Deletion of Slc1a4 Suppresses Single Mauthner Cell Axon Regeneration In Vivo through Growth-Associated Protein 43
Abstract
:1. Introduction
2. Results
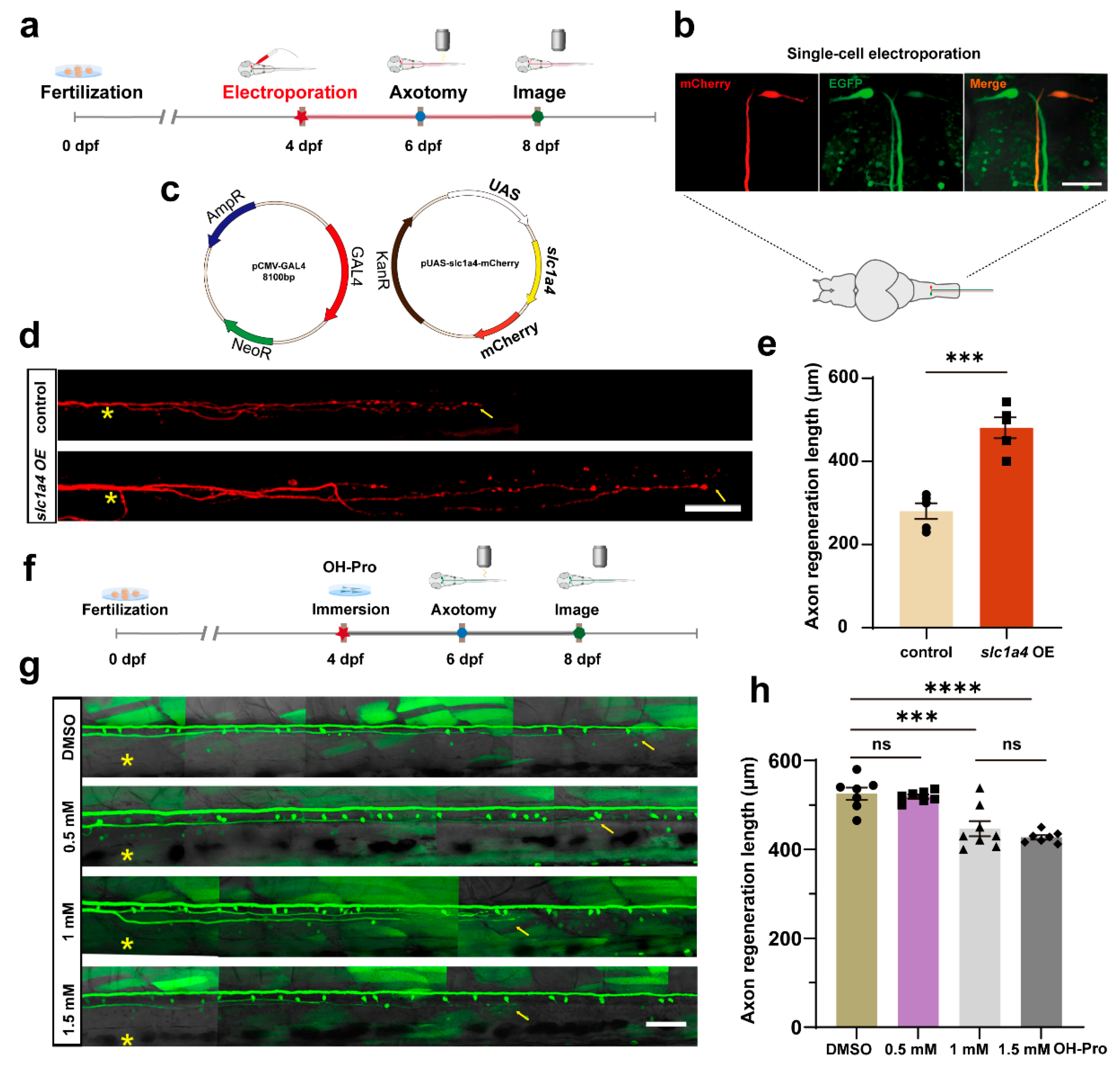
2.1. Slc1a4 Overexpression in Mauthner Cells Enhances Axon Regeneration In Vivo
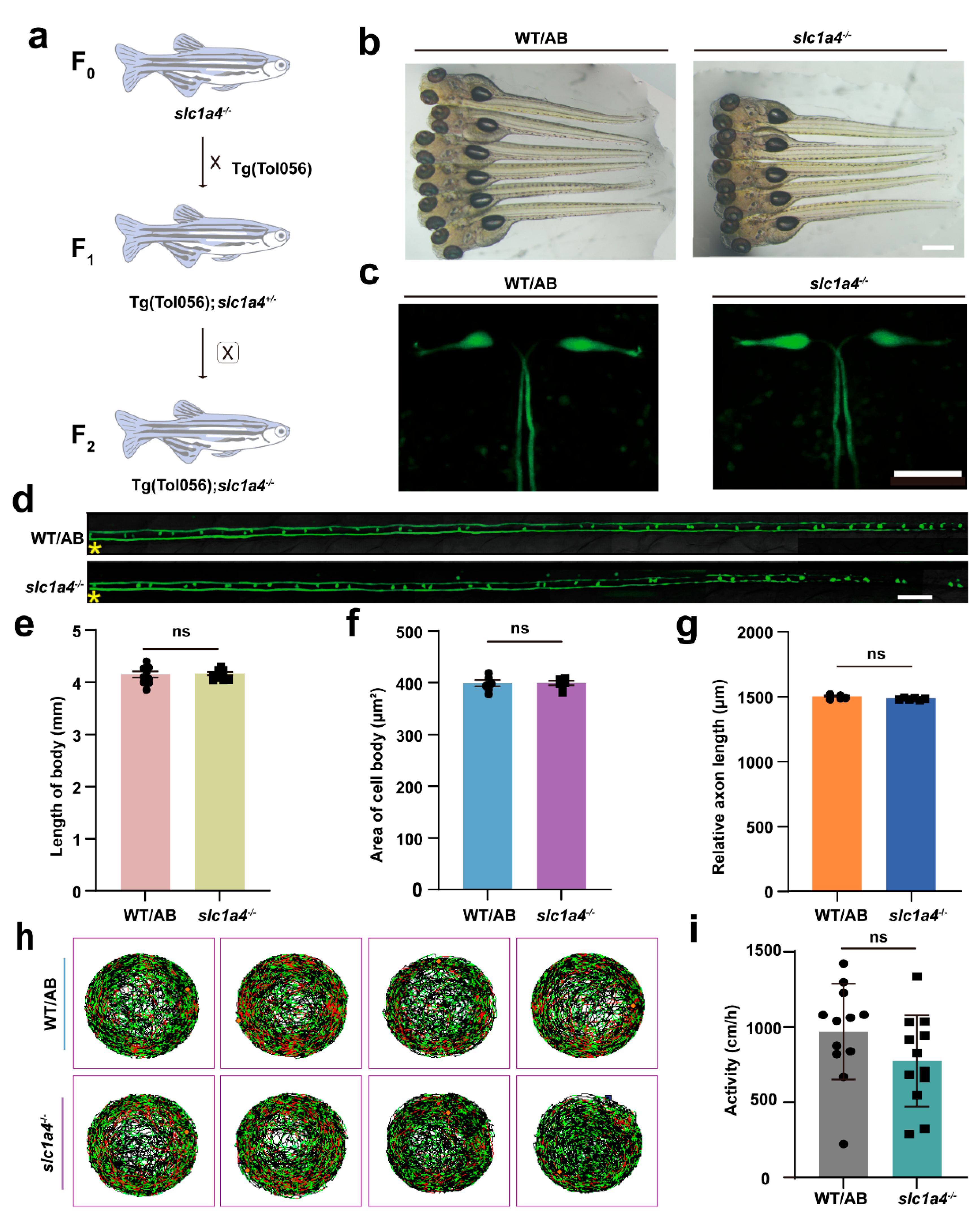
2.2. Identification of slc1a4 Mutant Zebrafish
2.3. Slc1a4 Deficiency Does Not Impact Mauthner Cell Development or Motor Function
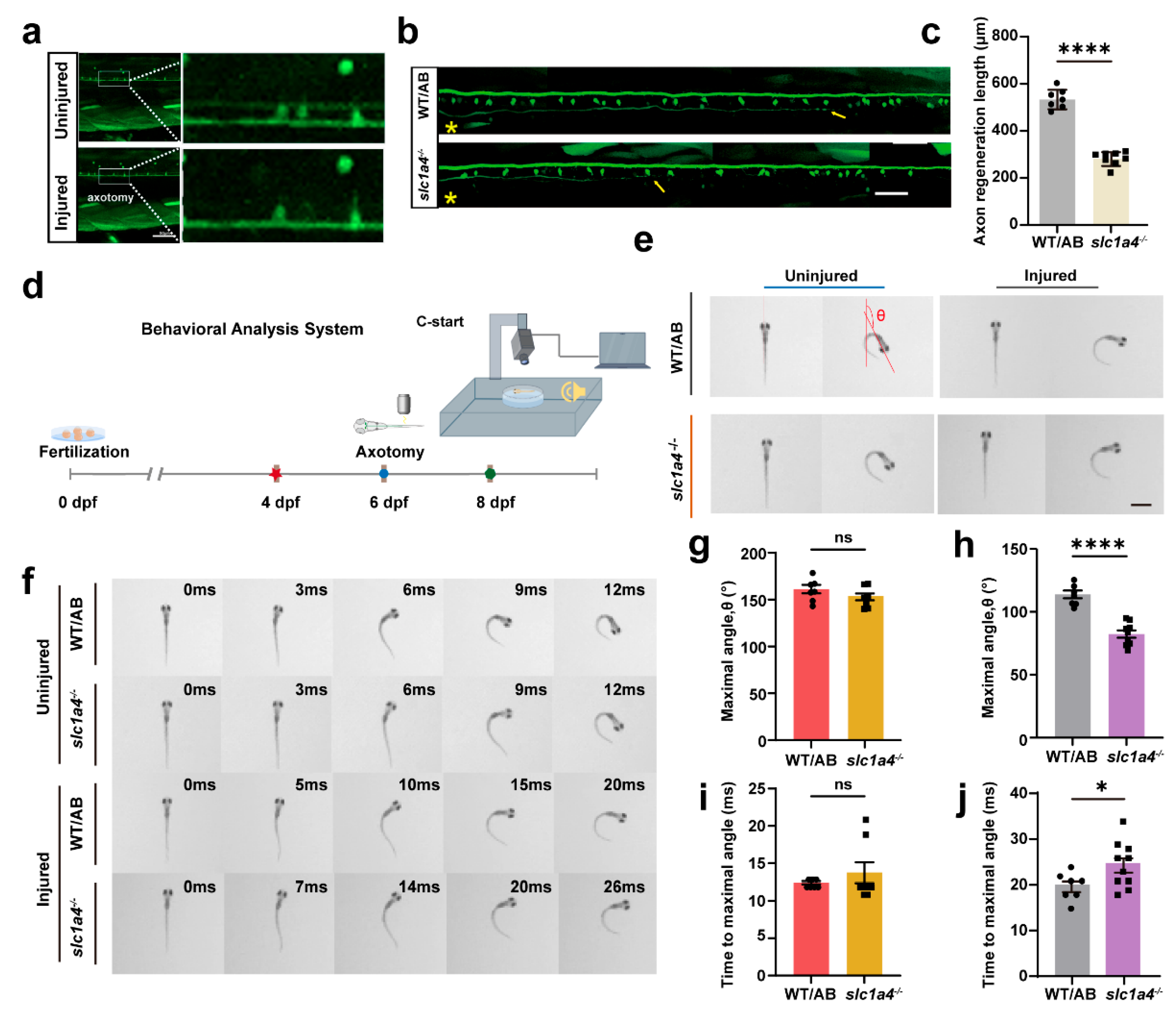
2.4. Slc1a4 Deficiency Suppresses Mauthner Cell Axon Regeneration and Associated Functions In Vivo
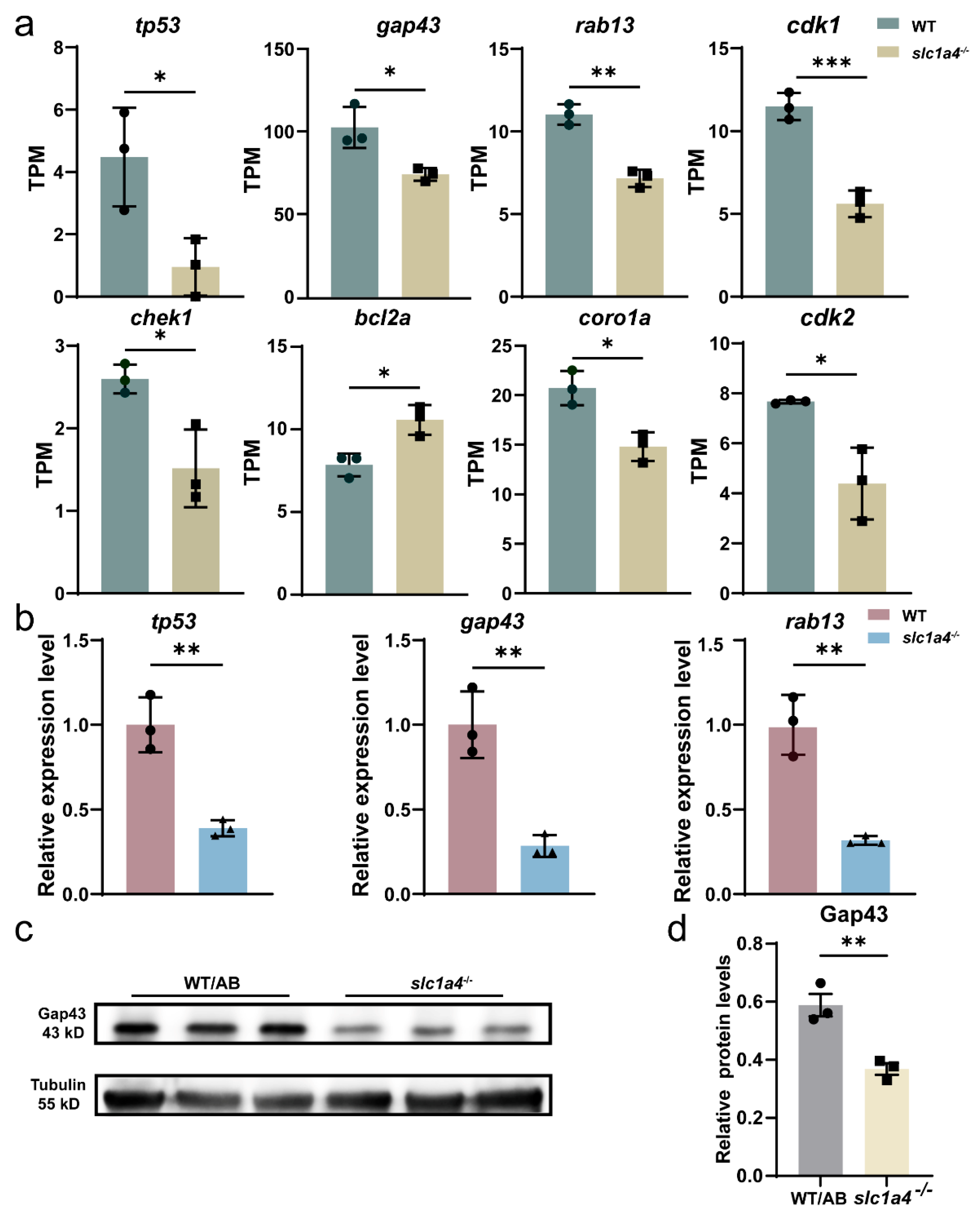
