Rapidly Evolved Genes in Three Reaumuria Transcriptomes and Potential Roles of Pentatricopeptide Repeat Superfamily Proteins in Endangerment of R. trigyna
Abstract
:1. Introduction
2. Results
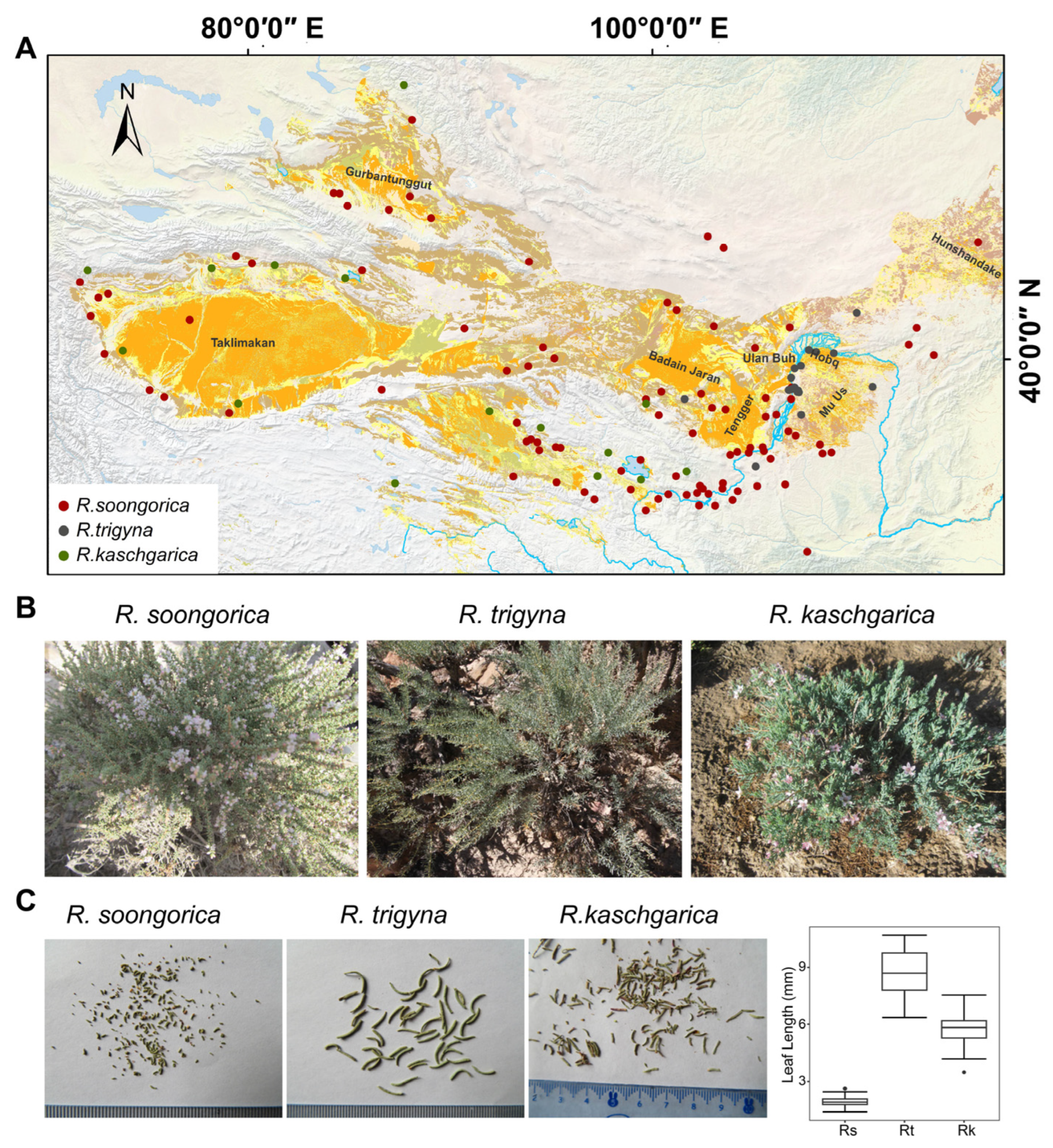
2.1. Assembly and Annotation of R. kaschgarica Transcriptome
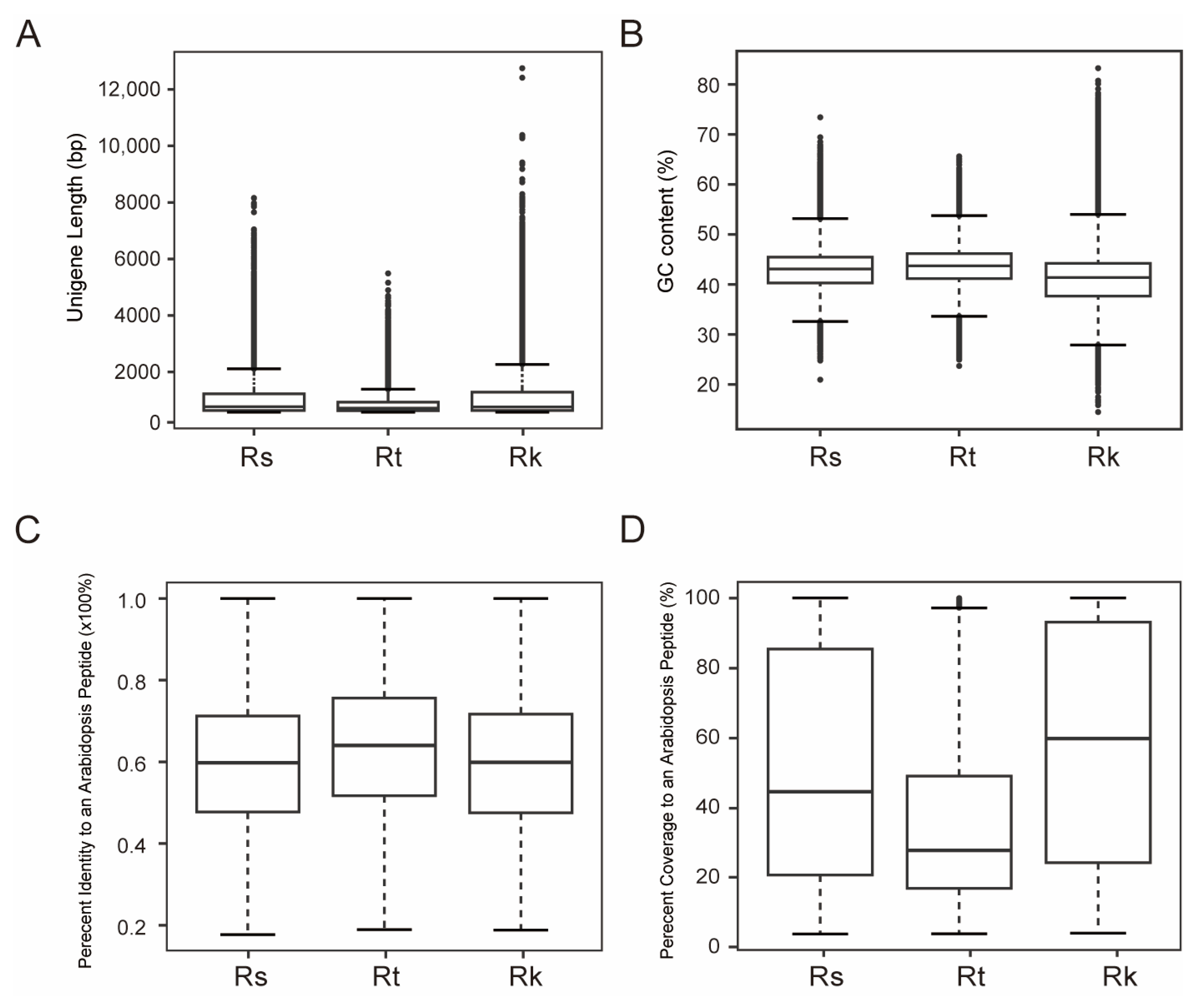
2.2. Characterizing Transcriptomes of R. soongorica, R. kaschgarica, and R. trigyna
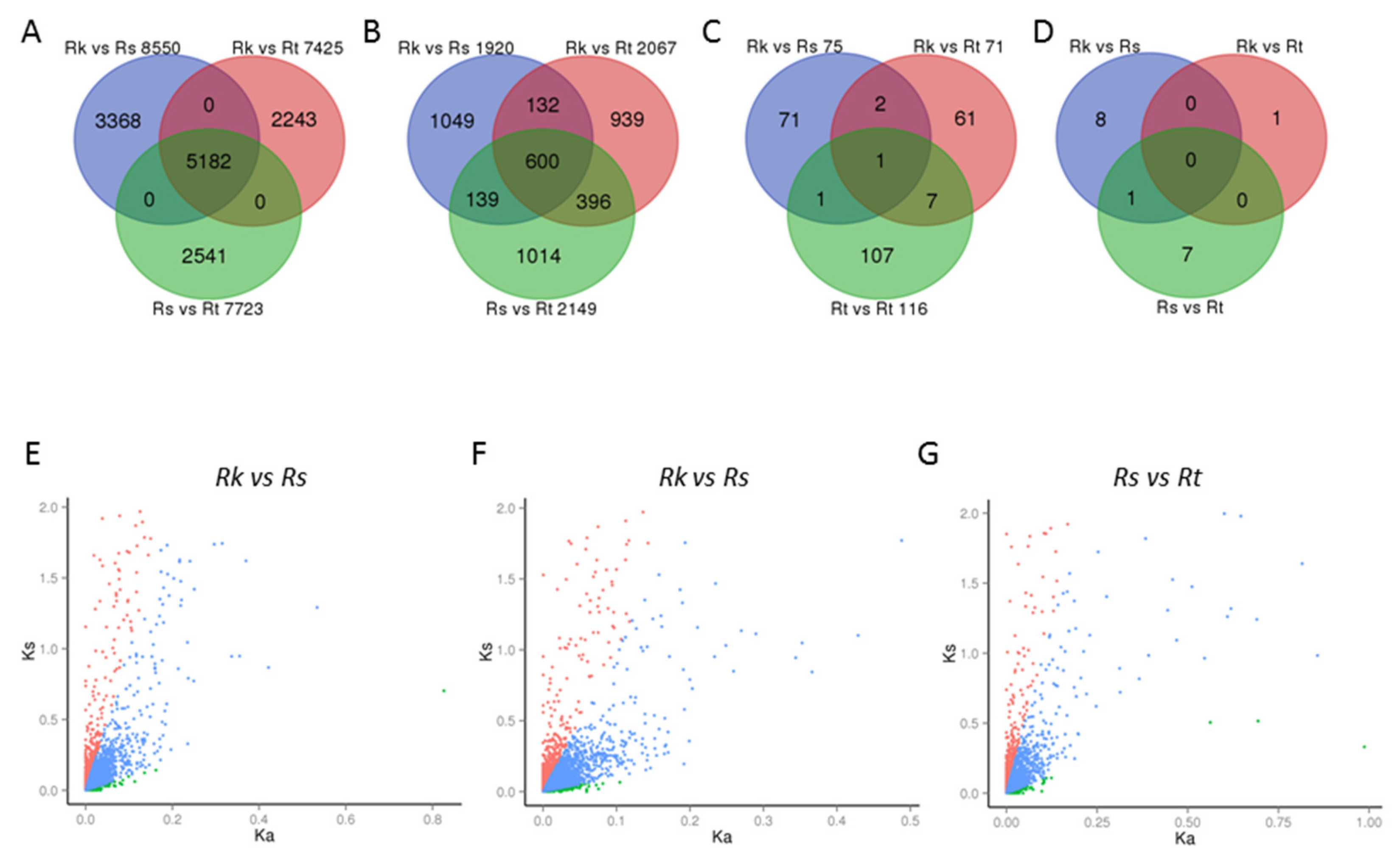
2.3. Comparative Evolutionary Analysis of Reaumuria Transcriptomes
3. Discussion
4. Materials and Methods
4.1. Plant Materials, RNA Extraction, Transcriptome Sequencing, and Analyses
4.2. Transcriptome Characterization Using Blastx Program Versus Arabidopsis Protein Database
4.3. Ortholog Grouping and Ka/Ks Calculation
4.4. GO Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Rk | R. kaschgarica |
| Rt | R. trigyna |
| Rs | R. soongorica |
| PPR | pentatricopeptide repeat |
| HSP | heat shock protein |
| Ka | non-synonymous nucleotide substitution |
| Ks | synonymous nucleotide substitution |
| NHX3 | sodium hydrogen exchanger 3 |
| TTG1 | transparent testa glabra 1 |
| ROS | reactive oxygen species |
| RFL9 | restorer of fertility like 9 |
| CMS | cytoplasmic male sterility |
References
- Zhang, M.; Hao, X.; Sanderson, S.C.; Vyacheslav, B.V.; Sukhorukov, A.P.; Zhang, X.I.A. Spatiotemporal evolution of Reaumuria (Tamaricaceae) in Central Asia: Insights from molecular biogeography. Phytotaxa 2014, 167, 89. [Google Scholar] [CrossRef]
- Yin, H.; Yan, X.; Shi, Y.; Qian, C.; Li, Z.; Zhang, W.; Wang, L.; Li, Y.; Li, X.; Chen, G.; et al. The role of East Asian monsoon system in shaping population divergence and dynamics of a constructive desert shrub Reaumuria soongarica. Sci. Rep. 2015, 5, 15823. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, M.; Li, X.; Cao, B.; Ma, X. Identification of differentially expressed genes in leaf of Reaumuria soongorica under PEG-induced drought stress by digital gene expression profiling. PLoS ONE 2014, 9, e94277. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.H.; Li, J.H.; Fang, X.W.; Wang, G.; Su, P.X. Photosynthetic activity of poikilochlorophyllous desiccation tolerant plant Reaumuria soongorica during dehydration and re-hydration. Photosynthetica 2008, 46, 547–551. [Google Scholar] [CrossRef]
