Serine Hydroxymethyltransferase 2 Deficiency in the Hematopoietic System Disrupts Erythropoiesis and Induces Anemia in Murine Models
Abstract
1. Introduction
2. Results
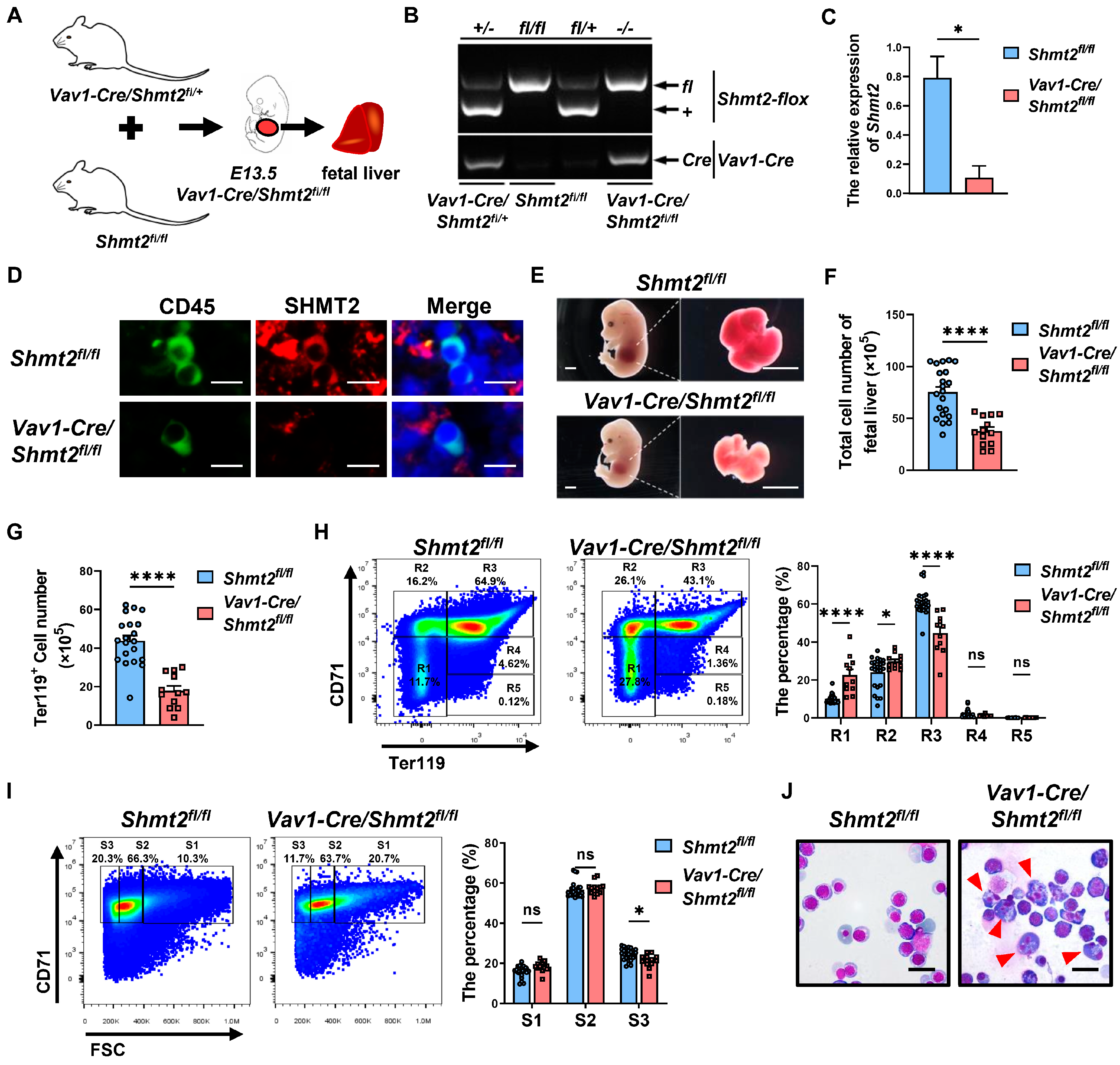
2.1. The Absence of the Shmt2 Gene within the Hematopoietic System Significantly Impairs the Development of Erythroid Cells in the Fetal Livers of Mice
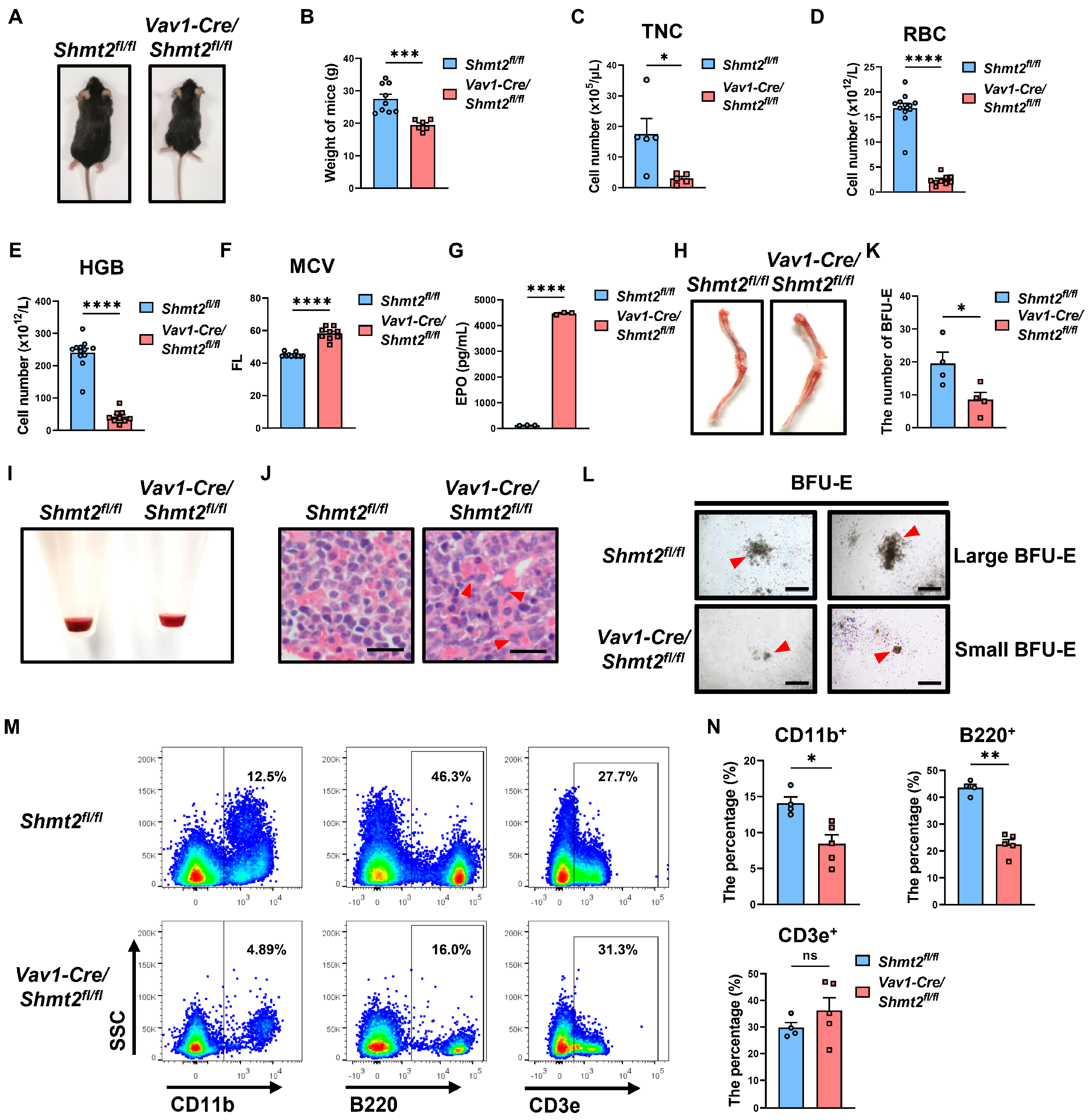
2.2. Anemia-like Symptoms in Adult Mice with the Hematopoietic Cell-Specific Deletion of Shmt2 Gene
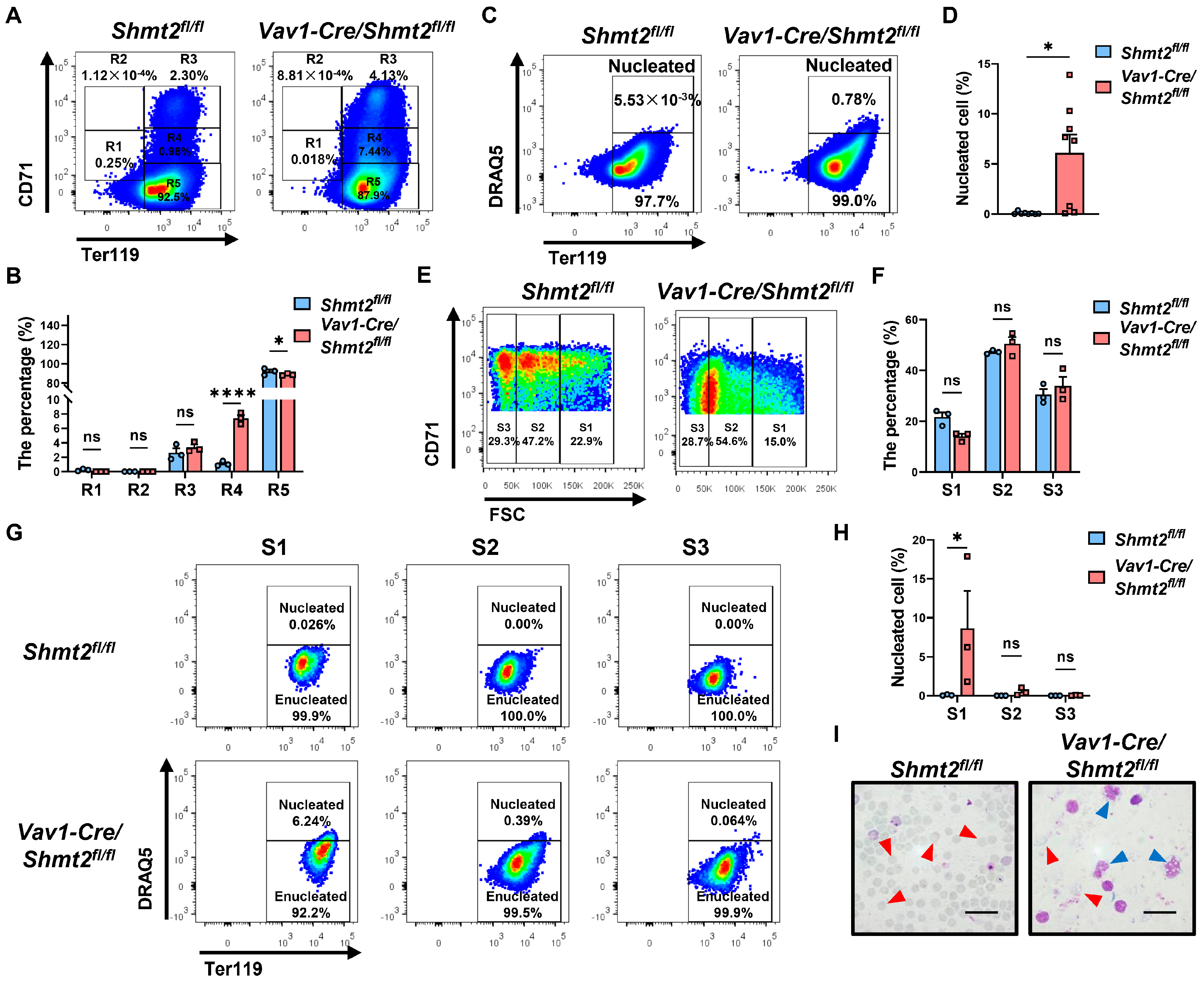
2.3. The Deletion of the Shmt2 Gene in the Hematopoietic System Impedes the Enucleation of Red Blood Cells in the Peripheral Blood of Adult Mice
2.4. The Absence of the Shmt2 Gene in the Hematopoietic System Hinders the Development of Erythroid Cells in the Bone Marrow of Adult Mice
2.5. Promotion of Erythroid-Related Gene Expression by Shmt2 Deletion in Proerythroblasts
2.6. Impaired Proliferation and Erythroid Differentiation in Basophilic Erythroblasts Due to Shmt2 Deletion
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Blood Routine Test
4.3. Determination of Serum EPO by ELISA
4.4. Quantitative Real-Time PCR
4.5. Immunofluorescence Staining
4.6. Flow Cytometry Analysis
4.7. Colony-Forming Unit Assay
4.8. Wright-Giemsa Staining
4.9. RNA Isolation and Library Preparation
