Understanding of Plant Salt Tolerance Mechanisms and Application to Molecular Breeding
Abstract
:1. Introduction
2. Salt Responses under Different Tissues and Organs of Plants
2.1. Perception and Response of Salt Stress by Roots
2.2. Perception and Response of Salt Stress by Stems
2.3. Perception and Response of Salt Stress by Leaves
2.4. Perception and Response of Salt Stress by Flowers
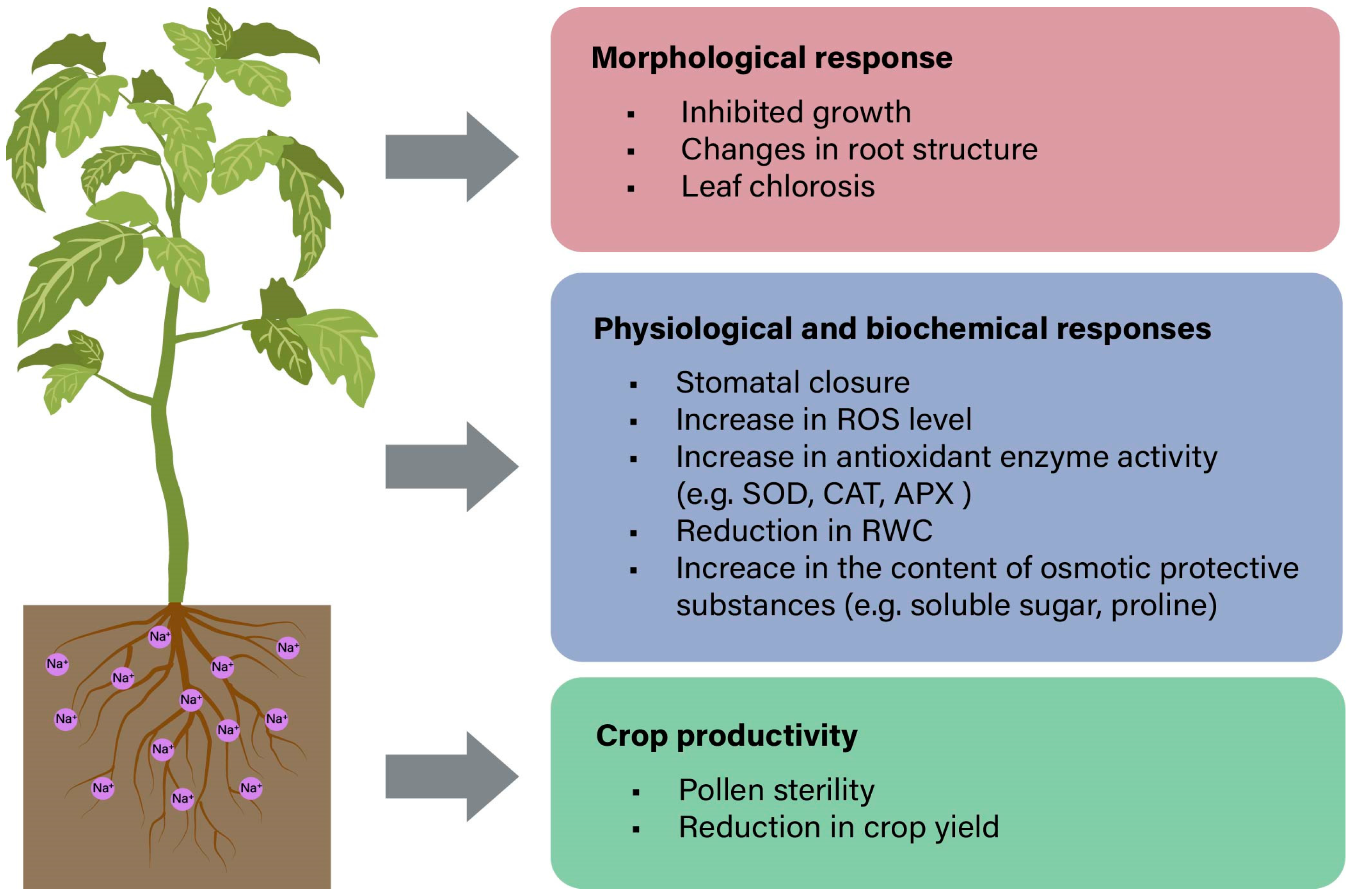
3. Plant Physiological Response to Salt Stress
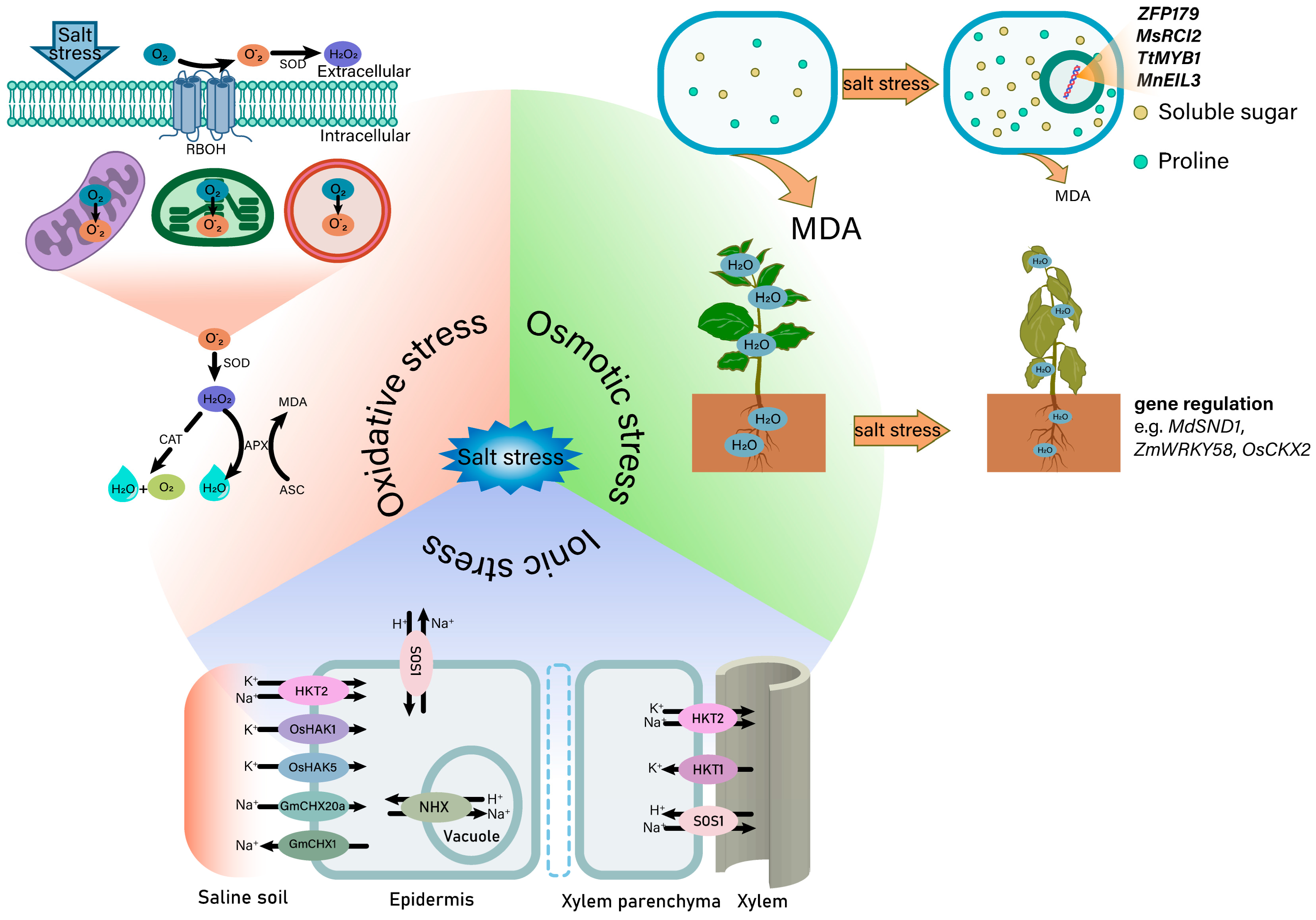
3.1. Salt-Induced Osmotic Stress
3.2. Salt-Induced Ionic Stress
3.3. Salt-Induced Oxidative Stress
4. Multiple Signaling Pathways Involved in Response to Salt Stress
4.1. Mechanisms of Calcium Influx and Signaling Pathway
4.2. Plant Salt Tolerance Mechanism Regulated by Hormonal Signaling
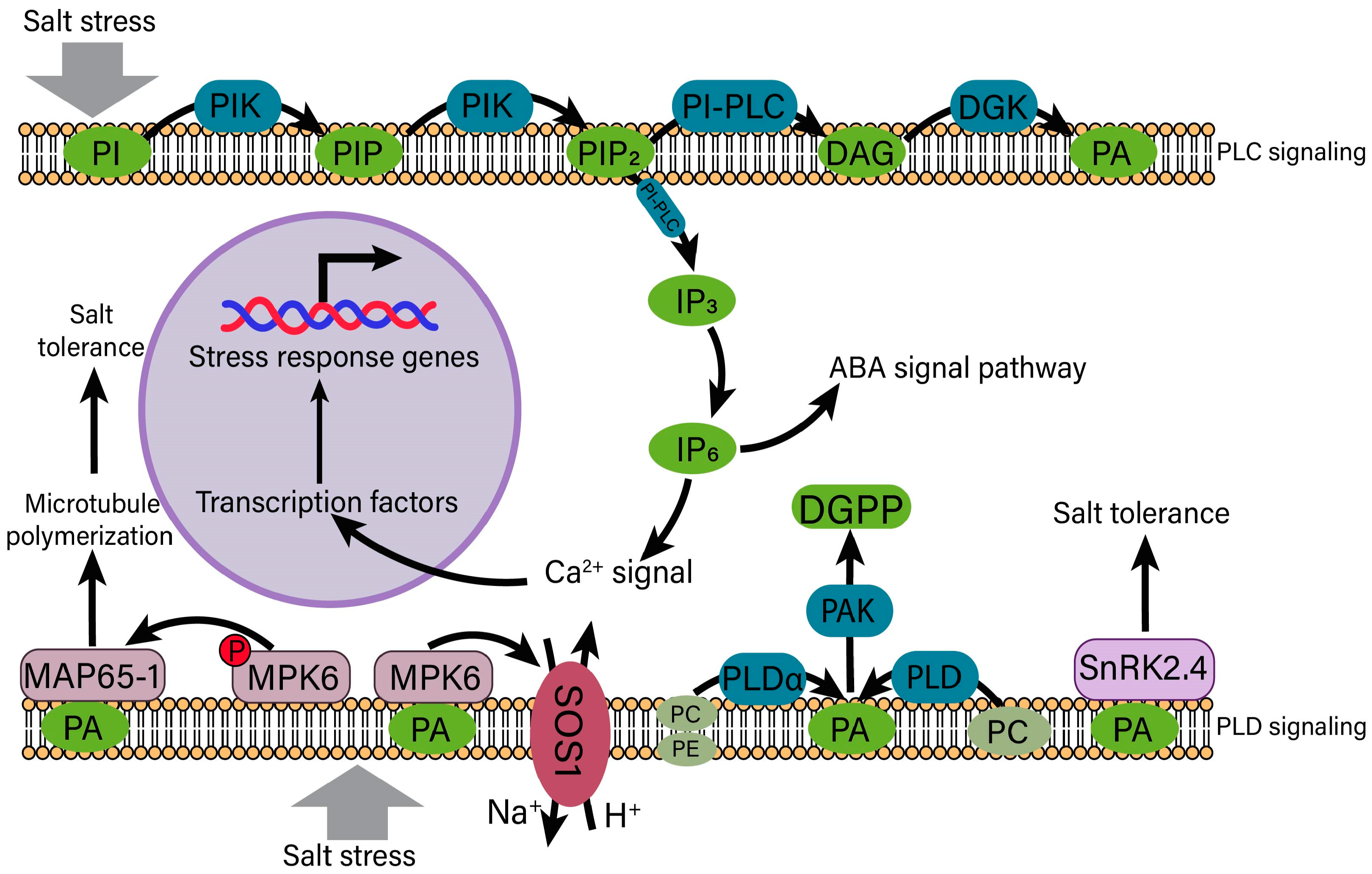
4.3. Phosphatidic Acid (PA) Signaling in the Regulation of Salt Stress
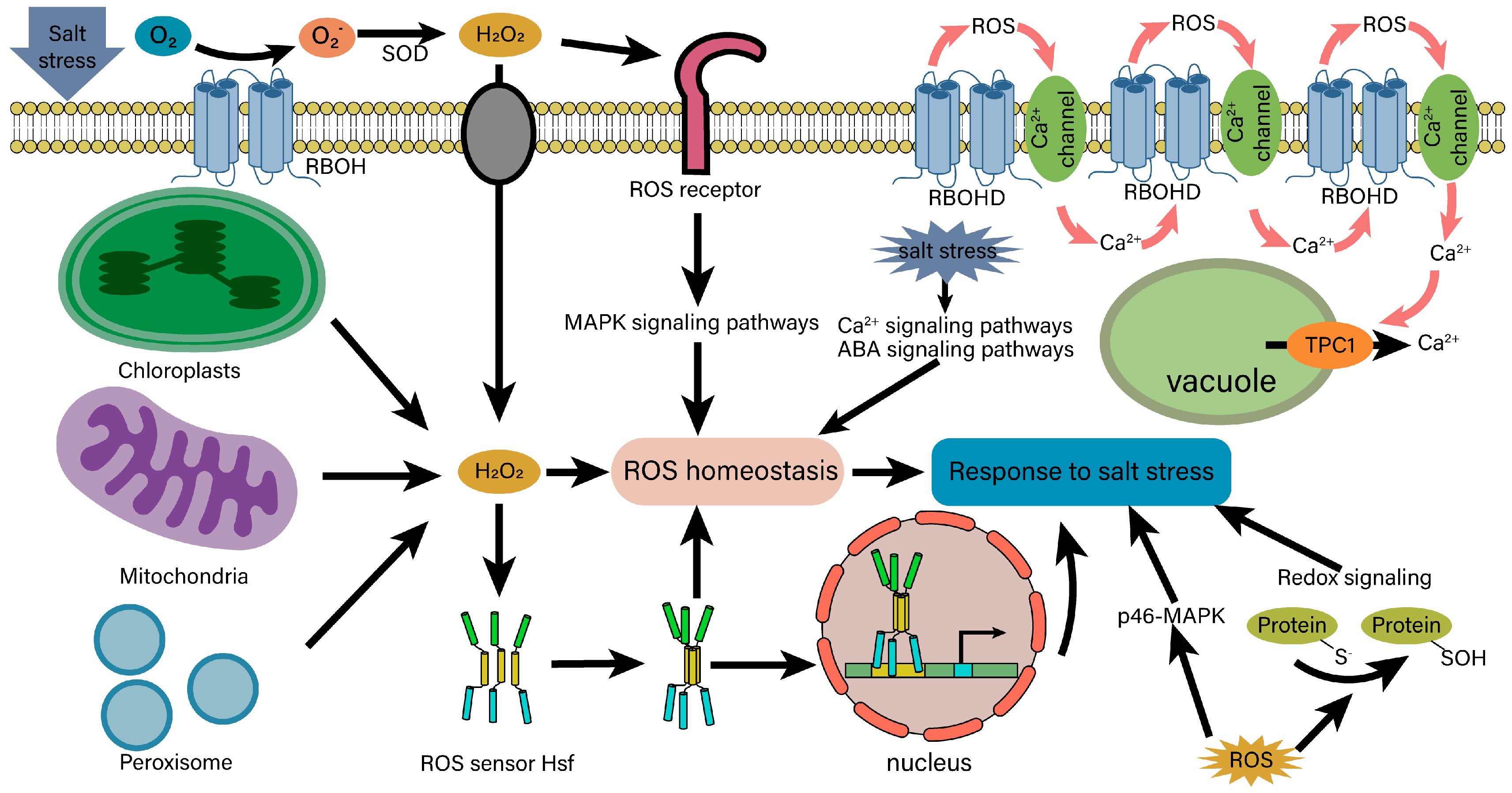
4.4. ROS Signaling and Homeostasis under Salt Stress
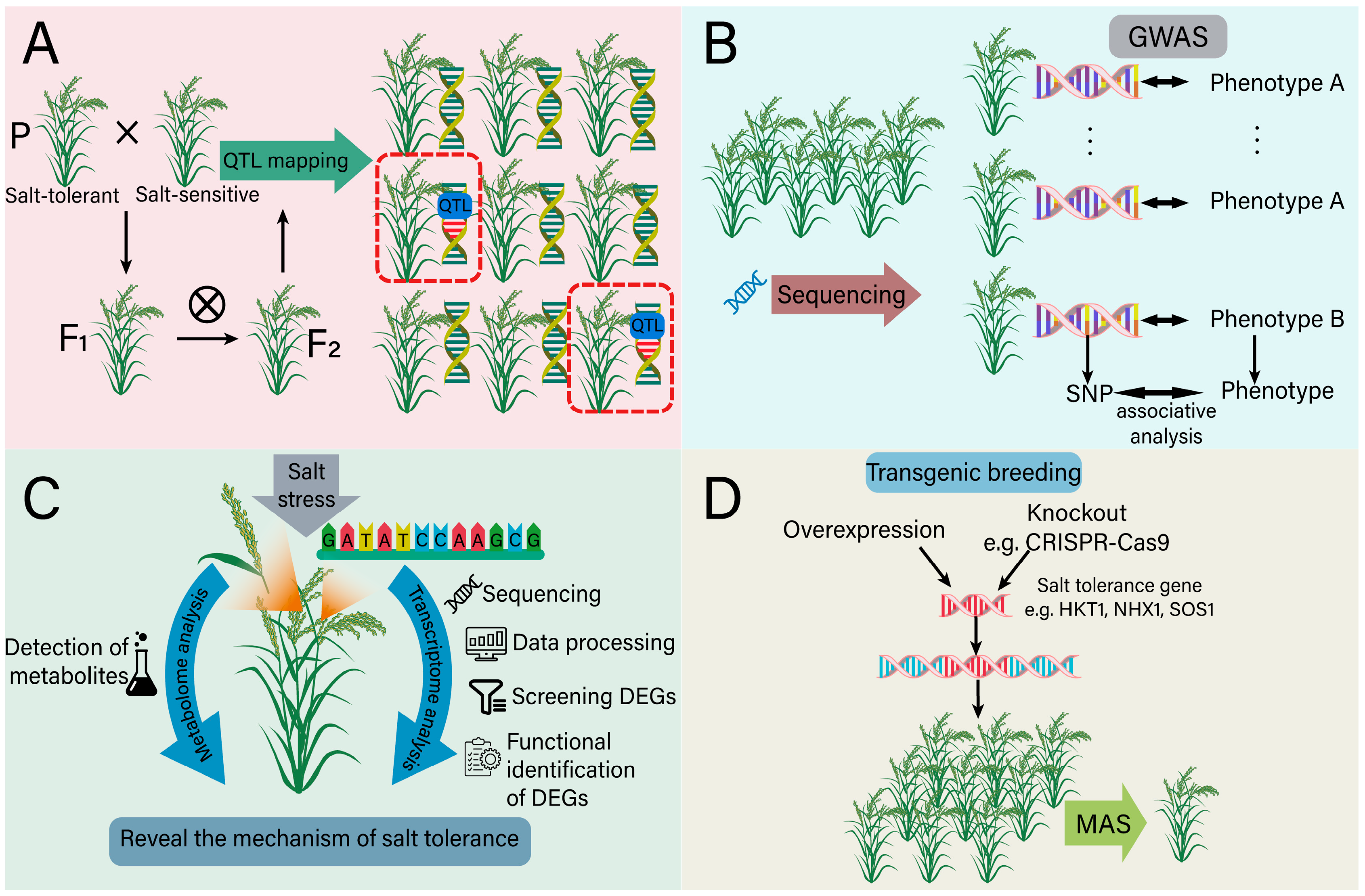
5. Molecular Breeding of Plants with Salt Tolerance
5.1. The Role of Genomics in Salt-Tolerant Breeding
5.2. The Role of Transcriptome in Salt-Tolerant Breeding
5.3. The Role of Metabolome in Salt-Tolerant Breeding
5.4. Transgenic Breeding
6. Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zhang, X.G.; Huang, B. Prediction of Soil Salinity with Soil-Reflected Spectra: A Comparison of Two Regression Methods. Sci. Rep. 2019, 9, 5067. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Meng, W.L.; Wang, Y.K.; Zhou, Y.M.; Wang, S.Y.; Qi, F.; Wang, N.N.; Ma, J. Comparative Analysis of Physiological, Hormonal and Transcriptomic Responses Reveal Mechanisms of Saline-Alkali Tolerance in Autotetraploid Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 16146. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, P.; Kumar, R. Soil Salinity: A Serious Environmental Issue and Plant Growth Promoting Bacteria as One of the Tools for Its Alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef]
- Dasgupta, S.; Hossain, M.M.; Huq, M.; Wheeler, D. Climate Change and Soil Salinity: The Case of Coastal Bangladesh. Ambio 2015, 44, 815–826. [Google Scholar] [CrossRef]
- Hailu, B.; Mehari, H. Impacts of Soil Salinity/Sodicity on Soil-Water Relations and Plant Growth in Dry Land Areas: A Review. J. Nat. Sci. Res. 2021, 12, 1–10. [Google Scholar] [CrossRef]
- Chourasia, K.N.; Lal, M.K.; Tiwari, R.K.; Dev, D.; Kardile, H.B.; Patil, V.U.; Kumar, A.; Vanishree, G.; Kumar, D.; Bhardwaj, V.; et al. Salinity Stress in Potato: Understanding Physiological, Biochemical and Molecular Responses. Life 2021, 11, 545. [Google Scholar] [CrossRef]
- Gong, Z.Z. Plant Abiotic Stress: New Insights into the Factors That Activate and Modulate Plant Responses. J. Integr. Plant Biol. 2021, 63, 429–430. [Google Scholar] [CrossRef]
- Pierzynski, G.M.; Vance, G.F.; Sims, J.T. Soils and Environmental Quality, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Mann, A.; Lata, C.; Kumar, N.; Kumar, A.; Kumar, A.; Sheoran, P. Halophytes as New Model Plant Species for Salt Tolerance Strategies. Front. Plant Sci. 2023, 14, 1137211. [Google Scholar] [CrossRef]
- Zhao, C.Z.; Zhang, H.; Song, C.P.; Zhu, J.K.; Shabala, S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Jez, J.M.; Lee, S.G.; Sherp, A.M. The next Green Movement: Plant Biology for the Environment and Sustainability. Science 2016, 353, 1241–1244. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, H.P. Plant Salt Response: Perception, Signaling, and Tolerance. Front. Plant Sci. 2023, 13, 1053699. [Google Scholar] [CrossRef]
- Hao, S.H.; Wang, Y.R.; Yan, Y.X.; Liu, Y.H.; Wang, J.Y.; Chen, S. A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance. Horticulturae 2021, 7, 132. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Guo, Y. Unraveling Salt Stress Signaling in Plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Zelm, E.; Zhang, Y.X.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Chen, H.F.; Zhang, Q.; Cai, H.M.; Xu, F.S. Ethylene Mediates Alkaline-Induced Rice Growth Inhibition by Negatively Regulating Plasma Membrane H+-ATPase Activity in Roots. Front. Plant Sci. 2017, 8, 1839. [Google Scholar] [CrossRef]
- Zhang, Q.D.; Gong, M.; Xu, X.; Li, H.H.; Deng, W. Roles of Auxin in the Growth, Development, and Stress Tolerance of Horticultural Plants. Cells 2022, 11, 2761. [Google Scholar] [CrossRef]
- Zou, Y.T.; Zhang, Y.X.; Testerink, C. Root Dynamic Growth Strategies in Response to Salinity. Plant Cell Environ. 2022, 45, 695–704. [Google Scholar] [CrossRef]
- Garrido-Vargas, F.; Godoy, T.; Tejos, R.; O’Brien, J.A. Overexpression of the Auxin Receptor AFB3 in Arabidopsis Results in Salt Stress Resistance and the Modulation of NAC4 and SZF1. Int. J. Mol. Sci. 2020, 21, 9528. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Koevoets, I.T.; Mol, S.; Hoefsloot, H.; Feron, R.; Tester, M.A.; Keurentjes, J.J.B.; Korte, A.; Haring, M.A.; Boer, G.J.; et al. Genetic Components of Root Architecture Remodeling in Response to Salt Stress. Plant Cell 2017, 29, 3198–3213. [Google Scholar] [CrossRef]
- Song, Y.S.; Li, S.M.; Sui, Y.; Zheng, H.X.; Han, G.L.; Sun, X.; Yang, W.J.; Wang, H.L.; Zhuang, K.Y.; Kong, F.Y.; et al. SbbHLH85, a bHLH Member, Modulates Resilience to Salt Stress by Regulating Root Hair Growth in Sorghum. Theor. Appl. Genet. 2022, 135, 201–216. [Google Scholar] [CrossRef]
- Fu, J.Y.; Zhu, C.Y.; Wang, C.; Liu, L.J.; Shen, Q.Q.; Xu, D.B.; Wang, Q. Maize Transcription Factor ZmEREB20 Enhanced Salt Tolerance in Transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 159, 257–267. [Google Scholar] [CrossRef]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of Shape During Stress: A Key Role for Auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef]
- Galvan-Ampudia, C.S.; Julkowska, M.M.; Darwish, E.; Gandullo, J.; Korver, R.A.; Brunoud, G.; Haring, M.A.; Munnik, T.; Vernoux, T.; Testerink, C. Halotropism Is a Response of Plant Roots to Avoid a Saline Environment. Curr. Biol. 2013, 23, 2044–2050. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Zhou, J.H.; Li, Y.X.; Quan, R.D.; Wang, J.; Huang, R.F.; Qin, H. Salt Stress Promotes Abscisic Acid Accumulation to Affect Cell Proliferation and Expansion of Primary Roots in Rice. Int. J. Mol. Sci. 2021, 22, 10892. [Google Scholar] [CrossRef]
- Lynch, J.P. Steep, Cheap and Deep: An Ideotype to Optimize Water and N Acquisition by Maize Root Systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef]
- Sun, Y.; Li, R.M.; Zhang, H.X.; Ye, J.J.; Li, C.Z. Proteomic Analysis of the Inflorescence Stem Mechanical Strength Difference in Herbaceous Peonies (Paeonia lactiflora Pall.). ACS Omega 2022, 7, 34801–34809. [Google Scholar] [CrossRef]
- Tatongjai, S.; Kraichak, E.; Kermanee, P. Comparative Anatomy and Salt Management of Sonneratia caseolaris (L.) Engl. (Lythraceae) Grown in Saltwater and Freshwater. PeerJ 2021, 9, e10962. [Google Scholar] [CrossRef]
- Qin, R.D.; Hu, Y.M.; Chen, H.; Du, Q.G.; Yang, J.; Li, W.X. MicroRNA408 Negatively Regulates Salt Tolerance by Affecting Secondary Cell Wall Development in Maize. Plant Physiol. 2023, 192, 1569–1583. [Google Scholar] [CrossRef]
- Dong, Z.X.; Alam, M.K.; Xie, M.L.; Yang, L.; Liu, J.; Helal, M.M.U.; Huang, J.Y.; Cheng, X.H.; Liu, Y.Y.; Tong, C.B.; et al. Mapping of a Major QTL Controlling Plant Height Using a High-Density Genetic Map and QTL-Seq Methods Based on Whole-Genome Resequencing in Brassica napus. G3 2021, 11, jkab118. [Google Scholar] [CrossRef]
- Scofield, S.; Murison, A.; Jones, A.; Fozard, J.; Aida, M.; Band, L.R.; Bennett, M.; Murray, J.A.H. Coordination of Meristem and Boundary Functions by Transcription Factors in the SHOOT MERISTEMLESS Regulatory Network. Development 2018, 145, dev157081. [Google Scholar] [CrossRef]