- Bai, J.; Gong, C.-M.; Chen, K.; Kang, H.-M.; Wang, G. Examination of Antioxidative System’s Responses in the Different Phases of Drought Stress and during Recovery in Desert Plant Reaumuria soongorica (Pall.) Maxim. J. Plant Biol. 2009, 52, 417–425. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, T.; Li, X.; Wang, G. Protective mechanism of desiccation tolerance in Reaumuria soongorica: Leaf abscission and sucrose accumulation in the stem. Sci. China Ser. C. 2007, 50, 15–21. [Google Scholar] [CrossRef]
- Hao, X.; Zhang, M.; Wang, S.; Zhang, X. Classification and Distribution of the Genus Reaumuria L. in Tamaricaceae. Arid. Zone Res. 2014, 31, 838–843. [Google Scholar]
- Shi, S.; Wang, Y.; Zhou, J.; Zhou, H. Endogenous hormone contents and their habitat differentia of Reaumuria trigyna and R. soongorica in different salt habitats. J. Appl. Ecol. 2011, 22, 350–356. [Google Scholar]
- Zhang, Y.; Wang, Y. Genetic Diversity of Endangered Shrub Reaumuria trigyna Population Detected by RAPD and ISSR Markers. Sci. Silvae Sin. 2008, 44, 43–47. [Google Scholar]
- Ma, S.-M.; Zhang, M.-L.; Ni, J.; Xi, C. Modelling the geographic distributions of endemic genera in the eastern Central Asian desert. Nord. J. Bot. 2012, 30, 372–384. [Google Scholar] [CrossRef]
- Dang, Z.; Zheng, L.; Wang, J.; Gao, Z.; Wu, S.; Qi, Z.; Wang, Y.C. Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. BMC Genomics 2013, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, H.; Zhang, F.; Xi, J.; Gu, H. Ions Secretion in Wild Reaumuria Soongorica under Natural Saline-Alkali Conditions. Acta Pedol. Sin. 2004, 41, 774–779. [Google Scholar]
- Ramadan, T. Ecophysiology of salt excretion in the xero-halophyte Reaumuria hirtella. N. Phytol. 1998, 139, 273–281. [Google Scholar] [CrossRef]
- Yan, X.U.E.; Yingchun, W. Study on Characters of Ions Secretion from Reaumuria trigyna. J. Desert Res. 2008, 28, 437–442. [Google Scholar]