4.10. RNA Sequencing and Differentially Expressed Genes Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ashorobi, D.; Munakomi, S. Myelophthisic Anemia; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Stabler, S.P. Clinical practice. Vitamin B12 deficiency. N. Engl. J. Med. 2013, 368, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Green, R.; Datta Mitra, A. Megaloblastic anemias: Nutritional and other causes. Med. Clin. N. Am. 2017, 101, 297–317. [Google Scholar] [CrossRef]
- Green, R. Vitamin B12 deficiency from the perspective of a practicing hematologist. Blood 2017, 129, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Socha, D.S.; DeSouza, S.I.; Flagg, A.; Sekeres, M.; Rogers, H.J. Severe megaloblastic anemia: Vitamin deficiency and other causes. Clevel. Clin. J. Med. 2020, 87, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Hesdorffer, C.S.; Longo, D.L. Drug-induced megaloblastic anemia. N. Engl. J. Med. 2015, 373, 1649–1658. [Google Scholar] [CrossRef]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef]
- Ye, J.; Fan, J.; Venneti, S.; Wan, Y.-W.; Pawel, B.R.; Zhang, J.; Finley, L.W.; Lu, C.; Lindsten, T.; Cross, J.R.; et al. Serine Catabolism Regulates Mitochondrial Redox Control during Hypoxia. Cancer Discov. 2014, 4, 1406–1417. [Google Scholar] [CrossRef]
- Shimshek, D.; Kim, J.; Hübner, M.; Spergel, D.; Buchholz, F.; Casanova, E.; Stewart, A.; Seeburg, P.; Sprengel, R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 2002, 32, 19–26. [Google Scholar] [CrossRef]
