The Putative Role of TIM-3 Variants in Polyendocrine Autoimmunity: Insights from a WES Investigation
Abstract
1. Introduction
2. Results
2.1. Identification and Structural Characterization of TIM-3 Variants
2.2. Evaluation of TIM-3 Expression on PBMCs
2.3. Exon 2 TIM-3 Sequencing in APS Patients’ DNAs
3. Discussion
4. Materials and Methods
4.1. Patients
4.1.1. Case History
4.1.2. Study Population
4.2. Autoantibody Screening
4.3. Molecular Studies
4.3.1. AIRE Gene Screening
4.3.2. Screening for the Presence of C1858T PTPN22
4.3.3. Whole-Exome Sequencing
4.3.4. Structural Modeling and Simulation
4.3.5. Screening for the Presence of Exon 2 TIM-3 Variants
4.4. Functional Studies
4.4.1. Confocal Microscopy Analysis
4.4.2. RT-qPCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Nomenclature
References
- Betterle, C.; Sabbadin, C.; Scaroni, C.; Presotto, F. Autoimmune Polyendocrine Syndromes (APS) or Multiple Autoimmune Syndromes (MAS). In Polyendocrine Disorders and Endocrine Neoplastic Syndromes; Colao, A., Jaffrain-Rea, M.-L., Beckers, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–50. [Google Scholar]
- Betterle, C.; Furmaniak, J.; Sabbadin, C.; Scaroni, C.; Presotto, F. Type 3 autoimmune polyglandular syndrome (APS-3) or type 3 multiple autoimmune syndrome (MAS-3): An expanding galaxy. J. Endocrinol. Investig. 2023, 46, 643–665. [Google Scholar] [CrossRef] [PubMed]
- Cudini, A.; Nardella, C.; Bellacchio, E.; Palma, A.; Delfino, D.V.; Betterle, C.; Cappa, M.; Fierabracci, A. Analysis of the AIRE Gene Promoter in Patients Affected by Autoimmune Polyendocrine Syndromes. Int. J. Mol. Sci. 2024, 25, 2656. [Google Scholar] [CrossRef] [PubMed]
- Fierabracci, A. Type 1 Diabetes in Autoimmune Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy Syndrome (APECED): A “Rare” Manifestation in a “Rare” Disease. Int. J. Mol. Sci. 2016, 17, 1106. [Google Scholar] [CrossRef]
- Frommer, L.; Kahaly, G.J. Autoimmune Polyendocrinopathy. J. Clin. Endocrinol. Metab. 2019, 104, 4769–4782. [Google Scholar] [CrossRef]
- Smith, T.J.; Hegedüs, L. Graves’ Disease. N. Engl. J. Med. 2016, 375, 1552–1565. [Google Scholar] [CrossRef]