2.5. RNA-Seq Revealed That Slc1a4 May Influence Mauthner Axons’ Regeneration through P53 Signaling Pathway
2.6. Slc1a4 Deletion May Inhibit Mauthner Axons’ Regeneration via Gap43 Suppression
3. Discussion
4. Materials and Methods
4.1. Zebrafish Strains and Maintenance
4.2. Genome Editing
4.3. Single-Cell Electroporation
4.4. Two-Photon Axotomy
4.5. In Vivo Imaging
4.6. Quantitative Real-Time PCR
4.7. Protein Extraction and Western Blotting
4.8. Drug Treatment
4.9. Escape Behavior Assay
4.10. Statistical Analysis
4.11. RNA-Seq
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Middleton, J.; Tran, Y.; Craig, A. Relationship Between Quality of Life and Self-Efficacy in Persons With Spinal Cord Injuries. Arch. Phys. Med. Rehabil. 2007, 88, 1643–1648. [Google Scholar] [CrossRef] [PubMed]
- Functional Role of a Specialized Class of Spinal Commissural Inhibitory Neurons during Fast Escapes in Zebrafish|Journal of Neuroscience. Available online: https://www.jneurosci.org/content/29/21/6780.short (accessed on 25 June 2024).
- Fawcett, J.W. The Struggle to Make CNS Axons Regenerate: Why Has It Been so Difficult? Neurochem. Res. 2020, 45, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Hilton, B.J.; Husch, A.; Schaffran, B.; Lin, T.; Burnside, E.R.; Dupraz, S.; Schelski, M.; Kim, J.; Müller, J.A.; Schoch, S.; et al. An Active Vesicle Priming Machinery Suppresses Axon Regeneration upon Adult CNS Injury. Neuron 2022, 110, 51–69.e7. [Google Scholar] [CrossRef]
- Curcio, M.; Bradke, F. Axon Regeneration in the Central Nervous System: Facing the Challenges from the Inside. Annu. Rev. Cell Dev. Biol. 2018, 34, 495–521. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, A.; Bradke, F. Spatial and Temporal Arrangement of Neuronal Intrinsic and Extrinsic Mechanisms Controlling Axon Regeneration. Curr. Opin. Neurobiol. 2017, 42, 118–127. [Google Scholar] [CrossRef]
- Chondroitinase ABC Promotes Functional Recovery after Spinal Cord Injury|Nature. Available online: https://www.nature.com/articles/416636a (accessed on 10 July 2024).