- Liu, Y.B.; Wang, G.; Liu, J.; Zhao, X.; Tan, H.J.; Li, X.R. Anatomical, morphological and metabolic acclimation in the resurrection plant Reaumuria soongorica during dehydration and rehydration. J. Arid. Environ. 2007, 70, 183–194. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, J.; Zhao, J.; Comes, H.P.; Li, P.; Fu, C.; Xie, X.; Lu, R.; Xu, W.; Feng, Y.; et al. Genomic insights on the contribution of balancing selection and local adaptation to the long-term survival of a widespread living fossil tree, Cercidiphyllum japonicum. New Phytol. 2020, 228, 1674–1689. [Google Scholar] [CrossRef]
- Xu, C.; Wang, J.C.; Sun, L.; Zhuang, L.H.; Guo, Z.J.; Ding, Q.S.; Ma, D.N.; Song, L.Y.; Li, J.; Tang, H.C.; et al. Genome-Wide Identification of Pentatricopeptide Repeat (PPR) Gene Family and Multi-Omics Analysis Provide New Insights into the Albinism Mechanism of Kandelia obovata Propagule Leaves. Plant Cell Environ. 2024. [Google Scholar] [CrossRef]
- Zheng, S.; Dong, J.; Lu, J.; Li, J.; Jiang, D.; Yu, H.; Ye, S.; Bu, W.; Liu, Z.; Zhou, H.; et al. A cytosolic pentatricopeptide repeat protein is essential for tapetal plastid development by regulating OsGLK1 transcript levels in rice. New Phytol. 2022, 234, 1678–1695. [Google Scholar] [CrossRef]
- Meng, L.; Du, M.; Zhu, T.; Li, G.; Ding, Y.; Zhang, Q. PPR proteins in plants: Roles, mechanisms, and prospects for rice research. Front. Plant Sci. 2024, 15, 1416742. [Google Scholar] [CrossRef]
- Li, X.; Sun, M.; Liu, S.; Teng, Q.; Li, S.; Jiang, Y. Functions of PPR Proteins in Plant Growth and Development. Int. J. Mol. Sci. 2021, 22, 11274. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yan, X.; Zhao, P.; Yin, H.; Zhao, X.; Xiao, H.; Li, X.; Chen, G.; Ma, X.-F. Transcriptomic Analysis of a Tertiary Relict Plant, Extreme Xerophyte Reaumuria soongorica to Identify Genes Related to Drought Adaptation. PLoS ONE 2013, 8, e63993. [Google Scholar] [CrossRef]
- Dorn, K.M.; Fankhauser, J.D.; Wyse, D.L.; Marks, M.D. De novo assembly of the pennycress (Thlaspi arvense) transcriptome provides tools for the development of a winter cover crop and biodiesel feedstock. Plant J. 2013, 75, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, J.; Zhao, X.; Chen, G.; Ma, X.F. Different Sets of Post-Embryonic Development Genes Are Conserved or Lost in Two Caryophyllales Species (Reaumuria soongorica and Agriophyllum squarrosum). PLoS ONE 2016, 11, e0148034. [Google Scholar] [CrossRef]