- Ralli, M.; Angeletti, D.; Fiore, M.; D’Aguanno, V.; Lambiase, A.; Artico, M.; de Vincentiis, M.; Greco, A. Hashimoto’s thyroiditis: An update on pathogenic mechanisms, diagnostic protocols, therapeutic strategies, and potential malignant transformation. Autoimmun. Rev. 2020, 19, 102649. [Google Scholar] [CrossRef] [PubMed]
- Gianchecchi, E.; Palombi, M.; Fierabracci, A. The putative role of the C1858T polymorphism of protein tyrosine phosphatase PTPN22 gene in autoimmunity. Autoimmun. Rev. 2013, 12, 717–725. [Google Scholar] [CrossRef]
- Vang, T.; Congia, M.; Macis, M.D.; Musumeci, L.; Orrú, V.; Zavattari, P.; Nika, K.; Tautz, L.; Taskén, K.; Cucca, F.; et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat. Genet. 2005, 37, 1317–1319. [Google Scholar] [CrossRef]
- Houcken, J.; Degenhart, C.; Bender, K.; König, J.; Frommer, L.; Kahaly, G.J. PTPN22 and CTLA-4 Polymorphisms Are Associated With Polyglandular Autoimmunity. J. Clin. Endocrinol. Metab. 2018, 103, 1977–1984. [Google Scholar] [CrossRef]
- Downes, K.; Marcovecchio, M.L.; Clarke, P.; Cooper, J.D.; Ferreira, R.C.; Howson, J.M.; Jolley, J.; Nutland, S.; Stevens, H.E.; Walker, N.M.; et al. Plasma concentrations of soluble IL-2 receptor α (CD25) are increased in type 1 diabetes and associated with reduced C-peptide levels in young patients. Diabetologia 2014, 57, 366–372. [Google Scholar] [CrossRef]
- Khan, S.; Mandal, R.K.; Jawed, A.; Dar, S.A.; Wahid, M.; Panda, A.K.; Areeshi, M.Y.; Ahmed Khan, M.E.; Haque, S. TNF-α -308 G > A (rs1800629) Polymorphism is Associated with Celiac Disease: A Meta-analysis of 11 Case-Control Studies. Sci. Rep. 2016, 6, 32677. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, M.; Kaczmarczyk, A.; Bischofs, C.; Kahaly, G.J. The Proinflammatory Cytokine TNF-α -308 AA Genotype is Associated with Polyglandular Autoimmunity. Immunol. Investig. 2009, 38, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Grover, P.; Goel, P.N.; Greene, M.I. Regulatory T Cells: Regulation of Identity and Function. Front. Immunol. 2021, 12, 750542. [Google Scholar] [CrossRef]
- Bauer, S.; Groh, V.; Wu, J.; Steinle, A.; Phillips, J.H.; Lanier, L.L.; Spies, T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 1999, 285, 727–729. [Google Scholar] [CrossRef] [PubMed]
