their close relationship, highlighted by the several morpho-molecular synapomorphies shared between their LVR L subregions.
- Two notable groups of species in the subfamily Sehirinae were found to be unrelated. These groups challenged the classification currently in use for this subfamily. One group displayed ochetostethan facies of spermatheca, which significantly differed in regard to their morpho-molecular data from species representing the sehiran facies of spermatheca within the subfamily. These findings indicate that the subfamily may need to be divided into at least two tribes. However, further supportive analyses incorporating alternative mitochondrial and nuclear genes must be conducted to address this further.
- The subfamily Cephalocteinae displayed a clear correlation with the species of the tribe Geotomini across several morpho-molecular data. It does not possess any distinctive morpho-molecular autapomorphy and exhibits the same type and facies of spermatheca as representatives of the tribe. Therefore, the subfamily may be classified as part of the tribe Geotomini or as a distinct tribe within the subfamily Cydninae.
- The relationship between two groups from the tribe Cydnini (C. aterrimus and C. piceus) suggests they are distantly related. C. aterrimus was found to be closely related to the tribe Geotomini, while C. piceus (the consensus species of the remaining Cydnini) appeared to be the closest relative to the subfamily Amnestinae. These findings imply that these two groups are not phylogenetically related. Therefore, it is likely inappropriate to categorise them as belonging to the same tribe.
- The subfamily status of Amaurocorinae was confirmed based on the morpho-molecular autapomorphy in the LA subregion of the LVR L, despite its relation to species of the subfamilies Cydninae and Sehirinae, as well as those of the families Thyreocoridae and Parastrachiidae in terms of morpho-molecular LVR L characters.
- The Amnestinae is the most distinct subfamily in the Cydnidae family due to its numerous morpho-molecular autapomorphies. Additionally, the group’s species do not show morpho-molecular synapomorphies with other subfamilies within the Cydnidae or closely related families, such as Thyreocoridae and Parastrachiidae.
- The subfamily Garsauriinae shared only one morpho-molecular synapomorphy with the other studied taxa, specifically with the family Thaumastellidae. Additionally, no synapomorphies were found in this subfamily with any other taxa of Cydnidae, Thyreocoridae, or Parastrachiidae. Therefore, the relationship of this subfamily to others within the family Cydnidae remains unclear.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Lis, J.A. Molecular Apomorphies in the Secondary and Tertiary Structures of Length-Variable Regions (LVRs) of 18S rRNA Shed Light on the Systematic Position of the Family Thaumastellidae (Hemiptera: Heteroptera: Pentatomoidea). Int. J. Mol. Sci. 2023, 24, 7758. [Google Scholar] [CrossRef]
- Lis, B.; Domagała, P.J.; Lis, J.A. Tribe Acalyptaini (Hemiptera: Tingidae: Tinginae) Revisited: Can Apomorphies in Secondary and Tertiary Structures of 18S rRNA Length-Variable Regions (LVRs) Support Tribe Validity? Insects 2023, 14, 600. [Google Scholar] [CrossRef]
- Song, N.; Li, H.; Cai, W.; Yan, F.; Wang, J.; Song, F. Phylogenetic relationships of Hemiptera inferred from mitochondrial and nuclear genes. Mitochondrial DNA 2016, 27, 4380–4389. [Google Scholar] [CrossRef] [PubMed]
- Lis, J.A.; Ziaja, D.; Lis, B.; Gradowska, P.A. Non-monophyly of the “cydnoid” complex within Pentatomoidea (Hemiptera: Heteroptera) revealed by Bayesian phylogenetic analysis of nuclear rDNA sequences. Arthropod. Syst. Phylogeny 2017, 75, 481–496. [Google Scholar] [CrossRef]