- Yuan, F.; Lyu, M.J.; Leng, B.Y.; Zheng, G.Y.; Feng, Z.T.; Li, P.H.; Zhu, X.G.; Wang, B.S. Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 2015, 38, 1637–1657. [Google Scholar] [CrossRef]
- Ma, C.L.; Haslbeck, M.; Babujee, L.; Jahn, O.; Reumann, S. Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 2006, 141, 47–60. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, W.T. Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 2013, 587, 2584–2590. [Google Scholar] [CrossRef]
- Suh, J.Y.; Kim, S.J.; Oh, T.R.; Cho, S.K.; Yang, S.W.; Kim, W.T. Arabidopsis Tóxicos en Levadura 78 (AtATL78) mediates ABA-dependent ROS signaling in response to drought stress. Biochem. Bioph Res. Commun. 2016, 469, 8–14. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Ann. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, N.; Hattori, M.; Andres, C.; Iida, K.; Lurin, C.; Schmitz-Linneweber, C.; Sugita, M.; Small, I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008, 25, 1120–1128. [Google Scholar] [CrossRef]
- Arnal, N.; Quadrado, M.; Simon, M.; Mireau, H. A restorer-of-fertility like pentatricopeptide repeat gene directs ribonucleolytic processing within the coding sequence of rps3-rpl16 and orf240a mitochondrial transcripts in Arabidopsis thaliana. Plant J. 2014, 78, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Bentolila, S.; Chateigner-Boutin, A.L.; Hanson, M.R. Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Physiol. 2005, 139, 2006–2016. [Google Scholar] [CrossRef] [PubMed]
- Stoll, K.; Jonietz, C.; Schleicher, S.; des Francs-Small, C.C.; Small, I.; Binder, S. In Arabidopsis thaliana distinct alleles encoding mitochondrial RNA PROCESSING FACTOR 4 support the generation of additional 5’ termini of ccmB transcripts. Plant Mol. Biol. 2017, 93, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Zehrmann, A.; van der Merwe, J.A.; Verbitskiy, D.; Brennicke, A.; Takenaka, M. Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion 2008, 8, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Zehrmann, A.; Verbitskiy, D.; Hartel, B.; Brennicke, A.; Takenaka, M. RNA editing competence of trans-factor MEF1 is modulated by ecotype-specific differences but requires the DYW domain. FEBS Lett. 2010, 584, 4181–4186. [Google Scholar] [CrossRef] [PubMed]