- Silver, J.; Miller, J.H. Regeneration beyond the Glial Scar. Nat. Rev. Neurosci. 2004, 5, 146–156. [Google Scholar] [CrossRef]
- Liu, K.; Tedeschi, A.; Park, K.K.; He, Z. Neuronal Intrinsic Mechanisms of Axon Regeneration. Annu. Rev. Neurosci. 2011, 34, 131–152. [Google Scholar] [CrossRef]
- Wu, S.; Xu, J.; Dai, Y.; Yu, B.; Zhu, J.; Mao, S. Insight into Protein Synthesis in Axon Regeneration. Exp. Neurol. 2023, 367, 114454. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, B.; Chen, X.; Shi, X.; Li, S.; Zhang, Q.; Zhang, C.; Piao, F. Improvement of Diabetes-Induced Spinal Cord Axon Injury with Taurine via Nerve Growth Factor-Dependent Akt/mTOR Pathway. Amino Acids 2024, 56, 32. [Google Scholar] [CrossRef]
- Kaplan, E.; Zubedat, S.; Radzishevsky, I.; Valenta, A.C.; Rechnitz, O.; Sason, H.; Sajrawi, C.; Bodner, O.; Konno, K.; Esaki, K.; et al. ASCT1 (Slc1a4) Transporter Is a Physiologic Regulator of Brain <span Class=. Proc. Natl. Acad. Sci. USA 2018, 115, 9628–9633. [Google Scholar] [CrossRef]
- Barut, B.A.; Zon, L.I. Realizing the Potential of Zebrafish as a Model for Human Disease. Physiol. Genom. 2000, 2, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Zebrafish: An Emerging Model System for Human Disease and Drug Discovery—Kari—2007—Clinical Pharmacology & Therapeutics—Wiley Online Library. Available online: https://ascpt.onlinelibrary.wiley.com/doi/full/10.1038/sj.clpt.6100223 (accessed on 10 July 2024).
- The Zebrafish Reference Genome Sequence and Its Relationship to the Human Genome|Nature. Available online: https://www.nature.com/articles/nature12111 (accessed on 25 June 2024).
- Cells|Free Full-Text|Know How to Regrow—Axon Regeneration in the Zebrafish Spinal Cord. Available online: https://www.mdpi.com/2073-4409/10/6/1404 (accessed on 10 July 2024).
- Gonzalez, D.; Allende, M.L. Current Advances in Comprehending Dynamics of Regenerating Axons and Axon–Glia Interactions after Peripheral Nerve Injury in Zebrafish. Int. J. Mol. Sci. 2021, 22, 2484. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-H.; Shi, Z.-J.; Li, Y.; Pan, B.; Yuan, S.-Y.; Shi, L.-L.; Hao, Y.; Cao, F.-J.; Feng, S.-Q. Bioinformatic Identification of Key Candidate Genes and Pathways in Axon Regeneration after Spinal Cord Injury in Zebrafish. Neural Regen. Res. 2020, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Narushima, M.; Oda, Y. In Vivo Imaging of Functional Inhibitory Networks on the Mauthner Cell of Larval Zebrafish. J. Neurosci. 2002, 22, 3929–3938. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Sessions, S.K.; Kimmel, R.J. Morphogenesis and Synaptogenesis of the Zebrafish Mauthner Neuron. J. Comp. Neurol. 1981, 198, 101–120. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Shi, L.; Cai, Y.; Hu, B. Wolfram Syndrome 1b Mutation Suppresses Mauthner-Cell Axon Regeneration via ER Stress Signal Pathway. Acta Neuropathol. Commun. 2022, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Becker, C.G. Axonal Regeneration in Zebrafish. Curr. Opin. Neurobiol. 2014, 27, 186–191. [Google Scholar] [CrossRef]