- Xie, Q.; Bu, W.; Zheng, L. The Bayesian phylogenetic analysis of the 18S rRNA sequences from the main lineages of Trichophora (Insecta: Heteroptera: Pentatomomorpha). Mol. Phylogenet. Evol. 2005, 34, 448–451. [Google Scholar] [CrossRef] [PubMed]
- Li, H.M.; Deng, R.Q.; Wang, J.W.; Chen, Z.Y.; Jia, F.L.; Wang, X.Z. A preliminary phylogeny of the Pentatomomorpha (Hemiptera: Heteroptera) based on nuclear 18S rDNA and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2005, 37, 313–326. [Google Scholar] [CrossRef]
- Li, H.M.; Deng, R.Q.; Wang, X.Z. Phylogenetic relationships of the Pentatomomorpha (Hemiptera: Heteroptera) inferred from nuclear 18S rDNA sequences. Zool. Res. 2006, 27, 307–316. [Google Scholar]
- Li, M.; Tian, Y.; Zhao, Y.; Bu, W. Higher Level Phylogeny and the First Divergence Time Estimation of Heteroptera (Insecta: Hemiptera) Based on Multiple Genes. PLoS ONE 2012, 7, e32152. [Google Scholar] [CrossRef]
- Weirauch, C.; Schuh, R.T.; Cassis, G.; Wheeler, W.C. Revisiting habitat and lifestyle transitions in Heteroptera (Insecta: Hemiptera): Insights from a combined morphological and molecular phylogeny. Cladistics 2019, 35, 67–105. [Google Scholar] [CrossRef]
- Roca-Cusachs, M.; Schwertner, C.F.; Kim, J.; Eger, J.; Grazia, J.; Jung, S. Opening Pandora’s box: Molecular phylogeny of the stink bugs (Hemiptera: Heteroptera: Pentatomidae) reveals great incongruences in the current classification. Syst. Entomol. 2022, 41, 36–51. [Google Scholar] [CrossRef]
- Grazia, J.; Schuh, R.T.; Wheeler, W.C. Phylogenetic relationships of family groups in Pentatomoidea based on morphology and DNA sequences (Insecta: Heteroptera). Cladistics 2008, 24, 932–976. [Google Scholar] [CrossRef]
- Bianchi, F.M.; Barão, K.R.; Grassi, A.; Ferrari, A. A milestone for Pentatomoidea: Grazia et al. 2008—What do we know and where can we go? Zootaxa 2021, 4958, 406–429. [Google Scholar] [CrossRef]
- Wang, Y.H.; Cui, Y.; Rédei, D.; Baňař, P.; Xie, Q.; Štys, P.; Damgaard, J.; Chen, P.P.; Yi, W.B.; Wang, Y.; et al. Phylogenetic divergences of the true bugs (Insecta: Hemiptera:Heteroptera), with emphasis on the aquatic lineages: The last piece of the aquatic insect jigsaw originated in the Late Permian/EarlyTriassic. Cladistics 2016, 32, 390–405. [Google Scholar] [CrossRef]
- Ouvrard, D.; Campbell, B.C.; Bourgoin, T.; Chan, K.L. 18S rRNA Secondary Structure and Phylogenetic Position of Peloridiidae (Insecta, Hemiptera). Mol. Phylogenet. Evol. 2000, 16, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, Y.; Rédei, D.; Xie, Q.; Bu, W. Secondary structure models of 18S and 28S rRNAs of the true bugs based on complete rDNA sequences of Eurydema maracandica Oshanin, 1871 (Heteroptera, Pentatomidae). ZooKeys 2013, 319, 363–377. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Yu, S.S.; Wang, Y.H.; Wu, Y.H.; Li, X.R.; Men, X.Y.; Zhang, Y.W.; Rédei, D.; Xie, Q.; Bu, W.J. The evolutionary position of Lestoniidae revealed by molecular autapomorphies in the secondary structure of rRNA besides phylogenetic reconstruction (Insecta: Hemiptera: Heteroptera). Zool. J. Linn. Soc. 2016, 177, 750–763. [Google Scholar] [CrossRef]
- Xie, Q.; Tian, Y.; Zheng, L.; Bu, W. 18S rRNA hyper-elongation and the phylogeny of Euhemiptera (Insecta: Hemiptera). Mol. Phylogenet. Evol. 2008, 47, 463–471. [Google Scholar] [CrossRef]
- Schuh, R.T.; Weirauch, C. True Bugs of the World (Hemiptera: Heteroptera): Classification and Natural History, 2nd ed.; Monograph Series 8; Siri Scientific Press: Rochdale, UK, 2020; 767p. [Google Scholar]
- Pluot-Sigwalt, D.; Lis, J.A. Morphology of the spermatheca in the Cydnidae (Hemiptera: Heteroptera): Bearing of its diversity on classification and phylogeny. Eur. J. Entomol. 2008, 105, 279–312. [Google Scholar] [CrossRef]