- Geddy, R.; Brown, G.G. Genes encoding pentatricopeptide repeat (PPR) proteins are not conserved in location in plant genomes and may be subject to diversifying selection. BMC Genomics 2007, 8, 130. [Google Scholar] [CrossRef]
- Fujii, S.; Small, I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011, 191, 37–47. [Google Scholar] [CrossRef]
- Ruberti, C.; Costa, A.; Pedrazzini, E.; Lo Schiavo, F.; Zottini, M. FISSION1A, an Arabidopsis Tail-Anchored Protein, Is Localized to Three Subcellular Compartments. Mol. Plant. 2014, 7, 1393–1396. [Google Scholar] [CrossRef]
- Scott, I.; Tobin, A.K.; Logan, D.C. BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 2006, 57, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, J. Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 2009, 57, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-C.; Hu, J.-P. FISSION1A and FISSION1B Proteins Mediate the Fission of Peroxisomes and Mitochondria in Arabidopsis. Mol. Plant. 2008, 1, 1036–1047. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Mei, C.; Liang, S.; Yu, Y.T.; Lu, K.; Wu, Z.; Wang, X.F.; Zhang, D.P. Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol. Biol. 2015, 88, 369-85. [Google Scholar] [CrossRef]
- Qiu, T.; Zhao, X.; Feng, H.; Qi, L.; Yang, J.; Peng, Y.L.; Zhao, W. OsNBL3, a mitochondrion-localized pentatricopeptide repeat protein, is involved in splicing nad5 intron 4 and its disruption causes lesion mimic phenotype with enhanced resistance to biotic and abiotic stresses. Plant Biotechnol. J. 2021, 19, 2277–2290. [Google Scholar] [CrossRef]
- Luo, Z.; Xiong, J.; Xia, H.; Wang, L.; Hou, G.; Li, Z.; Li, J.; Zhou, H.; Li, T.; Luo, L. Pentatricopeptide Repeat Gene-Mediated Mitochondrial RNA Editing Impacts on Rice Drought Tolerance. Front. Plant Sci. 2022, 13, 926285. [Google Scholar] [CrossRef]
- Austin, R.S.; Hiu, S.; Waese, J.; Ierullo, M.; Pasha, A.; Wang, T.T.; Fan, J.; Foong, C.; Breit, R.; Desveaux, D.; et al. New BAR tools for mining expression data and exploring Cis-elements in Arabidopsis thaliana. Plant J. 2016, 88, 490–504. [Google Scholar] [CrossRef]