- Ide, M.; Dittmar, M.; Wurm, M.; Kanitz, M.; Kahaly, G.J. Polymorphisms of MICA microsatellites in thyroidal autoimmunity. Med. Klin. 2007, 102, 11–15. [Google Scholar] [CrossRef]
- Tomer, Y.; Menconi, F. Type 1 diabetes and autoimmune thyroiditis: The genetic connection. Thyroid 2009, 19, 99–102. [Google Scholar] [CrossRef]
- Dittmar, M.; Kahaly, G.J. Immunoregulatory and susceptibility genes in thyroid and polyglandular autoimmunity. Thyroid 2005, 15, 239–250. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Oftedal, B.E.; Wolff, A.B.; Husebye, E.S. AIRE-mutations and autoimmune disease. Curr. Opin. Immunol. 2016, 43, 8–15. [Google Scholar] [CrossRef]
- Eriksson, D.; Røyrvik, E.C.; Aranda-Guillén, M.; Berger, A.H.; Landegren, N.; Artaza, H.; Hallgren, Å.; Grytaas, M.A.; Ström, S.; Bratland, E.; et al. GWAS for autoimmune Addison’s disease identifies multiple risk loci and highlights AIRE in disease susceptibility. Nat. Commun. 2021, 12, 959. [Google Scholar] [CrossRef]
- Oftedal, B.E.; Assing, K.; Baris, S.; Safgren, S.L.; Johansen, I.S.; Jakobsen, M.A.; Babovic-Vuksanovic, D.; Agre, K.; Klee, E.W.; Majcic, E.; et al. Dominant-negative heterozygous mutations in AIRE confer diverse autoimmune phenotypes. iScience 2023, 26, 106818. [Google Scholar] [CrossRef]
- Oftedal, B.E.; Hellesen, A.; Erichsen, M.M.; Bratland, E.; Vardi, A.; Perheentupa, J.; Kemp, E.H.; Fiskerstrand, T.; Viken, M.K.; Weetman, A.P.; et al. Dominant Mutations in the Autoimmune Regulator AIRE Are Associated with Common Organ-Specific Autoimmune Diseases. Immunity 2015, 42, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Retterer, K.; Juusola, J.; Cho, M.T.; Vitazka, P.; Millan, F.; Gibellini, F.; Vertino-Bell, A.; Smaoui, N.; Neidich, J.; Monaghan, K.G.; et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016, 18, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415, 536–541. [Google Scholar] [CrossRef]
- Moonla, C.; Polprasert, C.; Komvilaisak, P.; Rattanathammethee, T.; Kongkiatkamon, S.; Wudhikarn, K.; Kobbuaklee, S.; Boonyabaramee, P.; Tangcheewinsirikul, N.; Pakakasama, S.; et al. Germline HAVCR2 mutations and their relation to the clinical spectrum of subcutaneous panniculitis-like T-cell lymphoma and hemophagocytic lymphohistiocytosis: Results from a multicenter study and meta-analysis. Haematologica 2023, 108, 2743–2752. [Google Scholar] [CrossRef] [PubMed]