- Scofield, S.; Dewitte, W.; Nieuwland, J.; Murray, J.A.H. The Arabidopsis Homeobox Gene SHOOT MERISTEMLESS Has Cellular and Meristem-Organisational Roles with Differential Requirements for Cytokinin and CYCD3 Activity. Plant J. 2013, 75, 53–66. [Google Scholar] [CrossRef]
- Cao, X.W.; Du, Q.W.; Guo, Y.H.; Wang, Y.; Jiao, Y.L. Condensation of STM Is Critical for Shoot Meristem Maintenance and Salt Tolerance in Arabidopsis. Mol. Plant 2023, 16, 1445–1459. [Google Scholar] [CrossRef]
- Dabravolski, S.A.; Isayenkov, S.V. The Regulation of Plant Cell Wall Organisation under Salt Stress. Front. Plant Sci. 2023, 14, 1118313. [Google Scholar] [CrossRef]
- Oliveira, D.M.; Mota, T.R.; Salatta, F.V.; Sinzker, R.C.; Končitíková, R.; Kopečný, D.; Simister, R.; Silva, M.; Goeminne, G.; Morreel, K.; et al. Cell Wall Remodeling under Salt Stress: Insights into Changes in Polysaccharides, Feruloylation, Lignification, and Phenolic Metabolism in Maize. Plant Cell Environ. 2020, 43, 2172–2191. [Google Scholar] [CrossRef]
- Chen, K.Q.; Song, M.R.; Guo, Y.N.; Liu, L.F.; Xue, H.; Dai, H.Y.; Zhang, Z.H. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 2019, 17, 2341–2355. [Google Scholar] [CrossRef]
- Lou, T.X.; Lv, S.L.; Wang, J.H.; Wang, D.L.Y.; Lin, K.Q.; Zhang, X.; Zhang, B.; Guo, Z.J.; Yi, Z.; Li, Y.X. Cell Size and Xylem Differentiation Regulating Genes from Salicornia Europaea Contribute to Plant Salt Tolerance. Plant Cell Environ. 2024, 47, 2640–2659. [Google Scholar] [CrossRef]
- Xiong, D.L.; Flexas, J. Leaf Economics Spectrum in Rice: Leaf Anatomical, Biochemical, and Physiological Trait Trade-Offs. J. Exp. Bot. 2018, 69, 5599–5609. [Google Scholar] [CrossRef]
- Hong, E.; Xia, X.Z.; Ji, W.; Li, T.Y.; Xu, X.Y.; Chen, J.R.; Chen, X.; Zhu, X.T. Effects of High Temperature Stress on the Physiological and Biochemical Characteristics of Paeonia ostii. Int. J. Mol. Sci. 2023, 24, 11180. [Google Scholar] [CrossRef]
- Karaba, A.; Dixit, S.; Greco, R.; Aharoni, A.; Trijatmiko, K.R.; Marsch-Martinez, N.; Krishnan, A.; Nataraja, K.N.; Udayakumar, M.; Pereira, A. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc. Natl. Acad. Sci. USA 2007, 104, 15270–15275. [Google Scholar] [CrossRef]
- Antonova, E.V.; Shimalina, N.S.; Korotkova, A.M.; Kolosovskaya, E.V.; Gerasimova, S.V.; Khlestkina, E.K. Germination and Growth Characteristics of Nud Knockout and Win1 Knockout Barley Lines under Salt Stress. Plants 2024, 13, 1169. [Google Scholar] [CrossRef]
- Wang, J.J.; Liu, Y.T.; Hu, S.P.; Xu, J.; Nian, J.Q.; Cao, X.P.; Chen, M.M.; Cen, J.S.; Liu, X.; Zhang, Z.H.; et al. LEAF TIP RUMPLED 1 Regulates Leaf Morphology and Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 8818. [Google Scholar] [CrossRef] [PubMed]
- Baisakh, N.; RamanaRao, M.V.; Rajasekaran, K.; Subudhi, P.; Janda, J.; Galbraith, D.; Vanier, C.; Pereira, A. Enhanced Salt Stress Tolerance of Rice Plants Expressing a Vacuolar H+-ATPase Subunit C1 (SaVHAc1) Gene from the Halophyte Grass Spartina Alterniflora Löisel. Plant Biotechnol. J. 2012, 10, 453–464. [Google Scholar] [CrossRef]
- Tao, J.J.; Wei, W.; Pan, W.J.; Lu, L.; Li, Q.T.; Ma, J.B.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. An Alfin-like gene from Atriplex hortensis enhances salt and drought tolerance and abscisic acid response in transgenic Arabidopsis. Sci. Rep. 2018, 8, 2707. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.F.; Lv, B.; Li, J.; Luo, L.Q.; Lu, S.C.; Zhang, X.; Ma, H.; Ming, F. The NAC Family Transcription Factor OsNAP Confers Abiotic Stress Response through the ABA Pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef]
- Chu, X.Q.; Wang, C.; Chen, X.B.; Lu, W.J.; Li, H.; Wang, X.L.; Hao, L.L.; Guo, X.Q. The Cotton WRKY Gene GhWRKY41 Positively Regulates Salt and Drought Stress Tolerance in Transgenic Nicotiana Benthamiana. PLoS ONE 2015, 10, e0143022. [Google Scholar] [CrossRef]
- Hossain, Z.; Mandal, A.K.A.; Datta, S.K.; Biswas, A.K. Development of NaCl-Tolerant Strain in Chrysanthemum morifolium Ramat. through in Vitro Mutagenesis. Plant Biol. 2006, 8, 450–461. [Google Scholar] [CrossRef]
- Park, H.J.; Gámez-Arjona, F.M.; Lindahl, M.; Aman, R.; Villalta, I.; Cha, J.-Y.; Carranco, R.; Lim, C.J.; García, E.; Bressan, R.A.; et al. S-Acylated and Nucleus-Localized SALT OVERLY SENSITIVE3/CALCINEURIN B-LIKE4 Stabilizes GIGANTEA to Regulate Arabidopsis Flowering Time under Salt Stress. Plant Cell 2023, 35, 298–317. [Google Scholar] [CrossRef]
- Dong, L.D.; Hou, Z.H.; Li, H.Y.; Li, Z.B.; Fang, C.; Kong, L.P.; Li, Y.L.; Du, H.; Li, T.; Wang, L.S.; et al. Agronomical Selection on Loss-of-Function of GIGANTEA Simultaneously Facilitates Soybean Salt Tolerance and Early Maturity. J. Integr. Plant Biol. 2022, 64, 1866–1882. [Google Scholar] [CrossRef]
- Alsubaie, B.; Kharabian-Masouleh, A.; Furtado, A.; Al-Dossary, O.; Al-Mssallem, I.; Henry, R.J. Highly sex specific gene expression in Jojoba. BMC Plant Biol. 2023, 23, 440. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.X.; Zhao, F.Y.; Zhang, X.L.; Li, W.; Huang, J.S.; Pei, X.Y.; Ren, X.; Liu, Y.G.; He, K.L.; et al. Roles of S-Adenosylmethionine and Its Derivatives in Salt Tolerance of Cotton. Int. J. Mol. Sci. 2023, 24, 9517. [Google Scholar] [CrossRef]
- Xia, D.N.; Guan, L.L.; Yin, Y.; Wang, Y.X.; Shi, H.Y.; Li, W.Y.; Zhang, D.K.; Song, R.; Hu, T.X.; Zhan, X.Q. Genome-Wide Analysis of MBF1 Family Genes in Five Solanaceous Plants and Functional Analysis of SlER24 in Salt Stress. Int. J. Mol. Sci. 2023, 24, 13965. [Google Scholar] [CrossRef]
- Khan, I.; Muhammad, A.; Chattha, M.U.; Skalicky, M.; Bilal Chattha, M.; Ahsin Ayub, M.; Rizwan Anwar, M.; Soufan, W.; Hassan, M.U.; Rahman, M.A.; et al. Mitigation of Salinity-Induced Oxidative Damage, Growth, and Yield Reduction in Fine Rice by Sugarcane Press Mud Application. Front. Plant Sci. 2022, 13, 840900. [Google Scholar] [CrossRef]
- Li, H.P.; Kong, F.R.; Tang, T.T.; Luo, Y.L.; Gao, H.R.; Xu, J.; Xing, G.M.; Li, L.Z. Physiological and Transcriptomic Analyses Revealed That Humic Acids Improve Low-Temperature Stress Tolerance in Zucchini (Cucurbita pepo L.) Seedlings. Plants 2023, 12, 548. [Google Scholar] [CrossRef]
- Wei, H.; Movahedi, A.; Liu, G.Y.; Li, Y.X.; Liu, S.W.; Yu, C.M.; Chen, Y.H.; Zhong, F.; Zhang, J. Comprehensive Analysis of Carotenoid Cleavage Dioxygenases Gene Family and Its Expression in Response to Abiotic Stress in Poplar. Int. J. Mol. Sci. 2022, 23, 1418. [Google Scholar] [CrossRef]
- Zhang, B.B.; Du, H.; Yang, S.K.; Wu, X.L.; Liu, W.X.; Guo, J.; Xiao, Y.S.; Peng, F.T. Physiological and Transcriptomic Analyses of the Effects of Exogenous Lauric Acid on Drought Resistance in Peach (Prunus persica (L.) Batsch). Plants 2023, 12, 1492. [Google Scholar] [CrossRef]
- Cui, J.; Li, J.L.; Dai, C.H.; Li, L.P. Transcriptome and Metabolome Analyses Revealed the Response Mechanism of Sugar Beet to Salt Stress of Different Durations. Int. J. Mol. Sci. 2022, 23, 9599. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, W.; Fu, H.J.; Li, L.T.; Ruan, X.Y.; Zhang, X.Y. Effects of Salinity Stress on Growth and Physiological Parameters and Related Gene Expression in Different Ecotypes of Sesuvium Portulacastrum on Hainan Island. Genes 2023, 14, 1336. [Google Scholar] [CrossRef]
- Chen, K.Q.; Guo, Y.N.; Song, M.R.; Liu, L.F.; Xue, H.; Dai, H.Y.; Zhang, Z.H. Dual Role of MdSND1 in the Biosynthesis of Lignin and in Signal Transduction in Response to Salt and Osmotic Stress in Apple. Hortic. Res. 2020, 7, 204. [Google Scholar] [CrossRef]
- Cai, R.H.; Zhao, Y.; Wang, Y.F.; Lin, Y.X.; Peng, X.J.; Li, Q.; Chang, Y.W.; Jiang, H.Y.; Xiang, Y.; Cheng, B.J. Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tissue Organ. Cult. 2014, 119, 565–577. [Google Scholar] [CrossRef]