- Hammani, K.; Okuda, K.; Tanz, S.K.; Chateigner-Boutin, A.L.; Shikanai, T.; Small, I. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell. 2009, 21, 3686–3699. [Google Scholar] [CrossRef]
- Liu, J.M.; Zhao, J.Y.; Lu, P.P.; Chen, M.; Guo, C.H.; Xu, Z.S.; Ma, Y.Z. The E-Subgroup Pentatricopeptide Repeat Protein Family in Arabidopsis thaliana and Confirmation of the Responsiveness PPR96 to Abiotic Stresses. Front. Plant Sci. 2016, 7, 1825. [Google Scholar] [CrossRef]
- Dewitte, W.; Scofield, S.; Alcasabas, A.A.; Maughan, S.C.; Menges, M.; Braun, N.; Collins, C.; Nieuwland, J.; Prinsen, E.; Sundaresan, V.; et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14537–14542. [Google Scholar] [CrossRef]
- Dewitte, W.; Riou-Khamlichi, C.; Scofield, S.; Healy, J.M.; Jacqmard, A.; Kilby, N.J.; Murray, J.A. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 2003, 15, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Baekelandt, A.; Pauwels, L.; Wang, Z.; Li, N.; De Milde, L.; Natran, A.; Vermeersch, M.; Li, Y.; Goossens, A.; Inzé, D.; et al. Arabidopsis Leaf Flatness Is Regulated by PPD2 and NINJA through Repression of CYCLIN D3 Genes. Plant Physiol. 2018, 178, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Sosso, D.; Mbelo, S.; Vernoud, V.; Gendrot, G.; Dedieu, A.; Chambrier, P.; Dauzat, M.; Heurtevin, L.; Guyon, V.; Takenaka, M.; et al. PPR2263, a DYW-Subgroup Pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell. 2012, 24, 676–691. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Capella-Gutierrez, S.; Shi, Y.; Zhao, X.; Chen, G.; Gabaldon, T.; Ma, X.-F. Transcriptomic analysis of a psammophyte food crop, sand rice (Agriophyllum squarrosum) and identification of candidate genes essential for sand dune adaptation. BMC Genomics 2014, 15, 872. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Qiao, Q.; Xue, L.; Wang, Q.; Sun, H.; Zhong, Y.; Huang, J.; Lei, J.; Zhang, T. Comparative Transcriptomics of Strawberries (Fragaria spp.) Provides Insights into Evolutionary Patterns. Front. Plant Sci. 2016, 7, 1839. [Google Scholar] [CrossRef]
- Zhu, S.; Tang, S.; Tan, Z.; Yu, Y.; Dai, Q.; Liu, T. Comparative transcriptomics provide insight into the morphogenesis and evolution of fistular leaves in Allium. BMC Genomics 2017, 18, 60. [Google Scholar] [CrossRef]