- DeKruyff, R.H.; Bu, X.; Ballesteros, A.; Santiago, C.; Chim, Y.L.; Lee, H.H.; Karisola, P.; Pichavant, M.; Kaplan, G.G.; Umetsu, D.T.; et al. T cell/transmembrane, Ig, and mucin-3 allelic variants differentially recognize phosphatidylserine and mediate phagocytosis of apoptotic cells. J. Immunol. 2010, 184, 1918–1930. [Google Scholar] [CrossRef]
- Tromp, S.A.M.; Gillissen, M.A.; Bernelot Moens, S.J.; van Leeuwen, E.M.M.; Jansen, M.H.; Koens, L.; Rutten, C.E.; Kuijpers, T.W. Treatment of an HLH-mimic disease based on HAVCR2 variants with absent TIM-3 expression. Blood Adv. 2022, 6, 4501–4505. [Google Scholar] [CrossRef]
- Fierabracci, A.; Belcastro, E.; Carbone, E.; Pagliarosi, O.; Palma, A.; Pacillo, L.; Giancotta, C.; Zangari, P.; Finocchi, A.; Cancrini, C.; et al. In Search for the Missing Link in APECED-like Conditions: Analysis of the AIRE Gene in a Series of 48 Patients. J. Clin. Med. 2022, 11, 3242. [Google Scholar] [CrossRef]
- Zhao, L.; Cheng, S.; Fan, L.; Zhang, B.; Xu, S. TIM-3: An update on immunotherapy. Int. Immunopharmacol. 2021, 99, 107933. [Google Scholar] [CrossRef]
- Freeman, G.J.; Casasnovas, J.M.; Umetsu, D.T.; DeKruyff, R.H. TIM genes: A family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol. Rev. 2010, 235, 172–189. [Google Scholar] [CrossRef]
- Nakayama, M.; Akiba, H.; Takeda, K.; Kojima, Y.; Hashiguchi, M.; Azuma, M.; Yagita, H.; Okumura, K. Tim-3 mediates phagocytosis of apoptotic cells and cross-presentation. Blood 2009, 113, 3821–3830. [Google Scholar] [CrossRef]
- Tang, D.; Lotze, M.T. Tumor immunity times out: TIM-3 and HMGB1. Nat. Immunol. 2012, 13, 808–810. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Zhu, C.; Kondo, Y.; Anderson, A.C.; Gandhi, A.; Russell, A.; Dougan, S.K.; Petersen, B.S.; Melum, E.; Pertel, T.; et al. Corrigendum: CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 2016, 536, 359. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Anderson, D.E.; Bregoli, L.; Hastings, W.D.; Kassam, N.; Lei, C.; Chandwaskar, R.; Karman, J.; Su, E.W.; Hirashima, M.; et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 2007, 318, 1141–1143. [Google Scholar] [CrossRef] [PubMed]