- Lis, J.A. Coxal combs in the Cydnidae sensu lato and three other related “cydnoid” families—Parastrachiidae, Thaumastellidae, Thyreocoridae (Hemiptera: Heteroptera): Functional, taxonomic, and phylogenetic significance. Zootaxa 2010, 2476, 53–64. [Google Scholar] [CrossRef]
- Rider, D.A.; Schwertner, C.F.; Vilímová, J.; Rédei, D.; Kment, P.; Thomas, D.B. Higher systematics of the Pentatomoidea. In Invasive Stink Bugs and Related Species (Pentatomoidea): Biology, Higher Systematics, Semiochemistry, and Management; McPherson, J.E., Ed.; CRC Press: Boca Raton, FL, USA, 2018; pp. 25–201. [Google Scholar] [CrossRef]
- Schaefer, C.W.; Dolling, W.R.; Tachikawa, S. The shieldbug genus Parastrachia and its position within the Pentatomoidea (Insecta: Hemiptera). Zool. J. Linn. Soc. 1988, 93, 283–311. [Google Scholar] [CrossRef]
- Sweet, M.H.; Schaefer, C.W. Parastrachiinae (Hemiptera: Cydnidae) raised to family level. Ann. Entomol. Soc. Am. 2002, 95, 442–448. [Google Scholar] [CrossRef]
- Lis, J.A.; Heyna, J. Metathoracic wing venation in Cydnidae (Hemiptera: Heteroptera) and its bearing on the classification of the family. Annal. Zool. 2001, 51, 429–465. [Google Scholar]
- Lis, J.A.; Schaefer, C.W. Tibial combs in the Cydnidae (Hemiptera: Heteroptera) and their functional, taxonomic and phylogenetic significance. J. Zool. Syst. Evol. Res. 2005, 43, 277–283. [Google Scholar] [CrossRef]
- Lis, J.A. The mesothoracic wing and its phylogenetic significance in Cydnidae (Hemiptera: Heteroptera: Pentatomoidea). Pol. J. Entomol. 2002, 71, 43–71. [Google Scholar]
- Lis, J.A. Pretarsal structures in the family Parastrachiidae (Hemiptera: Heteroptera: Pentatomoidea). Zootaxa 2010, 2693, 60–62. [Google Scholar] [CrossRef]
- Tachikawa, S.; Schaefer, C.W. The biology of Parastrachia japonensis (Hemiptera: Pentatomoidea: ?-idae). Ann. Entomol. Soc. Am. 1985, 78, 387–397. [Google Scholar] [CrossRef]
- Henry, T.J. Biodiversity of Heteroptera. In Insect Biodiversity; Footit, R.G., Adler, P.H., Eds.; Science and Society, Wiley-Blackwell: Chichester, UK; Hoboken, UK, 2009; pp. 233–263. [Google Scholar]
- Zhu, G.; Liu, G.; Bu, W.; Lis, J.A. Geographic distribution and niche divergence of two stinkbugs, Parastrachia japonensis and Parastrachia nagaensis. J. Insect Sci. 2013, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Matesco, V.C.; Bianchi, F.M.; Campos, L.A.; Grazia, J. Egg ultrastructure of two species of Galgupha Amyot & Serville, with a discussion of the eggs and oviposition patterns of thyreocorid and allied groups (Hemiptera: Heteroptera: Pentatomoidea: Thyreocoridae). Zootaxa 2012, 3247, 43–51. [Google Scholar] [CrossRef]
- Schwertner, C.F.; Nardi, C. Chapter 21. Burrower Bugs (Cydnidae). In True Bugs (Heteroptera) of the Neotropics; Panizzi, A.R., Grazia, J., Eds.; Springer: Dordrecht, The Netherland; Berlin/Heidelberg, Germany; New York, NY, USA; London, UK, 2015; pp. 639–680. [Google Scholar]
- Ye, F.; Kment, P.; Rédei, D.; Luo, J.Y.; Wang, Y.H.; Kuechler, S.M.; Zhang, W.W.; Chen, P.P.; Wu, H.Y.; Wu, Y.Z.; et al. Diversification of the phytophagous lineages of true bugs (Insecta: Hemiptera: Heteroptera) shortly after that of the flowering plants. Cladistics 2022, 38, 403–428. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, 232–235. [Google Scholar] [CrossRef]
- Lukasiak, P.; Antczak, M.; Ratajczak, T.; Szachniuk, M.; Popenda, M.; Adamiak, R.W.; Blazewicz, J. RNAssess–a web server for quality assessment of RNA 3D structures. Nucleic Acids Res. 2015, 43, W502–W506. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L.; DeLano, W. PyMOL, Version 2.4.0; Schrödinger, LLC.: New York, NY, USA, 2020. Available online: http://www.pymol.org/pymol(accessed on 14 August 2023).