| Number of Unigenes | Percentage (%) | |
|---|---|---|
| Nr | 27,325 | 43.59 |
| Nt | 13,142 | 20.96 |
| KO | 9330 | 14.88 |
| Swiss-Prot | 19,696 | 31.42 |
| Pfam | 18,959 | 30.24 |
| GO | 19,414 | 30.97 |
| KOG | 10,199 | 16.27 |
| Annotated in all databases | 4131 | 6.59 |
| Annotated in at least one database | 30,002 | 47.86 |
| Total Unigenes | 62,680 | 100 |
| Orthologous Groups | Genes | Gene Symbols | Ka/Ks | ||
|---|---|---|---|---|---|
| Rk vs. Rs | Rk vs. Rt | Rs vs. Rt | |||
| OG03284 | Rk|c19117_g1 Rs|Unigene52639_A_Rs Rt|Unigene3477_Rt | HSP20-like | 1.52 | 1.329 | 1.20 |
| OG14217 | Rk|c41245_g1 Rs|Unigene22663_A_Rs Rt|Unigene66872_Rt | PPR | 1.02 | 0.69 | 1.20 |
| OG13726 | Rk|c17221_g1 Rs|CL3395.Contig2_A_Rs Rt|Unigene50964_Rt | ATL78 | 0.97 | 2.09 | 1.07 |
| OG10059 | Rs|Unigene46659_A_Rs Rk|c6241_g1 Rt|Unigene62804_Rt | BIGYIN/FIS1A | 0.22 | 0.0001 | 1.69 |
| OG14530 | Rs|CL10303.Contig1_A_Rs, Rt|Unigene64640_Rt | RFL9 | n.d. | n.d. | 1.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Cui, X.; Zhao, P. Rapidly Evolved Genes in Three Reaumuria Transcriptomes and Potential Roles of Pentatricopeptide Repeat Superfamily Proteins in Endangerment of R. trigyna. Int. J. Mol. Sci. 2024, 25, 11065. https://doi.org/10.3390/ijms252011065
Zhang R, Cui X, Zhao P. Rapidly Evolved Genes in Three Reaumuria Transcriptomes and Potential Roles of Pentatricopeptide Repeat Superfamily Proteins in Endangerment of R. trigyna. International Journal of Molecular Sciences. 2024; 25(20):11065. https://doi.org/10.3390/ijms252011065
Chicago/Turabian StyleZhang, Ruizhen, Xiaoyun Cui, and Pengshan Zhao. 2024. "Rapidly Evolved Genes in Three Reaumuria Transcriptomes and Potential Roles of Pentatricopeptide Repeat Superfamily Proteins in Endangerment of R. trigyna" International Journal of Molecular Sciences 25, no. 20: 11065. https://doi.org/10.3390/ijms252011065
APA StyleZhang, R., Cui, X., & Zhao, P. (2024). Rapidly Evolved Genes in Three Reaumuria Transcriptomes and Potential Roles of Pentatricopeptide Repeat Superfamily Proteins in Endangerment of R. trigyna. International Journal of Molecular Sciences, 25(20), 11065. https://doi.org/10.3390/ijms252011065