- Hastings, W.D.; Anderson, D.E.; Kassam, N.; Koguchi, K.; Greenfield, E.A.; Kent, S.C.; Zheng, X.X.; Strom, T.B.; Hafler, D.A.; Kuchroo, V.K. TIM-3 is expressed on activated human CD4+ T cells and regulates Th1 and Th17 cytokines. Eur. J. Immunol. 2009, 39, 2492–2501. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Joller, N.; Kuchroo, V.K. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 2016, 44, 989–1004. [Google Scholar] [CrossRef]
- Rakova, J.; Truxova, I.; Holicek, P.; Salek, C.; Hensler, M.; Kasikova, L.; Pasulka, J.; Holubova, M.; Kovar, M.; Lysak, D.; et al. TIM-3 levels correlate with enhanced NK cell cytotoxicity and improved clinical outcome in AML patients. Oncoimmunology 2021, 10, 1889822. [Google Scholar] [CrossRef]
- Sánchez-Fueyo, A.; Tian, J.; Picarella, D.; Domenig, C.; Zheng, X.X.; Sabatos, C.A.; Manlongat, N.; Bender, O.; Kamradt, T.; Kuchroo, V.K.; et al. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat. Immunol. 2003, 4, 1093–1101. [Google Scholar] [CrossRef]
- Xu, J.; Yang, Y.; Liu, X.; Wang, Y. The -1541 C>T and +4259 G>T of TIM-3 polymorphisms are associated with rheumatoid arthritis susceptibility in a Chinese Hui population. Int. J. Immunogenet. 2011, 38, 513–518. [Google Scholar] [CrossRef]
- Yaghoobi, E.; Abedian, S.; Babani, O.; Izad, M. TIM-3 Rs10515746 (A/C) and Rs10053538 (C/A) Gene Polymorphisms and Risk of Multiple Sclerosis. Iran. J. Public Health 2016, 45, 644–649. [Google Scholar]
- Guo, L.; Yang, X.; Xia, Q.; Zhen, J.; Zhuang, X.; Peng, T. Expression of human T cell immunoglobulin domain and mucin-3 (TIM-3) on kidney tissue from systemic lupus erythematosus (SLE) patients. Clin. Exp. Med. 2014, 14, 383–388. [Google Scholar] [CrossRef]
- Koguchi, K.; Anderson, D.E.; Yang, L.; O’Connor, K.C.; Kuchroo, V.K.; Hafler, D.A. Dysregulated T cell expression of TIM3 in multiple sclerosis. J. Exp. Med. 2006, 203, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Hosomi, S.; Yamagami, H.; Watanabe, K.; Kamata, N.; Sogawa, M.; Machida, H.; Okazaki, H.; Tanigawa, T.; Nagahara, H.; et al. Dysregulated upregulation of T-cell immunoglobulin and mucin domain-3 on mucosal T helper 1 cells in patients with Crohn’s disease. Scand. J. Gastroenterol. 