- Wagner, E. Untersuchungen über den taxonomischen Wert des Baues der Genitalien bei den Cydnidae (Hem. Het.). Acta Entomol. Musei Natl. Pragae 1963, 35, 73–115. [Google Scholar]
- Lis, J.A.; Heyna, J. Metathoracic wing stidulitrum of the Cydnidae (Hemiptera: Heteroptera). Pol. J. Entomol. 2001, 70, 221–245. [Google Scholar]
- Hua, J.; Li, M.; Dong, P.; Cui, Y.; Xie, Q.; Bu, W. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera). BMC Genom. 2008, 9, 610. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.H.; Duwal, R.K.; Lee, S.W. COI barcoding of true bugs (Insecta, Heteroptera). Mol. Ecol. Resour. 2011, 11, 266–270. [Google Scholar] [CrossRef]
- Tian, X.; Xie, Q.; Li, M.; Gao, C.; Cui, Y.; Xi, L.; Bu, W. Phylogeny of pentatomomorphan bugs (Hemiptera Heteroptera: Pentatomomorpha) based on six Hox gene fragments. Zootaxa 2011, 2888, 57–68. [Google Scholar] [CrossRef]
- Hosokawa, T.; Kikuchi, Y.; Nikoh, N.; Fukatsu, T. Polyphyly of Gut Symbionts in Stinkbugs of the Family Cydnidae. Appl. Environ. Microbiol. 2012, 78, 4758. [Google Scholar] [CrossRef]
- Song, N.; Liang, A.P.; Bu, C.P. A Molecular Phylogeny of Hemiptera Inferred from Mitochondrial Genome Sequences. PLoS ONE 2012, 7, e48778. [Google Scholar] [CrossRef]
- Raupach, M.J.; Hendrich, L.; Küchler, S.M.; Deister, F.; Morinière, J.; Gossner, M.M. Building-Up of a DNA Barcode Library for True Bugs (Insecta: Hemiptera: Heteroptera) of Germany Reveals Taxonomic Uncertainties and Surprises. PLoS ONE 2014, 9, e106940. [Google Scholar] [CrossRef]
- Yuan, M.L.; Zhang, Q.L.; Guo, Z.L.; Wang, J.; Shen, Y.Y. Comparative mitogenomic analysis of the superfamily Pentatomoidea (Insecta: Hemiptera: Heteroptera) and phylogenetic implications. BMC Genom. 2015, 16, 460. [Google Scholar] [CrossRef]
- Yuan, M.L.; Zhang, Q.L.; Guo, Z.L.; Wang, J.; Shen, Y.Y. The Complete Mitochondrial Genome of Corizus tetraspilus (Hemiptera: Rhopalidae) and Phylogenetic Analysis of Pentatomomorpha. PLoS ONE 2015, 10, e0129003. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Rédei, D.; Eger, J., Jr.; Wang, Y.H.; Wu, H.Y.; Carapezza, A.; Kment, P.; Cai, B.; Sun, X.Y.; Guo, P.L.; et al. Phylogeny and the colourful history of jewel bugs (Insecta: Hemiptera: Scutelleridae). Cladistics 2018, 34, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wu, Y.; Liu, Y.; Zhao, P.; Chen, Z.; Song, F.; Li, H.; Cai, W. Comparative Mitogenomics and Phylogenetic Analyses of Pentatomoidea (Hemiptera: Heteroptera). Genes 2021, 12, 1306. [Google Scholar] [CrossRef] [PubMed]
- Giribet, G.; Carranza, S.; Bagui, J.; Riutort, M.; Ribera, C. First Molecular Evidence for the Existence of a Tardigrada + Arthropoda Clade. Mol. Biol. Evol. 1996, 13, 76–84. [Google Scholar] [CrossRef]
- Whiting, M.F.; Carpernter, J.C.; Wheeler, Q.D.; Wheeler, W.C. The Strepsiptera problem: Phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst. Biol. 1997, 46, 1–68. [Google Scholar] [CrossRef] [PubMed]