- Ogilvy, S.; Metcalf, D.; Gibson, L.; Bath, M.L.; Harris, A.W.; Adamset, J.M. Promoter elements of vav drive transgene expression in vivo throughout the hemato-poietic compartment. Blood 1999, 94, 1855–1863. [Google Scholar] [CrossRef]
- de Boer, J.; Williams, A.; Skavdis, G.; Harker, N.; Coles, M.; Tolaini, M.; Norton, T.; Williams, K.; Roderick, K.; Potocnik, A.J.; et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003, 33, 314–325. [Google Scholar] [CrossRef]
- Kao, Y.-R.; Chen, J.; Kumari, R.; Ng, A.; Zintiridou, A.; Tatiparthy, M.; Ma, Y.; Aivalioti, M.M.; Moulik, D.; Sundaravel, S.; et al. An iron rheostat controls hematopoietic stem cell fate. Cell Stem Cell 2024, 31, 378–397. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Ohnishi, S.; Shitara, H.; Mito, T.; Yamaguchi, M.; Yonekawa, H.; Hashizume, O.; Ishikawa, K.; Nakada, K.; Hayashi, J.-I. Mice deficient in the Shmt2 gene have mitochondrial respiration defects and are embryonic lethal. Sci. Rep. 2018, 8, 425. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Mito, T.; Velagapudi, V.; Ishikawa, K.; Umehara, M.; Nakada, K.; Suomalainen, A.; Hayashi, J.-I. Disruption of the mouse Shmt2 gene confers embryonic anaemia via foetal liver-specific metabolomic disorders. Sci. Rep. 2019, 9, 16054. [Google Scholar] [CrossRef]
- Zhang, J.; Socolovsky, M.; Gross, A.W.; Lodish, H.F. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: Functional analysis by a flow cytometry–based novel culture system. Blood 2003, 102, 3938–3946. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.-B.; Wang, X.