2011, 46, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Guo, X.; Jiang, X.; Zhou, P.; Xiao, Y.; Zhou, T.; Chen, G.; Zhao, Z.; Xiao, H.; Hou, C.; et al. Dysregulated Tim-3 expression and its correlation with imbalanced CD4 helper T cell function in ulcerative colitis. Clin. Immunol. 2012, 145, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Z.; Xiao, Y.; Chen, H.; Zhou, Z. Altered expression of Tim family molecules and an imbalanced ratio of Tim-3 to Tim-1 expression in patients with type 1 diabetes. Front. Endocrinol. 2022, 13, 937109. [Google Scholar] [CrossRef]
- Boehne, C.; Behrendt, A.K.; Meyer-Bahlburg, A.; Boettcher, M.; Drube, S.; Kamradt, T.; Hansen, G. Tim-3 is dispensable for allergic inflammation and respiratory tolerance in experimental asthma. PLoS ONE 2021, 16, e0249605. [Google Scholar] [CrossRef]
- Eilers, L.F.; Kyle, W.B.; Allen, H.D.; Qureshi, A.M. Patent Ductus Arteriosus. Pediatr. Rev. 2021, 42, 632–634. [Google Scholar] [CrossRef]
- Neufeld, M.; Maclaren, N.; Blizzard, R. Autoimmune polyglandular syndromes. Pediatr. Ann. 1980, 9, 154–162. [Google Scholar] [CrossRef]
- Husebye, E.S.; Anderson, M.S.; Kämpe, O. Autoimmune Polyendocrine Syndromes. N. Engl. J. Med. 2018, 378, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Palma, A.; Gianchecchi, E.; Palombi, M.; Luciano, R.; Di Carlo, P.; Crinò, A.; Cappa, M.; Fierabracci, A. Analysis of the autoimmune regulator gene in patients with autoimmune non-APECED polyendocrinopathies. Genomics 2013, 102, 163–168. [Google Scholar] [CrossRef]
- Arena, A.; Belcastro, E.; Ceccacci, F.; Petrini, S.; Conti, L.A.; Pagliarosi, O.; Giorda, E.; Sennato, S.; Schiaffini, R.; Wang, P.; et al. Improvement of Lipoplexes With a Sialic Acid Mimetic to Target the C1858T PTPN22 Variant for Immunotherapy in Endocrine Autoimmunity. Front. Immunol. 2022, 13, 838331. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7.20.1–7.20.41. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Ittisoponpisan, S.; Islam, S.A.; Khanna, T.; Alhuzimi, E.; David, A.; Sternberg, M.J.E. Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated? J. Mol. Biol. 2019, 431, 2197–2212. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Biagini, T.; Chillemi, G.; Mazzoccoli, G.; Grottesi, A.; Fusilli, C.; Capocefalo, D.; Castellana, S.; Vescovi, A.L.; Mazza, T. Molecular dynamics recipes for genome research. Brief. Bioinform. 2017, 19, 853–862. [Google Scholar] [CrossRef]
- Cocciadiferro, D.; Mazza, T.; Vecchio, D.; Biagini, T.; Petrizzelli, F.; Agolini, E.; Villani, A.; Minervino, D.; Martinelli, D.; Rizzo, C.; et al. Exploiting in silico structural analysis to introduce emerging genotype-phenotype correlations in DHCR24-related sterol biosynthesis disorder: A case study. Front. Genet. 2023, 14, 1307934. [Google Scholar] [CrossRef]
- Biagini, T.; Petrizzelli, F.; Truglio, M.; Cespa, R.; Barbieri, A.; Capocefalo, D.; Castellana, S.; Tevy, M.F.; Carella, M.; Mazza, T. Are Gaming-Enabled Graphic Processing Unit Cards Convenient for Molecular Dynamics Simulation? Evol. Bioinform. Online 2019, 15, 1176934319850144. [Google Scholar] [CrossRef]
- Suh, D.; Feng, S.; Lee, H.; Zhang, H.; Park, S.J.; Kim, S.; Lee, J.; Choi, S.; Im, W. CHARMM-GUI Enhanced Sampler for various collective variables and enhanced sampling methods. Protein Sci. 2022, 31, e4446. [Google Scholar] [CrossRef] [PubMed]
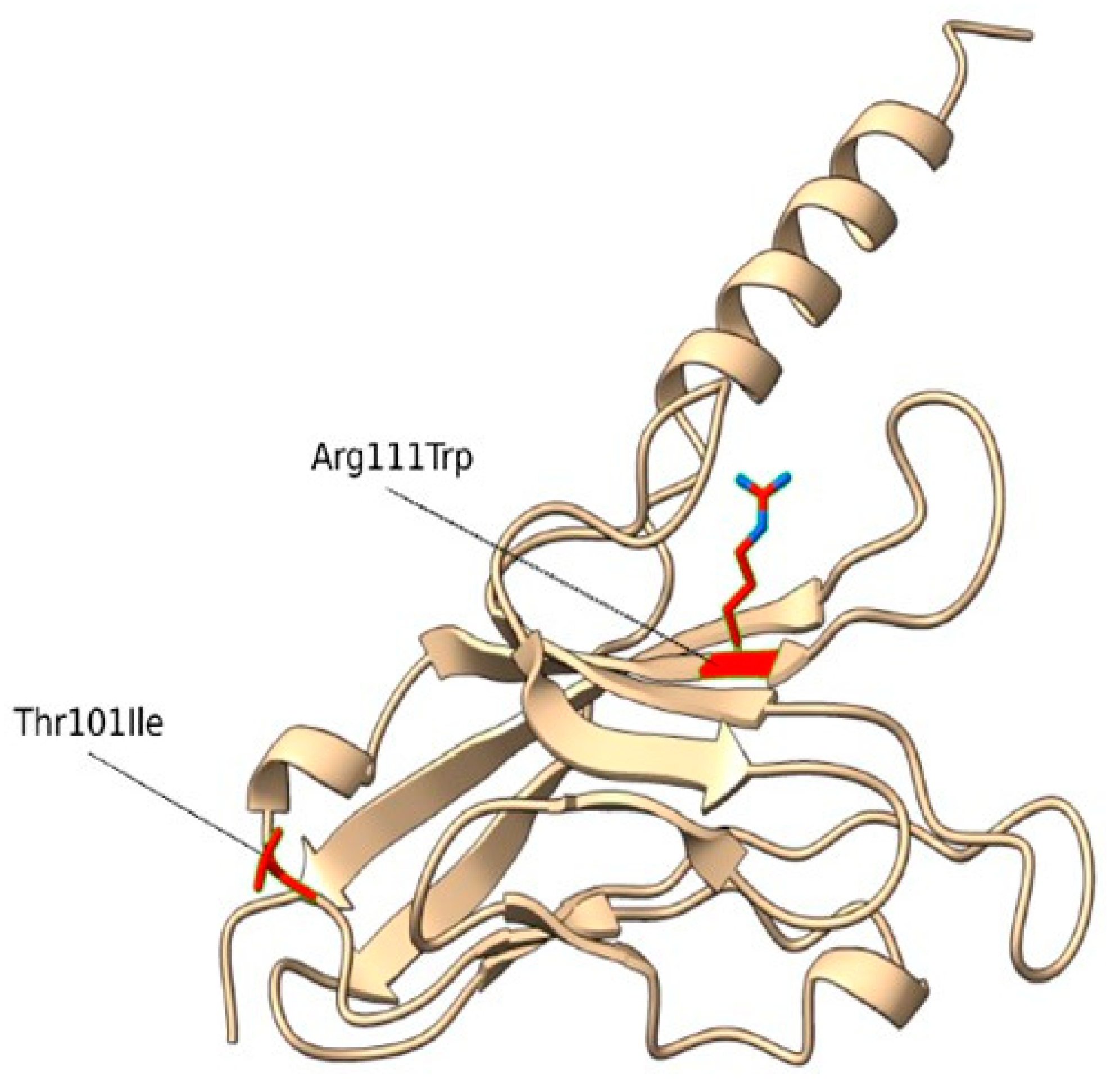
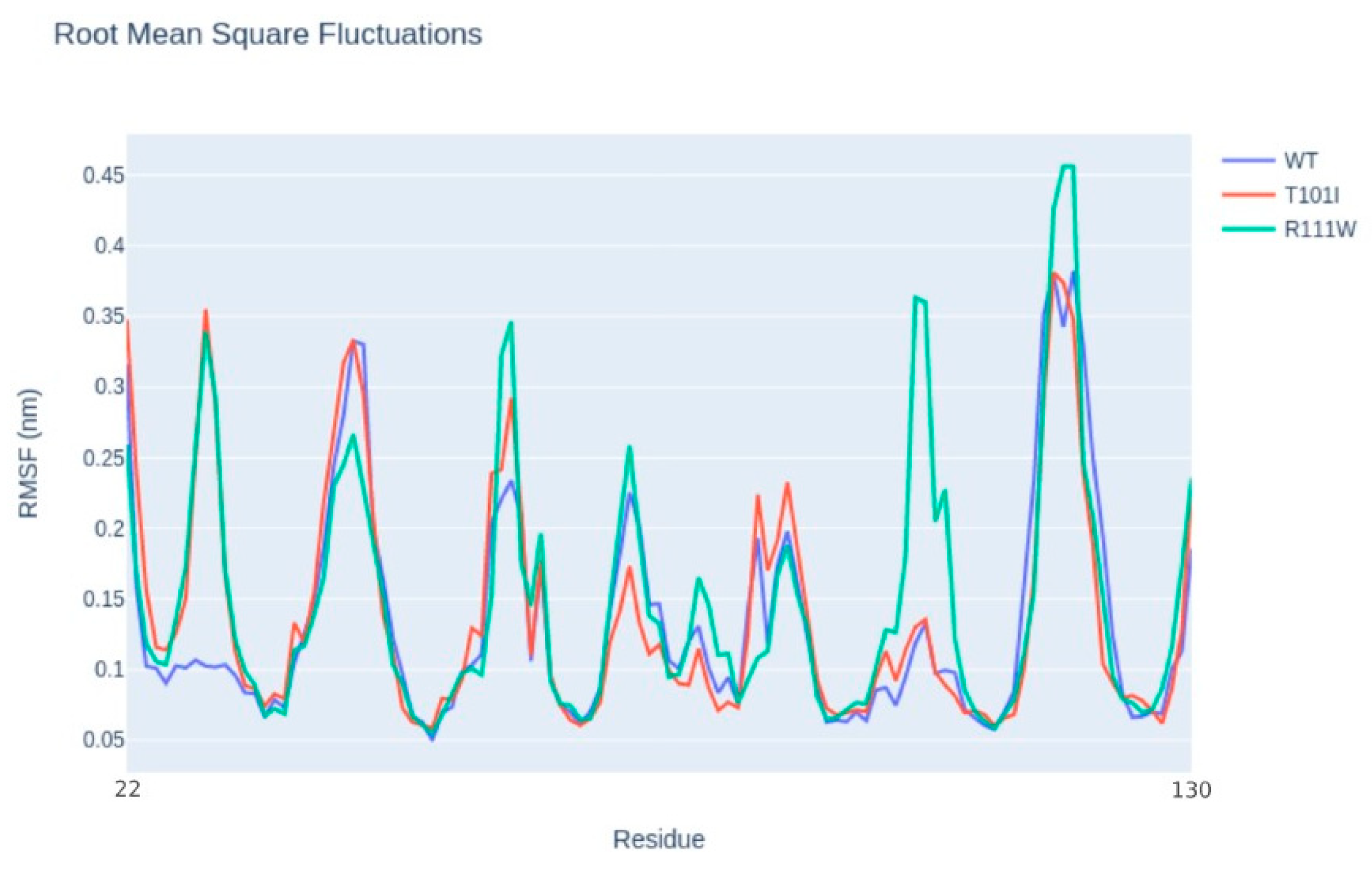
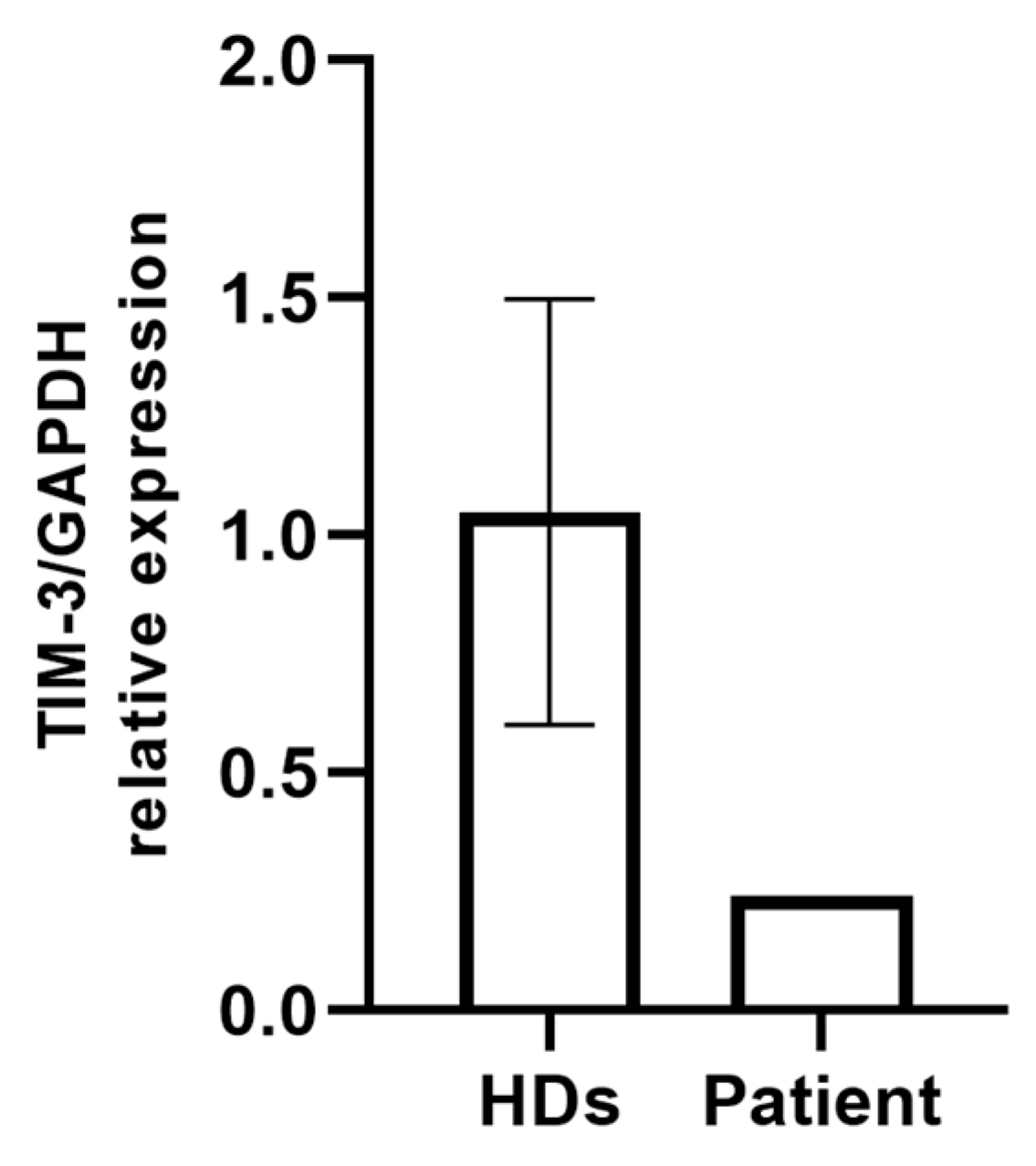
| Gene | OMIM Inheritance | HGVSC | HGVSP | Zygosity | Codon | dbSNP |
|---|---|---|---|---|---|---|
| HAVCR2 | AR | c.331C>T | p.Arg111Trp | HET | Cgg/Tgg | rs145478313 |
| HAVCR2 | AR | c.302C>T | p.Thr101Ile | HET | aCt/aTt | rs147827860 |
| Age | Clinical Manifestation/Therapy | Ab | Laboratory and Instrumental Parameters |
|---|---|---|---|
| 1.8 yrs | Patent ductus arteriosus Type 1 diabetes mellitus (insulin treatment started) IgA deficit | GADAb, IAA pos IA2Ab neg | IgA 29mg/dL (nr 36–165) |
| 3.5 yrs | Celiac disease (gluten-deprived diet) | EMA, TRGAb neg | |
| 5 yrs | Amplatzer device treatment of patent ductus arteriosus | ||
| 12 yrs | Added metformin treatment | ||