- Neefs, J.M.; van der Peer, Y.; De Rijk, P.; Chapelle, S.; De Wachter, R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993, 21, 3025–3049. [Google Scholar] [CrossRef] [PubMed]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Huang, Y.; Xiao, Y. 3dRNA v2.0: An Updated Web Server for RNA 3D Structure Prediction. Int. J. Mol. Sci. 2019, 20, 4116. [Google Scholar] [CrossRef] [PubMed]
- Popenda, M.; Szachniuk, M.; Antczak, M.; Purzycka, K.J.; Lukasiak, P.; Bartol, N.; Blazewicz, J.; Adamiak, R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012, 40, e112. [Google Scholar] [CrossRef] [PubMed]
- Biesiada, M.; Purzycka, K.J.; Szachniuk, M.; Blazewicz, J.; Adamiak, R.W. Automated RNA 3D structure prediction with RNAComposer. In RNA Structure Determination: Methods in Molecular Biology; Turner, D., Mathews, D., Eds.; Humana Press: New York, NY, USA, 2016; Volume 1490, pp. 199–215. [Google Scholar] [CrossRef]
- Xie, Q.; Tian, X.; Qin, Y.; Bu, W. Phylogenetic comparison of local length plasticity of the small subunit of nuclear rDNAs among all Hexapoda orders and the impact of hyper-length-variation on alignment. Mol. Phylogenet. Evol. 2009, 50, 310–316. [Google Scholar] [CrossRef]
- Hajdin, C.E.; Ding, F.; Dokholyan, N.V.; Weeks, K.M. On the significance of an RNA tertiary structure prediction. RNA 2010, 16, 1340–1349. [Google Scholar] [CrossRef]
- Parisien, M.; Cruz, J.A.; Westhof, E.; Major, F. New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA 2009, 15, 1875–1885. Available online: http://www.rnajournal.org/cgi/doi/10.1261/rna.1700409 (accessed on 5 May 2023). [CrossRef]
- Westhof, E.; Masquida, B.; Jossinet, F. Predicting and modeling RNA architecture. Cold Spring Harb. Perspect. Biol. 2011, 3, a003632. [Google Scholar] [CrossRef]
- Rother, M.; Rother, K.; Puton, T.; Bujnicki, J.M. ModeRNA: A tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011, 39, 4007–4022. [Google Scholar] [CrossRef]


| Family | Subfamily | Spermathecal Types/Facies (According to [19]) | Species | Number of Nucleotides | ||
|---|---|---|---|---|---|---|
| V4 | LVR L | L2 | ||||
| Cydnidae | Amaurocorinae | Amaurocorinae type | Amaurocoris curtus (Brullé, 1838) * | 316 | 73 | 6 |
| Amnestinae | Amnestinae type | Amnestus zacki Mayorga & Cervantes, 2009 * | 324 | 73 | 6 | |
| Garsauriinae | Garsauriinae type | Garsauria aradoides Walker, 1868 * | 317 | 74 | 6 | |
| Cephalocteinae | Cydnoid type Geotoman facies | Stibaropus indonesicus J.A. Lis, 1991 * | 317 | 74 | 4 | |
| Cydninae | Cydnoid type Geotoman facies | Fromundus pygmaeus (Dallas, 1851) ** | 317 | 74 | 4 | |
| Macroscytus badius (Walker, 1867) ** | 317 | 74 | 4 | |||
| Cyrtomenus emarginatus Stål, 1862 ** | 317 | 74 | 4 | |||
| Rhytidoporus indentatus Uhler, 1877 ** | 317 | 74 | 4 | |||
| Cydnoid type Adrisan facies | Adrisa romani J.A. Lis, 1994 * | 316 | 73 | 4 | ||
| Cydnoid type Scoparipan facies | Pseudoscoparipes fraterculus J.A. Lis, 1994 * | 316 | 73 | 4 | ||