- Joshi, R.; Sahoo, K.K.; Tripathi, A.K.; Kumar, R.; Gupta, B.K.; Pareek, A.; Singla-Pareek, S.L. Knockdown of an Inflorescence Meristem-Specific Cytokinin Oxidase—OsCKX2 in Rice Reduces Yield Penalty under Salinity Stress Condition. Plant Cell Environ. 2018, 41, 936–946. [Google Scholar] [CrossRef]
- Sun, S.J.; Guo, S.Q.; Yang, X.; Bao, Y.M.; Tang, H.J.; Sun, H.; Huang, J.; Zhang, H.S. Functional Analysis of a Novel Cys2/His2-Type Zinc Finger Protein Involved in Salt Tolerance in Rice. J. Exp. Bot. 2010, 61, 2807–2818. [Google Scholar] [CrossRef]
- Li, C.X.; Song, T.T.; Zhan, L.F.; Cong, C.L.; Xu, H.H.; Dong, L.; Cai, H. Overexpression of MsRCI2A, MsRCI2B, and MsRCI2C in Alfalfa (Medicago sativa L.) Provides Different Extents of Enhanced Alkali and Salt Tolerance Due to Functional Specialization of MsRCI2s. Front. Plant Sci. 2021, 12, 702195. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.H.; Shi, L.X.; Tian, H.; Tian, H.Z.; Zhang, J.; Zhao, F.S.; Zhang, Q.Q.; Zhang, S.Q.; Geng, G.D. Characterization and transformation of TtMYB1 transcription factor from Tritipyrum to improve salt tolerance in wheat. BMC Genom. 2024, 25, 163. [Google Scholar] [CrossRef]
- Liu, C.Y.; Li, J.; Zhu, P.P.; Yu, J.; Hou, J.M.; Wang, C.H.; Long, D.P.; Yu, M.D.; Zhao, A.C. Mulberry EIL3 Confers Salt and Drought Tolerances and Modulates Ethylene Biosynthetic Gene Expression. PeerJ 2019, 7, e6391. [Google Scholar] [CrossRef]
- Abrar, M.M.; Saqib, M.; Abbas, G.; Atiq-Ur-Rahman, M.; Mustafa, A.; Shah, S.A.A.; Mehmood, K.; Maitlo, A.A.; Ul-Hassan, M.; Sun, N.; et al. Evaluating the Contribution of Growth, Physiological, and Ionic Components Towards Salinity and Drought Stress Tolerance in Jatropha curcas. Plants 2020, 9, 1574. [Google Scholar] [CrossRef]
- Apell, H.J.; Roudna, M. Partial Reactions of the Na,K-ATPase: Determination of Activation Energies and an Approach to Mechanism. J. Membr. Biol. 2020, 253, 631–645. [Google Scholar] [CrossRef]
- Blumwald, E.; Aharon, G.S.; Apse, M.P. Sodium Transport in Plant Cells. Biochim. Biophys. Acta 2000, 1465, 140–151. [Google Scholar] [CrossRef]
- Zhu, J.K. Regulation of Ion Homeostasis under Salt Stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Wu, H.H.; Zhang, X.C.; Giraldo, J.P.; Shabala, S. It is not all about sodium: Revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil 2018, 431, 1–17. [Google Scholar] [CrossRef]
- Chen, G.; Hu, Q.D.; Luo, L.; Yang, T.Y.; Zhang, S.; Hu, Y.B.; Yu, L.; Xu, G.H. Rice Potassium Transporter OsHAK1 Is Essential for Maintaining Potassium-Mediated Growth and Functions in Salt Tolerance over Low and High Potassium Concentration Ranges. Plant Cell Environ. 2015, 38, 2747–2765. [Google Scholar] [CrossRef]
- Yang, T.Y.; Zhang, S.; Hu, Y.B.; Wu, F.C.; Hu, Q.D.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The Role of a Potassium Transporter OsHAK5 in Potassium Acquisition and Transport from Roots to Shoots in Rice at Low Potassium Supply Levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Rubio, F.; Nieves-Cordones, M.; Horie, T.; Shabala, S. Doing “business as Usual” Comes with a Cost: Evaluating Energy Cost of Maintaining Plant Intracellular K+ Homeostasis under Saline Conditions. New Phytol. 2020, 225, 1097–1104. [Google Scholar] [CrossRef] [PubMed]
- Asins, M.J.; Villalta, I.; Aly, M.M.; Olías, R.; Alvarez DE Morales, P.; Huertas, R.; Li, J.; Jaime-Pérez, N.; Haro, R.; Raga, V.; et al. Two Closely Linked Tomato HKT Coding Genes Are Positional Candidates for the Major Tomato QTL Involved in Na+/K+ Homeostasis. Plant Cell Environ. 2013, 36, 1171–1191. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Gilliham, M.; Hrmova, M. Plant High-Affinity Potassium (HKT) Transporters Involved in Salinity Tolerance: Structural Insights to Probe Differences in Ion Selectivity. Int. J. Mol. Sci. 2013, 14, 7660–7680. [Google Scholar] [CrossRef]
- Wang, T.; Chen, X.Y.; Ju, C.F.; Wang, C. Calcium Signaling in Plant Mineral Nutrition: From Uptake to Transport. Plant Commun. 2023, 4, 100678. [Google Scholar] [CrossRef]
- Sameeullah, M.; Yildirim, M.; Aslam, N.; Baloğlu, M.C.; Yucesan, B.; Lössl, A.G.; Saba, K.; Waheed, M.T.; Gurel, E. Plastidial Expression of 3β-Hydroxysteroid Dehydrogenase and Progesterone 5β-Reductase Genes Confer Enhanced Salt Tolerance in Tobacco. Int. J. Mol. Sci. 2021, 22, 11736. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Quintero, F.J.; Pardo, J.M.; Schumaker, K.S.; Zhu, J.K. Regulation of Vacuolar Na+/H+ Exchange in Arabidopsis Thaliana by the Salt-Overly-Sensitive (SOS) Pathway. J. Biol. Chem. 2004, 279, 207–215. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Britto, D.T. Sodium Transport in Plants: A Critical Review. New Phytol. 2011, 189, 54–81. [Google Scholar] [CrossRef]
- Jia, Q.; Li, M.W.; Zheng, C.W.; Xu, Y.Y.; Sun, S.; Li, Z.; Wong, F.L.; Song, J.L.; Lin, W.W.; Li, Q.H.; et al. The Soybean Plasma Membrane-Localized Cation/H+ Exchanger GmCHX20a Plays a Negative Role under Salt Stress. Physiol. Plant 2021, 171, 714–727. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genom. 2023, 51, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Abogadallah, G.M. Antioxidative Defense under Salt Stress. Plant Signal Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef]
- Gao, Y.L.; Long, R.C.; Kang, J.M.; Wang, Z.; Zhang, T.J.; Sun, H.; Li, X.; Yang, Q.C. Comparative Proteomic Analysis Reveals That Antioxidant System and Soluble Sugar Metabolism Contribute to Salt Tolerance in Alfalfa (Medicago sativa L.) Leaves. J. Proteome Res. 2019, 18, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS Signaling: The New Wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Venza, I.; Venza, M.; Visalli, M.; Lentini, G.; Teti, D.; d’Alcontres, F.S. ROS as Regulators of Cellular Processes in Melanoma. Oxid. Med. Cell Longev. 2021, 2021, 1208690. [Google Scholar] [CrossRef]
- Hongrapipat, J.; Kopecková, P.; Liu, J.H.; Prakongpan, S.; Kopecek, J. Combination chemotherapy and photodynamic therapy with fab’ fragment targeted HPMA copolymer conjugates in human ovarian carcinoma cells. Mol. Pharm. 2008, 5, 696–709. [Google Scholar] [CrossRef]
- Wang, B.F.; Zhang, Y.X.; Bi, Z.Z.; Liu, Q.E.; Xu, T.T.; Yu, N.; Cao, Y.R.; Zhu, A.K.; Wu, W.X.; Zhan, X.D.; et al. Impaired Function of the Calcium-Dependent Protein Kinase, OsCPK12, Leads to Early Senescence in Rice (Oryza sativa L.). Front. Plant Sci. 2019, 10, 52. [Google Scholar] [CrossRef]
- Li, S.Z.; Sun, M.T.; Miao, L.; Di, Q.H.; Lv, L.J.; Yu, X.C.; Yan, Y.; He, C.X.; Wang, J.; Shi, A.K.; et al. Multifaceted Regulatory Functions of CsBPC2 in Cucumber under Salt Stress Conditions. Hortic. Res. 2023, 10, uhad051. [Google Scholar] [CrossRef]
- Hernández, J.A.; Almansa, M.S. Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves. Physiol. Plant 2002, 115, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Verma, T.; Bhardwaj, S.; Raza, A.; Djalovic, I.; Prasad, P.V.; Kapoor, D. Mitigation of Salt Stress in Indian Mustard (Brassica juncea L.) by the Application of Triacontanol and Hydrogen Sulfide. Plant Signal Behav. 2023, 18, 2189371. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Z.; Guo, R.R.; Cheng, C.X.; Zhang, H.J.; Zhang, Y.C.; Wang, X.P. Overexpression of ALDH2B8, an Aldehyde Dehydrogenase Gene from Grapevine, Sustains Arabidopsis Growth upon Salt Stress and Protects Plants against Oxidative Stress. Plant Cell Tissue Organ. Cult. 2013, 114, 187–196. [Google Scholar] [CrossRef]