- Kim, D.; Lee, Y.-R.; Choi, T.-I.; Kim, S.-H.; Kang, H.-C.; Kim, C.-H.; Lee, S. Comparative Proteome Research in a Zebrafish Model for Vanishing White Matter Disease. Int. J. Mol. Sci. 2021, 22, 2707. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.-R.J.; Joung, J.K. Efficient Genome Editing in Zebrafish Using a CRISPR-Cas System. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Cui, N.; Faure, G.; Singh, A.; Macrae, R.; Zhang, F. Microfluidic Enrichment and Computational Analysis of Rare Sequences from Mixed Genomic Samples for Metagenomic Mining. CRISPR J. 2022, 5, 677–684. [Google Scholar] [CrossRef]
- Scheldeman, C.; Mills, J.D.; Siekierska, A.; Serra, I.; Copmans, D.; Iyer, A.M.; Whalley, B.J.; Maes, J.; Jansen, A.C.; Lagae, L.; et al. mTOR-Related Neuropathology in Mutant Tsc2 Zebrafish: Phenotypic, Transcriptomic and Pharmacological Analysis. Neurobiol. Dis. 2017, 108, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Banu, S.; Gaur, N.; Nair, S.; Ravikrishnan, T.; Khan, S.; Mani, S.; Bharathi, S.; Mandal, K.; Kuram, N.A.; Vuppaladadium, S.; et al. Understanding the Complexity of Epimorphic Regeneration in Zebrafish: A Transcriptomic and Proteomic Approach. Genomics 2021, 114, 110300. [Google Scholar] [CrossRef] [PubMed]
- How Stimulus Direction Determines the Trajectory of the Mauthner-Inthated Escape Response in a Teleost Fish|Journal of Experimental Biology|The Company of Biologists. Available online: https://journals.biologists.com/jeb/article/161/1/469/6421/How-Stimulus-Direction-Determines-the-Trajectory (accessed on 26 June 2024).
- Liu, Y.-C.; Bailey, I.; Hale, M.E. Alternative Startle Motor Patterns and Behaviors in the Larval Zebrafish (Danio Rerio). J. Comp. Physiol. A 2012, 198, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.C.; Reichl, J.; Song, M.Y.; Dearinger, A.D.; Moridzadeh, N.; Lu, E.D.; Pearce, K.; Esdin, J.; Glanzman, D.L. Habituation of the C-Start Response in Larval Zebrafish Exhibits Several Distinct Phases and Sensitivity to NMDA Receptor Blockade. PLoS ONE 2011, 6, e29132. [Google Scholar] [CrossRef] [PubMed]
- Herrero-Turrión, M.J.; Rodríguez-Martín, I.; López-Bellido, R.; Rodríguez, R.E. Whole-Genome Expression Profile in Zebrafish Embryos after Chronic Exposure to Morphine: Identification of New Genes Associated with Neuronal Function and Mu Opioid Receptor Expression. BMC Genomics 2014, 15, 874. [Google Scholar] [CrossRef]
- Yeo, M.-K.; Pak, S.-W. Exposing Zebrafish to Silver Nanoparticles during Caudal Fin Regeneration Disrupts Caudal Fin Growth and P53 Signaling. Mol. Cell. Toxicol. 2008, 4, 311–317. [Google Scholar]
- Hu, Z.; Holzschuh, J.; Driever, W. Loss of DDB1 Leads to Transcriptional P53 Pathway Activation in Proliferating Cells, Cell Cycle Deregulation, and Apoptosis in Zebrafish Embryos. PLoS ONE 2015, 10, e0134299. [Google Scholar] [CrossRef]
- He, Z.; Jin, Y. Intrinsic Control of Axon Regeneration. Neuron 2016, 90, 437–451. [Google Scholar] [CrossRef]
- Sun, F.; He, Z. Neuronal Intrinsic Barriers for Axon Regeneration in the Adult CNS. Curr. Opin. Neurobiol. 2010, 20, 510–518. [Google Scholar] [CrossRef]
- Mahar, M.; Cavalli, V. Intrinsic Mechanisms of Neuronal Axon Regeneration. Nat. Rev. Neurosci. 2018, 19, 323–337. [Google Scholar] [CrossRef]
- PTEN Deletion Enhances the Regenerative Ability of Adult Corticospinal Neurons—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/20694004/ (accessed on 2 October 2024).