| 13 yrs | Partial epileptic crisis Preclinical autoimmune thyroiditis | TPOAb pos TGAb neg | Ecography: thyroiditis Normal thyroid function EEG |
| 13.3 yrs | Autoimmune hepatitis Treated with cyclosporine and azathioprine | LKMAb, ASMA pos ANA, ANCA, AMA, ARA, anti-SLA/LP, anti-SP00, anti-gp210, APCA, TRGAb neg | ALT 606 U/L (nv < 33) AST 408 U/L (nv < 32) GGT 119 U/L (nv < 40) ALP, bilirubin, α1-antitrypsin, ceruloplasmin, cupruria, PT nv IgG 20.8 g/L Hepatic agobiopsy: chronic hepatitis Screening AIRE neg Screening C1858T PTPN22 neg |
| 14.8 yrs | Obesity Azathioprine, colecalcipherol, metformin, insulin | Anti-LC1 Ab pos | Macrocytosis due to azathioprine |
| 16.2 yrs | Anti-adrenal Ab, anti-SLA pos ASCA Ab neg | Neutropenia, leukopenia D hypovitaminosis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ariolli, A.; Agolini, E.; Mazza, T.; Petrizzelli, F.; Petrini, S.; D’Oria, V.; Cudini, A.; Nardella, C.; Pesce, V.; Comparcola, D.; et al. The Putative Role of TIM-3 Variants in Polyendocrine Autoimmunity: Insights from a WES Investigation. Int. J. Mol. Sci. 2024, 25, 10994. https://doi.org/10.3390/ijms252010994
Ariolli A, Agolini E, Mazza T, Petrizzelli F, Petrini S, D’Oria V, Cudini A, Nardella C, Pesce V, Comparcola D, et al. The Putative Role of TIM-3 Variants in Polyendocrine Autoimmunity: Insights from a WES Investigation. International Journal of Molecular Sciences. 2024; 25(20):10994. https://doi.org/10.3390/ijms252010994
Chicago/Turabian StyleAriolli, Andrea, Emanuele Agolini, Tommaso Mazza, Francesco Petrizzelli, Stefania Petrini, Valentina D’Oria, Annamaria Cudini, Caterina Nardella, Vanessa Pesce, Donatella Comparcola, and et al. 2024. "The Putative Role of TIM-3 Variants in Polyendocrine Autoimmunity: Insights from a WES Investigation" International Journal of Molecular Sciences 25, no. 20: 10994. https://doi.org/10.3390/ijms252010994
APA StyleAriolli, A., Agolini, E., Mazza, T., Petrizzelli, F., Petrini, S., D’Oria, V., Cudini, A., Nardella, C., Pesce, V., Comparcola, D., Cappa, M., & Fierabracci, A. (2024). The Putative Role of TIM-3 Variants in Polyendocrine Autoimmunity: Insights from a WES Investigation. International Journal of Molecular Sciences, 25(20), 10994. https://doi.org/10.3390/ijms252010994