- Li, W.H.; Li, P.; Chen, H.Y.; Zhong, J.L.; Liang, X.Q.; Wei, Y.F.; Zhang, L.H.; Wang, H.B.; Han, D.G. Overexpression of a Fragaria Vesca 1R-MYB Transcription Factor Gene (FvMYB114) Increases Salt and Cold Tolerance in Arabidopsis Thaliana. Int. J. Mol. Sci. 2023, 24, 5261. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Dong, X.; Wang, R.J.; Hao, F.; Zhang, H.; Zhang, Y.Y.; Lin, G.L. Exogenous Calcium Alleviates Oxidative Stress Caused by Salt Stress in Peanut Seedling Roots by Regulating the Antioxidant Enzyme System and Flavonoid Biosynthesis. Antioxidants 2024, 13, 233. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.Q.; Yang, Y.Q. How Plants Tolerate Salt Stress. Curr. Issues Mol. Biol. 2023, 45, 5914–5934. [Google Scholar] [CrossRef]
- Larisch, N.; Kirsch, S.A.; Schambony, A.; Studtrucker, T.; Böckmann, R.A.; Dietrich, P. The Function of the Two-Pore Channel TPC1 Depends on Dimerization of Its Carboxy-Terminal Helix. Cell Mol. Life Sci. 2016, 73, 2565–2581. [Google Scholar] [CrossRef]
- Brailoiu, E.; Churamani, D.; Cai, X.J.; Schrlau, M.G.; Brailoiu, G.C.; Gao, X.; Hooper, R.; Boulware, M.J.; Dun, N.J.; Marchant, J.S.; et al. Essential Requirement for Two-Pore Channel 1 in NAADP-Mediated Calcium Signaling. J. Cell Biol. 2009, 186, 201–209. [Google Scholar] [CrossRef]
- Baluska, F.; Samaj, J.; Wojtaszek, P.; Volkmann, D.; Menzel, D. Cytoskeleton-Plasma Membrane-Cell Wall Continuum in Plants. Emerging Links Revisited. Plant Physiol. 2003, 133, 482–491. [Google Scholar] [CrossRef]
- Zagorchev, L.; Kamenova, P.; Odjakova, M. The Role of Plant Cell Wall Proteins in Response to Salt Stress. Sci. World J. 2014, 2014, 764089. [Google Scholar] [CrossRef]
- Giridhar, M.; Meier, B.; Imani, J.; Kogel, K.H.; Peiter, E.; Vothknecht, U.C.; Chigri, F. Comparative Analysis of Stress-Induced Calcium Signals in the Crop Species Barley and the Model Plant Arabidopsis Thaliana. BMC Plant Biol. 2022, 22, 447. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Aleynova, O.A.; Ogneva, Z.V.; Suprun, A.R.; Ananev, A.A.; Nityagovsky, N.N.; Dneprovskaya, A.A.; Beresh, A.A.; Dubrovina, A.S. The Effect of Stress Hormones, Ultraviolet C, and Stilbene Precursors on Expression of Calcineurin B-like Protein (CBL) and CBL-Interacting Protein Kinase (CIPK) Genes in Cell Cultures and Leaves of Vitis amurensis Rupr. Plants 2023, 12, 1562. [Google Scholar] [CrossRef] [PubMed]
- Steinhorst, L.; He, G.F.; Moore, L.K.; Schültke, S.; Schmitz-Thom, I.; Cao, Y.B.; Hashimoto, K.; Andrés, Z.; Piepenburg, K.; Ragel, P.; et al. A Ca2+-Sensor Switch for Tolerance to Elevated Salt Stress in Arabidopsis. Dev. Cell 2022, 57, 2081–2094. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Raddatz, N.; Pardo, J.M.; Yun, D.J. HKT Sodium and Potassium Transporters in Arabidopsis Thaliana and Related Halophyte Species. Physiol. Plant 2021, 171, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.J.; Wang, C.W.; Xue, Y.; Liu, X.; Chen, S.; Song, C.P.; Yang, Y.Q.; Guo, Y. Calcium-Activated 14-3-3 Proteins as a Molecular Switch in Salt Stress Tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Gul, N.; Mir, M.A.; Asgher, M.; Al-Sulami, N.; Abulfaraj, A.A.; Qari, S. Role of Jasmonates, Calcium, and Glutathione in Plants to Combat Abiotic Stresses Through Precise Signaling Cascade. Front. Plant Sci. 2021, 12, 668029. [Google Scholar] [CrossRef]
- Ma, L.; Ye, J.M.; Yang, Y.Q.; Lin, H.X.; Yue, L.L.; Luo, J.; Long, Y.; Fu, H.Q.; Liu, X.N.; Zhang, Y.L.; et al. The SOS2-SCaBP8 Complex Generates and Fine-Tunes an AtANN4-Dependent Calcium Signature under Salt Stress. Dev. Cell 2019, 48, 697–709.e5. [Google Scholar] [CrossRef]
- Chérel, I.; Gaillard, I. The Complex Fine-Tuning of K+ Fluxes in Plants in Relation to Osmotic and Ionic Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 715. [Google Scholar] [CrossRef]
- Chen, G.J.; Zheng, D.F.; Feng, N.J.; Zhou, H.; Mu, D.W.; Zhao, L.M.; Shen, X.F.; Rao, G.S.; Meng, F.Y.; Huang, A.Q. Physiological mechanisms of ABA-induced salinity tolerance in leaves and roots of rice. Sci. Rep. 2022, 12, 8228. [Google Scholar] [CrossRef]
- Bai, Q.X.; Niu, Z.M.; Chen, Q.Y.; Gao, C.Y.; Zhu, M.J.; Bai, J.X.; Liu, M.J.; He, L.; Liu, J.Q.; Jiang, Y.Z.; et al. The C2H2 -Type Zinc Finger Transcription Factor OSIC1 Positively Regulates Stomatal Closure under Osmotic Stress in Poplar. Plant Biotechnol. J. 2023, 21, 943–960. [Google Scholar] [CrossRef]
- Zhang, X.L.; Jiang, L.; Xin, Q.; Liu, Y.; Tan, J.X.; Chen, Z.Z. Structural Basis and Functions of Abscisic Acid Receptors PYLs. Front. Plant Sci. 2015, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Loukehaich, R.; Wang, T.T.; Ouyang, B.; Ziaf, K.; Li, H.X.; Zhang, J.H.; Lu, Y.E.; Ye, Z.B. SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 2012, 63, 5593–5606. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular Basis of the Core Regulatory Network in ABA Responses: Sensing, Signaling and Transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Carianopol, C.S.; Chan, A.L.; Dong, S.W.; Provart, N.J.; Lumba, S.; Gazzarrini, S. An Abscisic Acid-Responsive Protein Interaction Network for Sucrose Non-Fermenting Related Kinase1 in Abiotic Stress Response. Commun. Biol. 2020, 3, 145. [Google Scholar] [CrossRef]
- Fàbregas, N.; Yoshida, T.; Fernie, A.R. Role of Raf-like Kinases in SnRK2 Activation and Osmotic Stress Response in Plants. Nat. Commun. 2020, 11, 6184. [Google Scholar] [CrossRef]
- Li, Y.Q.; Liu, Y.N.; Jin, L.B.; Peng, R.Y. Crosstalk between Ca2+ and Other Regulators Assists Plants in Responding to Abiotic Stress. Plants 2022, 11, 1351. [Google Scholar] [CrossRef]
- Niu, M.L.; Huang, Y.; Sun, S.T.; Sun, J.Y.; Cao, H.S.; Shabala, S.; Bie, Z.L. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J. Exp. Bot. 2017, 69, 3465–3476. [Google Scholar] [CrossRef]
- Korasick, D.A.; Jez, J.M.; Strader, L.C. Refining the Nuclear Auxin Response Pathway through Structural Biology. Curr. Opin. Plant Biol. 2015, 27, 22–28. [Google Scholar] [CrossRef]
- Wang, N.N.; Lin, Y.J.; Qi, F.; Xiaoyang, C.X.; Peng, Z.W.; Yu, Y.; Liu, Y.N.; Zhang, J.; Qi, X.; Deyholos, M.; et al. Comprehensive Analysis of Differentially Expressed Genes and Epigenetic Modification-Related Expression Variation Induced by Saline Stress at Seedling Stage in Fiber and Oil Flax, Linum usitatissimum L. Plants 2022, 11, 2053. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Z.Q. PIF4 and PIF4-Interacting Proteins: At the Nexus of Plant Light, Temperature and Hormone Signal Integrations. Int. J. Mol. Sci. 2021, 22, 10304. [Google Scholar] [CrossRef]
- Zhu, L.; Yin, T.; Zhang, M.J.; Yang, X.Y.; Wu, J.X.; Cai, H.B.; Yang, N.; Li, X.L.; Wen, K.; Chen, D.M.; et al. Genome-Wide Identification and Expression Pattern Analysis of the Kiwifruit GRAS Transcription Factor Family in Response to Salt Stress. BMC Genom. 2024, 25, 12. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yu, K.M.; Yan, J.B.; Shan, X.Y.; Xie, D.X. Jasmonate Perception: Ligand-Receptor Interaction, Regulation, and Evolution. Mol. Plant 2023, 16, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Nefissi Ouertani, R.; Arasappan, D.; Abid, G.; Ben Chikha, M.; Jardak, R.; Mahmoudi, H.; Mejri, S.; Ghorbel, A.; Ruhlman, T.A.; Jansen, R.K. Transcriptomic Analysis of Salt-Stress-Responsive Genes in Barley Roots and Leaves. Int. J. Mol. Sci. 2021, 22, 8155. [Google Scholar] [CrossRef] [PubMed]
- Sobrinho, T.G.; da Silva, A.A.R.; de Lima, G.S.; de Lima, V.L.A.; Borges, V.E.; Nunes, K.G.; Soares, L.A.D.A.; Saboya, L.M.F.; Gheyi, H.R.; Gomes, J.P.; et al. Foliar Applications of Salicylic Acid on Boosting Salt Stress Tolerance in Sour Passion Fruit in Two Cropping Cycles. Plants 2023, 12, 2023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, D.S.; Li, M.X.; Shi, L.X. Metabolic Profiles Reveal Changes in Wild and Cultivated Soybean Seedling Leaves under Salt Stress. PLoS ONE 2016, 11, e0159622. [Google Scholar] [CrossRef]
- Jayakannan, M.; Bose, J.; Babourina, O.; Rengel, Z.; Shabala, S. Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a GORK channel. J. Exp. Bot. 2013, 64, 2255–2268. [Google Scholar] [CrossRef]