- Smith, T.P.; Sahoo, P.K.; Kar, A.N.; Twiss, J.L. Intra-Axonal Mechanisms Driving Axon Regeneration. Brain Res. 2020, 1740, 146864. [Google Scholar] [CrossRef] [PubMed]
- Kiyoshi, C.; Tedeschi, A. Axon Growth and Synaptic Function: A Balancing Act for Axonal Regeneration and Neuronal Circuit Formation in CNS Trauma and Disease. Dev. Neurobiol. 2020, 80, 277–301. [Google Scholar] [CrossRef] [PubMed]
- Renthal, W.; Tochitsky, I.; Yang, L.; Cheng, Y.-C.; Li, E.; Kawaguchi, R.; Geschwind, D.H.; Woolf, C.J. Transcriptional Reprogramming of Distinct Peripheral Sensory Neuron Subtypes after Axonal Injury. Neuron 2020, 108, 128–144.e9. [Google Scholar] [CrossRef] [PubMed]
- Neural Regeneration Research. Available online: https://journals.lww.com/nrronline/fulltext/2021/16010/Neuroregeneration_and_functional_recovery_after.13.aspx/1000 (accessed on 12 July 2024).
- Mutations in SLC1A4, Encoding the Brain Serine Transporter, Are Associated with Developmental Delay, Microcephaly and Hypomyelination|Journal of Medical Genetics. Available online: https://jmg.bmj.com/content/52/8/541.short (accessed on 13 July 2024).
- Pujol-Giménez, J.; Mirzaa, G.; Blue, E.E.; Albano, G.; Miller, D.E.; Allworth, A.; Bennett, J.T.; Byers, P.H.; Chanprasert, S.; Chen, J.; et al. Dominant-Negative Variant in SLC1A4 Causes an Autosomal Dominant Epilepsy Syndrome. Ann. Clin. Transl. Neurol. 2023, 10, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Odeh, M.; Sajrawi, C.; Majcher, A.; Zubedat, S.; Shaulov, L.; Radzishevsky, A.; Mizrahi, L.; Chung, W.K.; Avital, A.; Hornemann, T.; et al. A New Type of Blood-Brain Barrier Aminoacidopathy Underlies Metabolic Microcephaly Associated with SLC1A4 Mutations. Brain J. Neurol. 2024, awae134. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.C.; Bayik, D.; Storevik, S.; Moreino, S.S.; Sprowls, S.A.; Han, J.; Augustsson, M.T.; Lauko, A.; Sravya, P.; Røsland, G.V.; et al. GAP43-Dependent Mitochondria Transfer from Astrocytes Enhances Glioblastoma Tumorigenicity. Nat. Cancer 2023, 4, 648–664. [Google Scholar] [CrossRef]
- Zuo, Z.; Fan, B.; Zhang, Z.; Liang, Y.; Chi, J.; Li, G. Interleukin-4 Protects Retinal Ganglion Cells and Promotes Axon Regeneration. Cell Commun. Signal. CCS 2024, 22, 236. [Google Scholar] [CrossRef]
- Hu, M.; Veldman, M.B. Intraocular Axon Regeneration in a Model of Penetrating Eye Injury. J. Ocul. Pharmacol. Ther. Off. J. Assoc. Ocul. Pharmacol. Ther. 2023, 39, 563–571. [Google Scholar] [CrossRef]
- Williams, R.R.; Venkatesh, I.; Pearse, D.D.; Udvadia, A.J.; Bunge, M.B. MASH1/Ascl1a Leads to GAP43 Expression and Axon Regeneration in the Adult CNS. PLoS ONE 2015, 10, e0118918. [Google Scholar] [CrossRef]
- Skene, J.H.P. Axonal Growth-Associated Proteins. Annu. Rev. Neurosci. 1989, 12, 127–156. [Google Scholar] [CrossRef]
- Curtis, R.; Green, D.; Lindsay, R.M.; Wilkin, G.P. Up-Regulation of GAP-43 and Growth of Axons in Rat Spinal Cord after Compression Injury. J. Neurocytol. 1993, 22, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Schaden, H.; Stuermer, C.A.O.; Bähr, M. Gap-43 Immunoreactivity and Axon Regeneration in Retinal Ganglion Cells of the Rat. J. Neurobiol. 1994, 25, 1570–1578. [Google Scholar] [CrossRef]
- Zhang, Y.; Bo, X.; Schoepfer, R.; Holtmaat, A.J.D.G.; Verhaagen, J.; Emson, P.C.; Lieberman, A.R.; Anderson, P.N. Growth-Associated Protein GAP-43 and L1 Act Synergistically to Promote Regenerative Growth of Purkinje Cell Axons in Vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 14883–14888. [Google Scholar] [CrossRef] [PubMed]