| Cydnoid type Cydnan facies | Chilocoris piceus Signoret, 1884 ** | 325 | 81 | 7 | ||
| Cydnus aterrimus (Forster, 1771) ** | 316 | 73 | 4 | |||
| Sehirinae | Cydnoid type Sehiran facies | Adomerus biguttatus (Linnaeus, 1758) * | 316 | 73 | 6 | |
| Cydnoid type Ochetostethan facies | Ochetostethomorpha secunda J.A. Lis & B. Lis, 2014 * | 316 | 73 | 4 | ||
| Parastrachiidae | – | – | Parastrachia japonensis (Scott, 1880) * | 316 | 73 | 6 |
| Thyreocoridae | Thyreocorinae | – | Thyreocoris scarabaeoides (Linnaeus, 1758) * | 316 | 74 | 6 |
| Thaumastellidae (outgroup) | – | – | Thaumastella elizabethae Jacobs, 1989 * | 318 | 75 | 3 |
| Region or Subregion | Number of Nucleotides | Family | Subfamily and Tribe |
|---|---|---|---|
| V4 | 315 | Thaumastellidae | – |
| 316 | Cydnidae | Amaurocorinae | |
| Cydninae: Geotomini s. lato [part] | |||
| Sehirinae: Sehirini s. lato [part] | |||
| Parastrachiidae | – | ||
| Thyreocoridae | Corimelaeninae | ||
| Thyreocorinae | |||
| 317 | Cydnidae | Garsauriinae | |
| Cephalocteinae | |||
| Cydninae: Geotomini s. lato [part] | |||
| 318 | Thaumastellidae | – | |
| 324 | Cydnidae | Amnestinae | |
| 325 | Cydnidae | Cydninae: Cydnini [part] | |
| LVR L | 72 | Thaumastellidae | – |
| 73 | Cydnidae | Amaurocorinae | |
| Amnestinae | |||
| Cydninae: Geotomini s. lato [part] | |||
| Cydninae: Cydnini [part] | |||
| Sehirinae: Sehirini s. lato | |||
| Parastrachiidae | – | ||
| 74 | Cydnidae | Garsauriinae | |
| Cephalocteinae | |||
| Cydninae: Geotomini s. lato [part] | |||
| Thyreocoridae | Corimelaeninae | ||
| Thyreocorinae | |||
| 75 | Thaumastellidae | – | |
| 81 | Cydnidae | Cydninae: Cydnini [part] | |
| L2 | 3 | Thaumastellidae | – |
| 4 | Cydnidae | Cephalocteinae | |
| Cydninae: Geotomini s. lato | |||
| Cydninae: Cydnini [part] | |||
| Sehirinae: Sehirini s. lato [part] | |||
| 6 | Cydnidae | Amaurocorinae | |
| Garsauriinae | |||
| Amnestinae | |||
| Sehirinae: Sehirini s. lato [part] | |||
| Parastrachiidae | – | ||
| Thyreocoridae | Corimelaeninae | ||
| Thyreocorinae | |||
| 7 | Cydnidae | Cydninae: Cydnini [part] |
| Taxon Group | Species | Total Length | Number of Nucleotides of the LVR L Subregions | |||||
|---|---|---|---|---|---|---|---|---|
| L2 | LA (A1 + A2) | LB (B1 + B2) | LC (C1 + C2) | LD (D1 + D2) | LE (E1 + E2) | |||
| Thaumastellidae (outgroup) | Thaumastella elizabethae * | 75 | 3 | 17 (9 + 8) | 11 (6 + 5) | 16 (8 + 8) | 8 (5 + 3) | 20 (10 + 10) |
| Cydnidae: Cydninae | Cyrtomenus emarginatus ** | 74 | 4 | 15 (8 + 7) | 18 (10 + 8) | 12 (6 + 6) | 9 (6 + 3) | 16 (8 + 8) |
| Cydnidae: Cydninae | Rhytidoporus indentatus ** | 74 | 4 | 15 (8 + 7) | 18 (10 + 8) | 10 (5 + 5) | 11 (7 + 4) | 16 (8 + 8) |
| Cydnidae: Cephalocteinae | Stibaropus indonesicus * | 74 | 4 | 14 (7 + 7) | 16 (9 + 7) | 15 (8 + 7) | 9 (6 + 3) | 16 (8 + 8) |
| Cydnidae: Cydninae | Fromundus pygmaeus ** | 74 | 4 | 14 (7 + 7) | 16 (9 + 7) | 15 (8 + 7) | 9 (6 + 3) | 16 (8 + 8) |