- Yu, Z.P.; Duan, X.B.; Luo, L.; Dai, S.J.; Ding, Z.J.; Xia, G.M. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Wang, B.K.; Wang, J.; Yang, T.; Wang, J.X.; Dai, Q.; Zhang, F.L.; Xi, R.; Yu, Q.H.; Li, N. The Transcriptional Regulatory Network of Hormones and Genes under Salt Stress in Tomato Plants (Solanum lycopersicum L.). Front. Plant Sci. 2023, 14, 1115593. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-Deficient Transgenic Arabidopsis Plants Show Multiple Developmental Alterations Indicating Opposite Functions of Cytokinins in the Regulation of Shoot and Root Meristem Activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.R.; Liu, R.J.; Zhang, H.W.; Wang, P.F.; Li, Y.L.; Wang, S.F.; Tang, S.Y.; Liu, C.Y.; et al. ABI4 Mediates Antagonistic Effects of Abscisic Acid and Gibberellins at Transcript and Protein Levels. Plant J. 2016, 85, 348–361. [Google Scholar] [CrossRef]
- Nishiyama, R.; Watanabe, Y.; Fujita, Y.; Le, D.T.; Kojima, M.; Werner, T.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Kakimoto, T.; et al. Analysis of Cytokinin Mutants and Regulation of Cytokinin Metabolic Genes Reveals Important Regulatory Roles of Cytokinins in Drought, Salt and Abscisic Acid Responses, and Abscisic Acid Biosynthesis. Plant Cell 2011, 23, 2169–2183. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhao, Y.; Gao, J.H.; Xiang, C.B.; Zhu, J.K. The ABA Receptor PYL9 Together with PYL8 Plays an Important Role in Regulating Lateral Root Growth. Sci. Rep. 2016, 6, 27177. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.A.; Rasmussen, A.; Traini, R.; Voß, U.; Sturrock, C.; Mooney, S.J.; Wells, D.M.; Bennett, M.J. Branching out in Roots: Uncovering Form, Function, and Regulation. Plant Physiol. 2014, 166, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Aroca, R.; Ruiz-Lozano, J.M.; Zamarreño, A.M.; Paz, J.A.; García-Mina, J.M.; Pozo, M.J.; López-Ráez, J.A. Arbuscular Mycorrhizal Symbiosis Influences Strigolactone Production under Salinity and Alleviates Salt Stress in Lettuce Plants. J. Plant Physiol. 2013, 170, 47–55. [Google Scholar] [CrossRef]
- Ren, C.G.; Kong, C.C.; Xie, Z.H. Role of Abscisic Acid in Strigolactone-Induced Salt Stress Tolerance in Arbuscular Mycorrhizal Sesbania Cannabina Seedlings. BMC Plant Biol. 2018, 18, 74. [Google Scholar] [CrossRef]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Qi, X.L.; Li, X.K.; Guo, H.M.; Guo, N.; Cheng, H.M. VdPLP, A Patatin-Like Phospholipase in Verticillium Dahliae, Is Involved in Cell Wall Integrity and Required for Pathogenicity. Genes. 2018, 9, 162. [Google Scholar] [CrossRef]
- Byrne, S.L.; Foito, A.; Hedley, P.E.; Morris, J.A.; Stewart, D.; Barth, S. Early Response Mechanisms of Perennial Ryegrass (Lolium perenne) to Phosphorus Deficiency. Ann. Bot. 2011, 107, 243–254. [Google Scholar] [CrossRef]
- Lemtiri-Chlieh, F.; MacRobbie, E.A.C.; Webb, A.A.R.; Manison, N.F.; Brownlee, C.; Skepper, J.N.; Chen, J.; Prestwich, G.D.; Brearley, C.A. Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc. Natl. Acad. Sci. USA 2003, 100, 10091–10095. [Google Scholar] [CrossRef]
- Escobar-Sepúlveda, H.F.; Trejo-Téllez, L.I.; Pérez-Rodríguez, P.; Hidalgo-Contreras, J.V.; Gómez-Merino, F.C. Diacylglycerol Kinases Are Widespread in Higher Plants and Display Inducible Gene Expression in Response to Beneficial Elements, Metal, and Metalloid Ions. Front. Plant Sci. 2017, 8, 129. [Google Scholar] [CrossRef]
- Caillaud, M.C. Anionic Lipids: A Pipeline Connecting Key Players of Plant Cell Division. Front. Plant Sci. 2019, 10, 419. [Google Scholar] [CrossRef] [PubMed]
- Colin, L.; Ruhnow, F.; Zhu, J.K.; Zhao, C.Z.; Zhao, Y.; Persson, S. The Cell Biology of Primary Cell Walls during Salt Stress. Plant Cell 2023, 35, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Chen, Q.H.; Li, X.Y.; Li, Y.Z. MAP65-1 Is Required for the Depolymerization and Reorganization of Cortical Microtubules in the Response to Salt Stress in Arabidopsis. Plant Sci. 2017, 264, 112–121. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; van der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 2012, 72, 436–449. [Google Scholar] [CrossRef]
- McLoughlin, F.; Testerink, C. Phosphatidic acid, a versatile water-stress signal in roots. Front. Plant Sci. 2013, 4, 525. [Google Scholar] [CrossRef]
- Rodas-Junco, B.A.; Racagni-Di-Palma, G.E.; Canul-Chan, M.; Usorach, J.; Hernández-Sotomayor, S.M.T. Link between Lipid Second Messengers and Osmotic Stress in Plants. Int. J. Mol. Sci. 2021, 22, 2658. [Google Scholar] [CrossRef]
- Zhou, H.Y.; He, Y.; Zhu, Y.S.; Li, M.Y.; Song, S.; Bo, W.H.; Li, Y.Y.; Pang, X.M. Comparative transcriptome profiling reveals cold stress responsiveness in two contrasting Chinese jujube cultivars. BMC Plant Biol. 2020, 20, 240. [Google Scholar] [CrossRef]
- Pitzschke, A.; Djamei, A.; Bitton, F.; Hirt, H. A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2009, 2, 120–137. [Google Scholar] [CrossRef]
- Li, C.H.; Wang, G.; Zhao, J.L.; Zhang, L.Q.; Ai, L.F.; Han, Y.F.; Sun, D.Y.; Zhang, S.W.; Sun, Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef]
- Rentel, M.C.; Lecourieux, D.; Ouaked, F.; Usher, S.L.; Petersen, L.; Okamoto, H.; Knight, H.; Peck, S.C.; Grierson, C.S.; Hirt, H.; et al. OXI1 Kinase Is Necessary for Oxidative Burst-Mediated Signalling in Arabidopsis. Nature 2004, 427, 858–861. [Google Scholar] [CrossRef]
- Evans, M.J.; Choi, W.G.; Gilroy, S.; Morris, R.J. A ROS-Assisted Calcium Wave Dependent on the AtRBOHD NADPH Oxidase and TPC1 Cation Channel Propagates the Systemic Response to Salt Stress. Plant Physiol. 2016, 171, 1771–1784. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.G.; Schroeder, J.I. NADPH Oxidase AtrbohD and AtrbohF Genes Function in ROS-Dependent ABA Signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Szymańska, K.P.; Polkowska-Kowalczyk, L.; Lichocka, M.; Maszkowska, J.; Dobrowolska, G. SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress. Int. J. Mol. Sci. 2019, 20, 143. [Google Scholar] [CrossRef]
- Han, J.P.; Köster, P.; Drerup, M.M.; Scholz, M.; Li, S.Z.; Edel, K.H.; Hashimoto, K.; Kuchitsu, K.; Hippler, M.; Kudla, J. Fine-Tuning of RBOHF Activity Is Achieved by Differential Phosphorylation and Ca2+ Binding. New Phytol. 2019, 221, 1935–1949. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Mittler, R. Could Heat Shock Transcription Factors Function as Hydrogen Peroxide Sensors in Plants? Ann. Bot. 2006, 98, 279–288. [Google Scholar] [CrossRef]
- Pei, X.; Zhang, Y.; Zhu, L.Y.; Zhao, D.X.; Lu, Y.Z.; Zheng, J. Physiological and Transcriptomic Analyses Characterized High Temperature Stress Response Mechanisms in Sorbus Pohuashanensis. Sci. Rep. 2021, 11, 10117. [Google Scholar] [CrossRef]
- Livanos, P.; Galatis, B.; Gaitanaki, C.; Apostolakos, P. Phosphorylation of a P38-like MAPK Is Involved in Sensing Cellular Redox State and Drives Atypical Tubulin Polymer Assembly in Angiosperms. Plant Cell Environ. 2014, 37, 1130–1143. [Google Scholar] [CrossRef]
- Fortunato, S.; Lasorella, C.; Dipierro, N.; Vita, F.; de Pinto, M.C. Redox Signaling in Plant Heat Stress Response. Antioxidants 2023, 12, 605. [Google Scholar] [CrossRef]
- Skoko, J.J.; Attaran, S.; Neumann, C.A. Signals Getting Crossed in the Entanglement of Redox and Phosphorylation Pathways: Phosphorylation of Peroxiredoxin Proteins Sparks Cell Signaling. Antioxidants 2019, 8, 29. [Google Scholar] [CrossRef]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in Molecular Marker Techniques and Their Applications in Plant Sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef]