- Kusik, B.W.; Hammond, D.R.; Udvadia, A.J. Transcriptional Regulatory Regions of Gap43 Needed in Developing and Regenerating Retinal Ganglion Cells. Dev. Dyn. 2010, 239, 482–495. [Google Scholar] [CrossRef]
- Carriel, V.; Garzón, I.; Campos, A.; Cornelissen, M.; Alaminos, M. Differential Expression of GAP-43 and Neurofilament during Peripheral Nerve Regeneration through Bio-Artificial Conduits: GAP-43 and Neurofilament during Nerve Regeneration. J. Tissue Eng. Regen. Med. 2017, 11, 553–563. [Google Scholar] [CrossRef]
- Kim, D.-C.; Kim, S.; Hwang, K.-S.; Kim, C.-H. P-Coumaric Acid Potently Down-Regulates Zebrafish Embryo Pigmentation: Comparison of in Vivo Assay and Computational Molecular Modeling with Phenylthiourea. Biomed. Sci. Lett. 2017, 23, 8–16. [Google Scholar] [CrossRef]
- Li, Z.; Ptak, D.; Zhang, L.; Walls, E.K.; Zhong, W.; Leung, Y.F. Phenylthiourea Specifically Reduces Zebrafish Eye Size. PLoS ONE 2012, 7, e40132. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-C.; Hsieh, Y.-S.; Chen, I.-H.; Wang, C.-H.; Chang, H.-W.; Yang, C.-C.; Ku, T.-H.; Yeh, S.-R.; Chuang, Y.-J. Combined Use of MS-222 (Tricaine) and Isoflurane Extends Anesthesia Time and Minimizes Cardiac Rhythm Side Effects in Adult Zebrafish. Available online: https://www.liebertpub.com/doi/10.1089/zeb.2010.0653 (accessed on 26 June 2024).
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ultrafast One-pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp—Chen—2023—iMeta—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/10.1002/imt2.107 (accessed on 25 June 2024).
- Rapid and Accurate Alignment of Nucleotide Conversion Sequencing Reads with HISAT-3N. Available online: https://genome.cshlp.org/content/31/7/1290 (accessed on 25 June 2024).
- Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype|Nature Biotechnology. Available online: https://www.nature.com/articles/s41587-019-0201-4 (accessed on 25 June 2024).
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data—ScienceDirect. Available online: https://www.sciencedirect.com/science/article/pii/S2666675821000667?via%3Dihub (accessed on 26 June 2024).
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]


| Primers | Sequences (5′-3′) |
|---|---|
| β-actin-qPCR-F | CATTGGCAATGAGCGTTTC |
| β-actin-qPCR-R | TACTCCTGCTTGCTGATCCAC |
| tp53-qPCR-F | TGGAGAGGAGGTCGGCAAAATCAA |
| tp53-qPCR-R | GACTGCGGGAACCTGAGCCTAAAT |
| gap43-qPCR-F | TGCTGCATCAGAAGAACTAA |
| gap43-qPCR-R | CCTCCGGTTTGATTCCATC |
| rab13-qPCT-F | GCATACTACAGAGGGGCCA |
| rab13-qPCT-R | CATTCGACTTACACCCGCTG |
| slc1a4-sgRNA-F | TAATACGACTCACTATAGGCGAAATCAACGGACACGCTGTTTTAGAGCTAGAAATAGC |
| slc1a4-sgRNA-R | AGCACCGACTCGGTGCCACT |
| slc1a4-qPCR-F | TACCATCATCCCTAGCCCGA |
| slc1a4-qPCR-R | ATGGAGGAGAAGAAGAGCGAAATCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Fan, D.; Zhou, J.; Zhao, Z.; Han, A.; Song, Z.; Tang, X.; Hu, B. Deletion of Slc1a4 Suppresses Single Mauthner Cell Axon Regeneration In Vivo through Growth-Associated Protein 43. Int. J. Mol. Sci. 2024, 25, 10950. https://doi.org/10.3390/ijms252010950
Li K, Fan D, Zhou J, Zhao Z, Han A, Song Z, Tang X, Hu B. Deletion of Slc1a4 Suppresses Single Mauthner Cell Axon Regeneration In Vivo through Growth-Associated Protein 43. International Journal of Molecular Sciences. 2024; 25(20):10950. https://doi.org/10.3390/ijms252010950
Chicago/Turabian StyleLi, Keqiang, Dinggang Fan, Junhui Zhou, Ziang Zhao, Along Han, Zheng Song, Xiahui Tang, and Bing Hu. 2024. "Deletion of Slc1a4 Suppresses Single Mauthner Cell Axon Regeneration In Vivo through Growth-Associated Protein 43" International Journal of Molecular Sciences 25, no. 20: 10950. https://doi.org/10.3390/ijms252010950