| Cydnidae: Cydninae | Macroscytus badius ** | 74 | 4 | 14 (7 + 7) | 19 (11 + 8) | 12 (6 + 6) | 9 (6 + 3) | 16 (8 + 8) |
| Cydnidae: Cydninae | Adrisa romani * | 73 | 4 | 14 (7 + 7) | 16 (9 + 7) | 14 (7 + 7) | 9 (6 + 3) | 16 (8 + 8) |
| Cydnidae: Cydninae | Pseudoscoparipes fraterculus * | 73 | 4 | 14 (7 + 7) | 16 (9 + 7) | 14 (7 + 7) | 9 (6 + 3) | 16 (8 + 8) |
| Cydnidae: Cydninae | Cydnus aterrimus ** | 73 | 4 | 14 (7 + 7) | 16 (9 + 7) | 10 (5 + 5) | 10 (7 + 3) | 19 (9 + 10) |
| Cydnidae: Sehirinae | Ochetostethomorpha secunda * | 73 | 4 | 12 (6 + 6) | 18 (10 + 8) | 14 (7 + 7) | 9 (6 + 3) | 16 (8 + 8) |
| Thyreocoridae | Thyreocoris scarabaeoides * | 74 | 6 | 14 (7 + 7) | 17 (10 + 7) | 14 (7 + 7) | 9 (6 + 3) | 14 (7 + 7) |
| Cydnidae: Amaurocorinae | Amaurocoris curtus * | 73 | 6 | 14 (7 + 7) | 16 (9 + 7) | 12 (6 + 6) | 11 (7 + 4) | 14 (7 + 7) |
| Cydnidae: Sehirinae | Adomerus biguttatus * | 73 | 6 | 12 (6 + 6) | 18 (10 + 8) | 14 (7 + 7) | 9 (6 + 3) | 14 (7 + 7) |
| Parastrachiidae | Parastrachia japonensis * | 73 | 6 | 12 (6 + 6) | 18 (10 + 8) | 14 (7 + 7) | 9 (6 + 3) | 14 (7 + 7) |
| Cydnidae: Garsauriinae | Garsauria aradoides * | 74 | 6 | 17 (9 + 8) | 11 (6 + 5) | 15 (8 + 7) | 10 (7 + 3) | 15 (7 + 8) |
| Cydnidae: Amnestinae | Amnestus zacki * | 73 | 6 | 11 (10 + 1) | 13 (7 + 6) | 12 (5 + 7) | 10 (5 + 5) | 21 (11 + 10) |
| Cydnidae: Cydninae | Chilocoris piceus ** | 81 | 7 | 14 (7 + 7) | 16 (9 + 7) | 15 (8 + 7) | 11 (7 + 4) | 18 (8 + 10) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lis, J.A.; Domagała, P.J. Inconsistencies in the Classification of the Family Cydnidae (Hemiptera: Heteroptera: Pentatomoidea) Revealed by Molecular Apomorphies in the Secondary and Tertiary Structures of 18S rRNA Length-Variable Region L (LVR L). Int. J. Mol. Sci. 2024, 25, 939. https://doi.org/10.3390/ijms25020939
Lis JA, Domagała PJ. Inconsistencies in the Classification of the Family Cydnidae (Hemiptera: Heteroptera: Pentatomoidea) Revealed by Molecular Apomorphies in the Secondary and Tertiary Structures of 18S rRNA Length-Variable Region L (LVR L). International Journal of Molecular Sciences. 2024; 25(2):939. https://doi.org/10.3390/ijms25020939
Chicago/Turabian StyleLis, Jerzy A., and Paweł J. Domagała. 2024. "Inconsistencies in the Classification of the Family Cydnidae (Hemiptera: Heteroptera: Pentatomoidea) Revealed by Molecular Apomorphies in the Secondary and Tertiary Structures of 18S rRNA Length-Variable Region L (LVR L)" International Journal of Molecular Sciences 25, no. 2: 939. https://doi.org/10.3390/ijms25020939
APA StyleLis, J. A., & Domagała, P. J. (2024). Inconsistencies in the Classification of the Family Cydnidae (Hemiptera: Heteroptera: Pentatomoidea) Revealed by Molecular Apomorphies in the Secondary and Tertiary Structures of 18S rRNA Length-Variable Region L (LVR L). International Journal of Molecular Sciences, 25(2), 939. https://doi.org/10.3390/ijms25020939