- Fan, X.R.; Jiang, H.Z.; Meng, L.J.; Chen, J.G. Gene Mapping, Cloning and Association Analysis for Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 11674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Liu, M.F.; He, J.B.; Wang, Y.F.; Xing, G.N.; Li, Y.; Yang, S.P.; Zhao, T.J.; Gai, J.Y. Marker-Assisted Breeding for Transgressive Seed Protein Content in Soybean [Glycine max (L.) Merr]. Theor. Appl. Genet. 2015, 128, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.R.; Dong, S.Y.; Bo, K.L.; Miao, H.; Li, C.X.; Zhang, Y.Y.; Zhang, S.P.; Gu, X.F. Identification of QTLs Controlling Salt Tolerance in Cucumber (Cucumis sativus L.) Seedlings. Plants 2021, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.; Kim, M.Y.; Kwon, H.; Yang, X.F.; Lee, S.H. Novel QTL identification and candidate gene analysis for enhancing salt tolerance in soybean (Glycine max (L.) Merr.). Plant Sci. 2021, 313, 111085. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, Z.T.; Chen, X.Q.; Li, X.R.; Shi, Y.J.; Xu, L.; Yu, C.Y.; Jing, B.; Li, W.W.; Xu, A.X.; et al. Identification Candidate Genes for Salt Resistance through Quantitative Trait Loci-Sequencing in Brassica napus L. J. Plant Physiol. 2024, 294, 154187. [Google Scholar] [CrossRef]
- Yuan, L.; Zhang, L.C.; Wei, X.; Wang, R.H.; Li, N.N.; Chen, G.L.; Fan, F.F.; Huang, S.Y.; Li, J.X.; Li, S.Q. Quantitative Trait Locus Mapping of Salt Tolerance in Wild Rice Oryza Longistaminata. Int. J. Mol. Sci. 2022, 23, 2379. [Google Scholar] [CrossRef]
- Yin, W.; Lu, T.; Chen, Z.; Lu, T.; Ye, H.; Mao, Y.; Luo, Y.; Lu, M.; Zhu, X.; Yuan, X.; et al. Quantitative trait locus mapping and candidate gene analysis for salt tolerance at bud stage in rice. Front. Plant Sci. 2023, 13, 1041081. [Google Scholar] [CrossRef]
- Zeng, P.; Zhu, P.W.; Qian, L.F.; Qian, X.M.; Mi, Y.X.; Lin, Z.F.; Dong, S.N.; Aronsson, H.; Zhang, H.S.; Cheng, J.P. Identification and Fine Mapping of qGR6.2, a Novel Locus Controlling Rice Seed Germination under Salt Stress. BMC Plant Biol. 2021, 21, 36. [Google Scholar] [CrossRef]
- Quan, R.D.; Wang, J.; Hui, J.; Bai, H.B.; Lyu, X.L.; Zhu, Y.X.; Zhang, H.W.; Zhang, Z.J.; Li, S.H.; Huang, R.F. Improvement of Salt Tolerance Using Wild Rice Genes. Front. Plant Sci. 2018, 8, 2269. [Google Scholar] [CrossRef]
- Hussain, B.; Lucas, S.J.; Ozturk, L.; Budak, H. Mapping QTLs conferring salt tolerance and micronutrient concentrations at seedling stagein wheat. Sci. Rep. 2017, 7, 15662. [Google Scholar] [CrossRef]
- Asif, M.A.; Garcia, M.; Tilbrook, J.; Brien, C.; Dowling, K.; Berger, B.; Schilling, R.K.; Short, L.; Trittermann, C.; Gilliham, M.; et al. Identification of Salt Tolerance QTL in a Wheat RIL Mapping Population Using Destructive and Non-Destructive Phenotyping. Funct. Plant Biol. 2021, 48, 131–140. [Google Scholar] [CrossRef]
- Guo, H.L.; Ding, W.W.; Chen, J.B.; Chen, X.; Zheng, Y.Q.; Wang, Z.Y.; Liu, J.X. Genetic Linkage Map Construction and QTL Mapping of Salt Tolerance Traits in Zoysiagrass (Zoysia japonica). PLoS ONE 2014, 9, e107249. [Google Scholar] [CrossRef]
- Guo, A.H.; Li, H.J.; Huang, Y.; Ma, X.Q.; Li, B.; Du, X.Q.; Cui, Y.N.; Zhao, N.; Hua, J.P. Yield-Related Quantitative Trait Loci Identification and Lint Percentage Hereditary Dissection under Salt Stress in Upland Cotton. Plant J. 2024, 119, 115–136. [Google Scholar] [CrossRef]
- Yang, X.H.; Xia, X.Z.; Zeng, Y.; Nong, B.X.; Zhang, Z.Q.; Wu, Y.Y.; Xiong, F.Q.; Zhang, Y.X.; Liang, H.F.; Deng, G.F.; et al. Identification of candidate genes for gelatinization temperature, gel consistency and pericarp color by GWAS in rice based on SLAF-sequencing. PLoS ONE 2018, 13, e0196690. [Google Scholar] [CrossRef]
- Chen, H.D.; Xie, W.B.; He, H.; Yu, H.H.; Chen, W.; Li, J.; Yu, R.B.; Yao, Y.; Zhang, W.H.; He, Y.Q.; et al. A High-Density SNP Genotyping Array for Rice Biology and Molecular Breeding. Mol. Plant 2014, 7, 541–553. [Google Scholar] [CrossRef]
- Kumar, V.; Singh, A.; Mithra, S.V.A.; Krishnamurthy, S.L.; Parida, S.K.; Jain, S.; Tiwari, K.K.; Kumar, P.; Rao, A.R.; Sharma, S.K.; et al. Genome-Wide Association Mapping of Salinity Tolerance in Rice (Oryza sativa). DNA Res. 2015, 22, 133–145. [Google Scholar] [CrossRef]
- Shi, Y.Y.; Gao, L.L.; Wu, Z.C.; Zhang, X.J.; Wang, M.M.; Zhang, C.S.; Zhang, F.; Zhou, Y.L.; Li, Z.K. Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol. 2017, 17, 92. [Google Scholar] [CrossRef]
- Lekklar, C.; Pongpanich, M.; Suriya-Arunroj, D.; Chinpongpanich, A.; Tsai, H.; Comai, L.; Chadchawan, S.; Buaboocha, T. Genome-Wide Association Study for Salinity Tolerance at the Flowering Stage in a Panel of Rice Accessions from Thailand. BMC Genom. 2019, 20, 76. [Google Scholar] [CrossRef]
- Xu, P.; Guo, Q.; Meng, S.; Zhang, X.G.; Xu, Z.Z.; Guo, W.Z.; Shen, X.L. Genome-Wide Association Analysis Reveals Genetic Variations and Candidate Genes Associated with Salt Tolerance Related Traits in Gossypium Hirsutum. BMC Genom. 2021, 22, 26. [Google Scholar] [CrossRef]
- Do, T.D.; Vuong, T.D.; Dunn, D.; Smothers, S.; Patil, G.; Yungbluth, D.C.; Chen, P.Y.; Scaboo, A.; Xu, D.; Carter, T.E.; et al. Mapping and Confirmation of Loci for Salt Tolerance in a Novel Soybean Germplasm, Fiskeby III. Theor. Appl. Genet. 2018, 131, 513–524. [Google Scholar] [CrossRef]
- Liang, X.; Li, J.; Yang, Y.; Jiang, C.; Guo, Y. Designing salt stress-resilient crops: Current progress and future challenges. J. Integr. Plant Biol. 2023, 66, 303–329. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, A.; Brickner, J.H. Mechanisms of Epigenetic Memory. Trends Genet. 2014, 30, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Liu, X.B.; Lin, J.C.; Liu, X.Y.; Wang, Z.Y.; Xin, M.M.; Yao, Y.Y.; Peng, H.R.; Zhou, D.X.; Ni, Z.F.; et al. Histone Acetyltransferase GCN5 Contributes to Cell Wall Integrity and Salt Stress Tolerance by Altering the Expression of Cellulose Synthesis Genes. Plant J. 2019, 97, 587–602. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Paszkowski, J. Epigenetic Memory in Plants. EMBO J. 2014, 33, 1987–1998. [Google Scholar] [CrossRef]
- Yung, W.S.; Wang, Q.W.; Huang, M.K.; Wong, F.L.; Liu, A.L.; Ng, M.S.; Li, K.P.; Sze, C.C.; Li, M.W.; Lam, H.M. Priming-Induced Alterations in Histone Modifications Modulate Transcriptional Responses in Soybean under Salt Stress. Plant J. 2022, 109, 1575–1590. [Google Scholar] [CrossRef]
- Hiz, M.C.; Canher, B.; Niron, H.; Turet, M. Transcriptome Analysis of Salt Tolerant Common Bean (Phaseolus vulgaris L.) under Saline Conditions. PLoS ONE 2014, 9, e92598. [Google Scholar] [CrossRef]
- Raza, A.; Tabassum, J.; Fakhar, A.Z.; Sharif, R.; Chen, H.; Zhang, C.; Ju, L.; Fotopoulos, V.; Siddique, K.H.M.; Singh, R.K.; et al. Smart Reprograming of Plants against Salinity Stress Using Modern Biotechnological Tools. Crit. Rev. Biotechnol. 2023, 43, 1035–1062. [Google Scholar] [CrossRef]
- McGettigan, P.A. Transcriptomics in the RNA-Seq Era. Curr. Opin. Chem. Biol. 2013, 17, 4–11. [Google Scholar] [CrossRef]
- Liu, X.; Yang, X.X.; Zhang, B. Transcriptome analysis and functional identification of GmMYB46 in soybean seedlings under salt stress. PeerJ 2021, 9, e12492. [Google Scholar] [CrossRef]
- Sui, N.; Wang, Y.; Liu, S.S.; Yang, Z.; Wang, F.; Wan, S.B. Transcriptomic and Physiological Evidence for the Relationship between Unsaturated Fatty Acid and Salt Stress in Peanut. Front. Plant Sci. 2018, 9, 7. [Google Scholar] [CrossRef]
- Xing, B.Y.; Gu, C.R.; Zhang, T.X.; Zhang, Q.Z.; Yu, Q.B.; Jiang, J.; Liu, G.F. Functional Study of BpPP2C1 Revealed Its Role in Salt Stress in Betula Platyphylla. Front. Plant Sci. 2020, 11, 617635. [Google Scholar] [CrossRef] [PubMed]
- Gul, H.S.; Ulfat, M.; Zafar, Z.U.; Haider, W.; Ali, Z.; Manzoor, H.; Afzal, S.; Ashraf, M.; Athar, H.U.R. Photosynthesis and Salt Exclusion Are Key Physiological Processes Contributing to Salt Tolerance of Canola (Brassica napus L.): Evidence from Physiology and Transcriptome Analysis. Genes 2022, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.P.; Zhang, D.Q. The Role of Long Noncoding RNAs in Plant Stress Tolerance. Methods Mol. Biol. 2017, 1631, 41–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Z.; Liu, M.; Zhao, M.G.; Chen, R.; Zhang, W.H. Identification and characterization of long non-coding RNAs involved inosmotic and salt stress in Medicago truncatulausing genome-wide high-throughputsequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef]
- Li, W.Q.; Zheng, W.J.; Peng, Y.; Shao, Y.; Liu, C.T.; Li, J.; Hu, Y.Y.; Zhao, B.R.; Mao, B.G. OsPMS1 Mutation Enhances Salt Tolerance by Suppressing ROS Accumulation, Maintaining Na+/K+ Homeostasis, and Promoting ABA Biosynthesis. Genes 2023, 14, 1621. [Google Scholar] [CrossRef]
- Wen, D.N.; Bao, L.R.; Huang, X.Z.; Qian, X.D.; Chen, E.Y.; Shen, B. OsABT Is Involved in Abscisic Acid Signaling Pathway and Salt Tolerance of Roots at the Rice Seedling Stage. Int. J. Mol. Sci. 2022, 23, 10656. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, B.; Zhang, P.; Han, Q.H.; Zhao, G.W.; Zhao, F.C. Comparative Transcriptome Analysis Reveals the Underlying Response Mechanism to Salt Stress in Maize Seedling Roots. Metabolites 2023, 13, 1155. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Li, D.H.; Zhou, R.; Wang, X.; Dossa, K.; Wang, L.H.; Zhang, Y.X.; Yu, J.Y.; Gong, H.H.; Zhang, X.R.; et al. Transcriptome and Metabolome Analyses of Two Contrasting Sesame Genotypes Reveal the Crucial Biological Pathways Involved in Rapid Adaptive Response to Salt Stress. BMC Plant Biol. 2019, 19, 66. [Google Scholar] [CrossRef]
- Zou, L.J.; Li, T.T.; Li, B.B.; He, J.; Liao, C.L.; Wang, L.Z.; Xue, S.Y.; Sun, T.; Ma, X.; Wu, Q.G. De Novo Transcriptome Analysis Provides Insights into the Salt Tolerance of Podocarpus Macrophyllus under Salinity Stress. BMC Plant Biol. 2021, 21, 489. [Google Scholar] [CrossRef]
- Wang, Y.X.; Huang, L.Y.; Du, F.P.; Wang, J.; Zhao, X.Q.; Li, Z.K.; Wang, W.S.; Xu, J.L.; Fu, B.Y. Comparative Transcriptome and Metabolome Profiling Reveal Molecular Mechanisms Underlying OsDRAP1-Mediated Salt Tolerance in Rice. Sci. Rep. 2021, 11, 5166. [Google Scholar] [CrossRef]
- Wang, W.C.; Pang, J.Y.; Zhang, F.H.; Sun, L.P.; Yang, L.; Siddique, K.H.M. Transcriptomic and metabolomics-based analysis of key biological pathways reveals the role of lipid metabolism in response to salt stress in the root system of Brassica napus. Plant Growth Regul. 2022, 97, 127–141. [Google Scholar] [CrossRef]
- Li, M.X.; Guo, R.; Jiao, Y.; Jin, X.F.; Zhang, H.Y.; Shi, L.X. Comparison of Salt Tolerance in Soja Based on Metabolomics of Seedling Roots. Front. Plant Sci. 2017, 8, 1101. [Google Scholar] [CrossRef] [PubMed]
- Vaziriyeganeh, M.; Khan, S.; Zwiazek, J.J. Transcriptome and Metabolome Analyses Reveal Potential Salt Tolerance Mechanisms Contributing to Maintenance of Water Balance by the Halophytic Grass Puccinellia Nuttalliana. Front. Plant Sci. 2021, 12, 760863. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.W.; Li, Z.; Dai, S.J.; Ding, H.F.; Wang, Q.G.; Li, X.B.; Ding, G.H.; Wang, P.F.; Guan, Y.N.; Liu, W. Integrative Analyses of Transcriptomics and Metabolomics upon Seed Germination of Foxtail Millet in Response to Salinity. Sci. Rep. 2020, 10, 13660. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A.; Jha, R.; Parida, S.K. Salinity Stress Response and “omics” Approaches for Improving Salinity Stress Tolerance in Major Grain Legumes. Plant Cell Rep. 2019, 38, 255–277. [Google Scholar] [CrossRef]
- Sugiyama, A. The Soybean Rhizosphere: Metabolites, Microbes, and beyond-A Review. J. Adv. Res. 2019, 19, 67–73. [Google Scholar] [CrossRef]
- Wang, X.S.; Yin, J.C.; Wang, J.; Li, J.H. Integrative analysis of transcriptome and metabolome revealed the mechanisms by which flavonoids and phytohormones regulated the adaptation of alfalfa roots to NaCl stress. Front. Plant Sci. 2023, 14, 1117868. [Google Scholar] [CrossRef]
- Mahajan, M.; Yadav, S.K. Overexpression of a Tea Flavanone 3-Hydroxylase Gene Confers Tolerance to Salt Stress and Alternaria Solani in Transgenic Tobacco. Plant Mol. Biol. 2014, 85, 551–573. [Google Scholar] [CrossRef]
- Kim, J.; Lee, W.J.; Vu, T.T.; Jeong, C.Y.; Hong, S.W.; Lee, H. High Accumulation of Anthocyanins via the Ectopic Expression of AtDFR Confers Significant Salt Stress Tolerance in Brassica napus L. Plant Cell Rep. 2017, 36, 1215–1224. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Y.; Zhu, C.Y.; Yao, X.Y.; Zheng, Z.; Tian, Z.N.; Cai, X. EkFLS Overexpression Promotes Flavonoid Accumulation and Abiotic Stress Tolerance in Plant. Physiol. Plant 2021, 172, 1966–1982. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Siahpoosh, M.R.; Roessner, U.; Udvardi, M.; Kopka, J. Plant Metabolomics Reveals Conserved and Divergent Metabolic Responses to Salinity. Physiol. Plant 2008, 132, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tahir Ul Qamar, M.; Yang, L.; Liang, J.C.; You, J.; Wang, L.H. Current Progress, Applications and Challenges of Multi-Omics Approaches in Sesame Genetic Improvement. Int. J. Mol. Sci. 2023, 24, 3105. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Tang, X.Q.; Yang, X.Y.; Zhang, H.X. Comprehensive transcriptome and metabolome profiling reveal metabolic mechanisms of Nitraria sibirica Pall. to salt stress. Sci. Rep. 2021, 11, 12878. [Google Scholar] [CrossRef] [PubMed]
- Usman, B.; Derakhshani, B.; Jung, K.H. Recent Molecular Aspects and Integrated Omics Strategies for Understanding the Abiotic Stress Tolerance of Rice. Plants 2023, 12, 2019. [Google Scholar] [CrossRef]
- Zhang, H.X.; Blumwald, E. Transgenic Salt-Tolerant Tomato Plants Accumulate Salt in Foliage but Not in Fruit. Nat. Biotechnol. 2001, 19, 765–768. [Google Scholar] [CrossRef]
- Katori, T.; Ikeda, A.; Iuchi, S.; Kobayashi, M.; Shinozaki, K.; Maehashi, K.; Sakata, Y.; Tanaka, S.; Taji, T. Dissecting the Genetic Control of Natural Variation in Salt Tolerance of Arabidopsis Thaliana Accessions. J. Exp. Bot. 2010, 61, 1125–1138. [Google Scholar] [CrossRef]
- Kumar, G.; Basu, S.; Singla-Pareek, S.L.; Pareek, A. Unraveling the contribution of OsSOS2 in conferring salinity and drought tolerance in a high-yielding rice. Physiol. Plant 2022, 174, e13638. [Google Scholar] [CrossRef]
- Ma, J.C.; Lu, J.; Xu, J.M.; Duan, B.B.; He, X.D.; Liu, J.Q. Genome-Wide Identification of WRKY Genes in the Desert Poplar Populus Euphratica and Adaptive Evolution of the Genes in Response to Salt Stress. Evol. Bioinform. Online 2015, 11, 47–55. [Google Scholar] [CrossRef]
- Liaqat, A.; Alfatih, A.; Jan, S.U.; Sun, L.Q.; Zhao, P.X.; Xiang, C.B. Transcription Elongation Factor AtSPT4-2 Positively Modulates Salt Tolerance in Arabidopsis Thaliana. BMC Plant Biol. 2023, 23, 49. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.H.; Yu, W.C.; Wang, L.P.; Lan, Q.G.; Wang, Y.; Chen, C.B.; Zhang, Y. Knocking Out the Transcription Factor OsNAC092 Promoted Rice Drought Tolerance. Biology 2022, 11, 1830. [Google Scholar] [CrossRef]
- Alam, M.S.; Kong, J.R.; Tao, R.F.; Ahmed, T.; Alamin, M.; Alotaibi, S.S.; Abdelsalam, N.R.; Xu, J.H. CRISPR/Cas9 Mediated Knockout of the OsbHLH024 Transcription Factor Improves Salt Stress Resistance in Rice (Oryza sativa L.). Plants 2022, 11, 1184. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.J.; Ma, N.; Wang, C.Q.; Fan, H.F.; Wang, M.X.; Zhang, J.; Cao, J.F.; Wang, D.M. A Golgi-Localized Sodium/Hydrogen Exchanger Positively Regulates Salt Tolerance by Maintaining Higher K+/Na+ Ratio in Soybean. Front. Plant Sci. 2021, 12, 638340. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Mukherjee, A.; Das, P.; Bandyopadhyay, S.; Chattopadhyay, D.; Chatterjee, J.; Majumder, A.L. A Salt-tolerant Chloroplastic FBPase from Oryza Coarctata Confers Improved Photosynthesis with Higher Yield and Multi-stress Tolerance to Indica Rice. Plant Cell Tissue Organ. Cult. 2021, 145, 561–578. [Google Scholar] [CrossRef]
- Ayadi, M.; Brini, F.; Masmoudi, K. Overexpression of a Wheat Aquaporin Gene, TdPIP2;1, Enhances Salt and Drought Tolerance in Transgenic Durum Wheat Cv. Maali. Int. J. Mol. Sci. 2019, 20, 2389. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.X.; He, X.L.; Zhao, B.C.; Zhou, C.J.; Liang, Y.Z.; Ge, R.C.; Shen, Y.Z.; Huang, Z.J. Overexpressing a Putative Aquaporin Gene from Wheat, TaNIP, Enhances Salt Tolerance in Transgenic Arabidopsis. Plant Cell Physiol. 2010, 51, 767–775. [Google Scholar] [CrossRef]
- Ma, Q.J.; Sun, M.H.; Kang, H.; Lu, J.; You, C.X.; Hao, Y.J. A CIPK Protein Kinase Targets Sucrose Transporter MdSUT2.2 at Ser254 for Phosphorylation to Enhance Salt Tolerance. Plant Cell Environ. 2019, 42, 918–930. [Google Scholar] [CrossRef]
- Yu, Y.C.; Xuan, Y.; Bian, X.F.; Zhang, L.; Pan, Z.Y.; Kou, M.; Cao, Q.H.; Tang, Z.H.; Li, Q.; Ma, D.F.; et al. Overexpression of Phosphatidylserine Synthase IbPSS1 Affords Cellular Na+ Homeostasis and Salt Tolerance by Activating Plasma Membrane Na+/H+ Antiport Activity in Sweet Potato Roots. Hortic. Res. 2020, 7, 131. [Google Scholar] [CrossRef]
- Han, X.L.; Chen, Z.J.; Li, P.D.; Xu, H.S.; Liu, K.; Zha, W.J.; Li, S.H.; Chen, J.X.; Yang, G.C.; Huang, J.L.; et al. Development of Novel Rice Germplasm for Salt-Tolerance at Seedling Stage Using CRISPR-Cas9. Sustainability 2022, 14, 2621. [Google Scholar] [CrossRef]
- Zhang, A.N.; Liu, Y.; Wang, F.M.; Li, T.F.; Chen, Z.H.; Kong, D.Y.; Bi, J.G.; Zhang, F.Y.; Luo, X.X.; Wang, J.H.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.B.; Wang, Z.P.; Wang, Z.Q.; Shi, J.P.; Liang, X.Y.; Song, W.B.; Chen, Q.J.; Lai, J.S.; Jiang, C.F. A Retrotransposon in an HKT1 Family Sodium Transporter Causes Variation of Leaf Na+ Exclusion and Salt Tolerance in Maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef]
- Teng, Y.T.; Lv, M.; Zhang, X.X.; Cai, M.H.; Chen, T. BEAR1, a bHLH Transcription Factor, Controls Salt Response Genes to Regulate Rice Salt Response. J. Plant Biol. 2022, 65, 217–230. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.J.; Xia, J.X. OsNAC45 Is Involved in ABA Response and Salt Tolerance in Rice. Rice 2020, 13, 79. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.; Cao, H.; Liu, B. OsmiR535, a Potential Genetic Editing Target for Drought and Salinity Stress Tolerance in Oryza sativa. Plants 2020, 9, 1337. [Google Scholar] [CrossRef] [PubMed]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a Transcription Factor Downstream of Ethylene and Auxin Signaling Pathways, Is Involved in Salt Stress Response and Lateral Root Development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef]
- Yang, Z.M.; Yang, Y.X.; Dai, Z.G.; Xie, D.W.; Tang, Q.; Cheng, C.H.; Xu, Y.; Liu, C.; Deng, C.H.; Chen, J.Q.; et al. Construction of a high-resolution genetic map and identification of quantitative trait loci for salt tolerance in jute (Corchous spp.). BMC Plant Biol. 2019, 19, 391. [Google Scholar] [CrossRef]



| Species | Parents | Traits | QTL | Reference |
|---|---|---|---|---|
| Rice (Oryza Longistaminata) | 9311 × wild rice | Salt injury score and water content of seedling | qSIS2, qWCSST2 | [167] |
| Rice (Oryza sativa) | Huazhan × Nekken2 | Germination ability under salt stress | qST12.3 | [168] |
| Rice (Oryza sativa) | Wujiaozhan × Nipponbare | Percentage of germination under salt stress | qGR6.2 | [169] |
| Rice (Oryza sativa) | Dongxiang/Ningjing 15 × Ningjing16 | Salt tolerance of rice at seedling stage | qST1.2, qST6 | [170] |
| Wheat (Triticum aestivum L.) | WTSD91 × WN-64 | Na+ exclusion ability | qSNAX.2A.1, qSNAX.2A.2 | [171] |
| Wheat (Triticum aestivum L.) | Excalibur×Kukri | Maintenance of shoot growth under salinity, Na+ accumulation, Cl− accumulation, K+/Na+ ratio | QG(1-5).asl-5A, QG(1-5).asl-7B, QNa.asl-2A, QCl.asl-2A, QCl.asl-3A, QK:Na.asl-2DS2 | [172] |
| Zoysiagrass (Zoysia Japonica) | Z105 × Z061 | Salt tolerance traits | qLF-1, qLF-2, qSCW-1 | [173] |
| Cotton (Gossypium hirsutum L.) | GX1135 × GX100-2 | Yield component traits under salt stress | qLY-Chr6-2, qBNP-Chr4-1, qBNP-Chr12-1, qBNP-Chr15-5, qLP-Chr19-2, qLP-Chr5-3, qLP-Chr13-1, qBW-Chr5-5 | [174] |
| Species | Omics Method | Biological Processes Associated with Salt Stress | Reference |
|---|---|---|---|
| Rice (Oryza sativa) | Transcriptome | ROS homeostasis, ABA signaling pathway and osmotic and ionic homeostasis | [196] |
| Rice (Oryza sativa) | Transcriptome | ABA signaling pathway | [197] |
| Maize (Zea mays L.) | Transcriptome | The MAPK signaling pathway—plant and plant hormone signal transduction | [198] |
| Sesame (Sesamum indicum L.) | Transcriptome | Oxidation-reduction process and oxidoreductase activity | [199] |
| Podocarpus Macrophyllus | Transcriptome | The carbohydrate, glutamine, and xyloglucan metabolic pathways | [200] |
| Rice (Oryza sativa) | Metabolome | Secondary metabolites such as aminoadipic acid, calactin and satratoxin H and glycerylphosphorylethanolamine | [201] |
| Canola (Brassica napus) | Metabolome | Lipid metabolism | [202] |
| Soja (Glycine soja) | Metabolome | Amino acid metabolism, fatty acid metabolism, sugar alcohol metabolism, carboxylic acids, the TCA cycle, antioxidants from secondary metabolism and nucleic acids | [203] |
| Halophytic Grass (Puccinellia nuttalliana) | Metabolome | Proline, dopamine, phosphatidylcholines and the enriched TCA cycle in leaves | [204] |
| Foxtail millet (Setaria italica L.) | Metabolome | The biosynthetic pathways of phenylpropanoids, flavonoids, lignin and lysophospholipids | [205] |
| Gene Name | Molecular Strategy | Functions | Reference |
|---|---|---|---|
| GmNHX5 | Overexpression | Maintaining higher K+/Na+ ratio | [223] |
| PcCFR | Overexpression | Keeping the photosynthetic cycle by unabated generation of RuBP and retaining better light harvesting capacity of the leaves under stress | [224] |
| TdPIP2 | Overexpression | Reducing water evaporation from leaves | [225] |
| TaNIP | Overexpression | Regulating the balance of Na+ and K+ | [226] |
| MdSUT2.2 | Overexpression | Scavenging of ROS and transporting sucrose | [227] |
| IbPSS1 | Overexpression | Maintaining Na+ homeostasis | [228] |
| OsRR22 | Knock-out | Increasing plant height and total fresh weight | [229,230] |
| ZmHKT1 | Knock-out | Promoting Na+ exclusion of leaves | [231] |
| BEARI | Knock-out | Regulating the expression of salt-responsive genes and ions transport | [232] |
| OsNAC45 | Overexpression and knock-out | Regulating germination and seedling growth | [233] |
| OsmiR535 | Overexpression and knock-out | Improving resistance to NaCl | [234] |
| AtNAC2 | Overexpression and knock-out | Promoting the development of lateral roots | [235] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Feng, C.; Wang, Y.; Yun, C.; Zou, X.; Cheng, N.; Zhang, W.; Jing, Y.; Li, H. Understanding of Plant Salt Tolerance Mechanisms and Application to Molecular Breeding. Int. J. Mol. Sci. 2024, 25, 10940. https://doi.org/10.3390/ijms252010940
Zhou Y, Feng C, Wang Y, Yun C, Zou X, Cheng N, Zhang W, Jing Y, Li H. Understanding of Plant Salt Tolerance Mechanisms and Application to Molecular Breeding. International Journal of Molecular Sciences. 2024; 25(20):10940. https://doi.org/10.3390/ijms252010940
Chicago/Turabian StyleZhou, Yuxia, Chen Feng, Yuning Wang, Chunxia Yun, Xinqing Zou, Nuo Cheng, Wenping Zhang, Yan Jing, and Haiyan Li. 2024. "Understanding of Plant Salt Tolerance Mechanisms and Application to Molecular Breeding" International Journal of Molecular Sciences 25, no. 20: 10940. https://doi.org/10.3390/ijms252010940
APA StyleZhou, Y., Feng, C., Wang, Y., Yun, C., Zou, X., Cheng, N., Zhang, W., Jing, Y., & Li, H. (2024). Understanding of Plant Salt Tolerance Mechanisms and Application to Molecular Breeding. International Journal of Molecular Sciences, 25(20), 10940. https://doi.org/10.3390/ijms252010940