-H.; Li, C.-Y.; Tian, H.-H.; Zhang, J.; Bi, J.-J.; Ren, G.-M.; Tao, S.-S.; Liu, X.; Zhang, W.; et al. GPS2 promotes erythroid differentiation by control of the stability of EKLF protein. Blood 2020, 135, 2302–2315. [Google Scholar] [CrossRef]
- Sui, Z.; Nowak, R.B.; Bacconi, A.; Kim, N.E.; Liu, H.; Li, J.; Wickrema, A.; An, X.-L.; Fowler, V.M. Tropomodulin3-null mice are embryonic lethal with anemia due to impaired erythroid terminal differentiation in the fetal liver. Blood 2014, 123, 758–767. [Google Scholar] [CrossRef]
- Li, H.; Natarajan, A.; Ezike, J.; Barrasa, M.I.; Le, Y.; Feder, Z.A.; Yang, H.; Ma, C.; Markoulaki, S.; Lodish, H.F. Rate of Progression through a Continuum of Transit-Amplifying Progenitor Cell States Regulates Blood Cell Production. Dev. Cell 2019, 49, 118–129.e7. [Google Scholar] [CrossRef]
- Gnanapragasam, M.N.; McGrath, K.E.; Catherman, S.; Xue, L.; Palis, J.; Bieker, J.J. EKLF/KLF1-regulated cell cycle exit is essential for erythroblast enucleation. Blood 2016, 128, 1631–1641. [Google Scholar] [CrossRef]
- Bryan, L.J.; Zakai, N.A. Why Is. My Patient Anemic? Hematol. Oncol. Clin. N. Am. 2012, 26, 205–230. [Google Scholar] [CrossRef]
- Collaborators GBDA. Prevalence, years lived with disability, and trends in anaemia burden by severity and cause, 1990–2021: Findings from the Global Burden of Disease Study 2021. Lancet Haematol. 2023, 10, e713–e734. [Google Scholar] [CrossRef]
- Hunt, J.M. Reversing productivity losses from iron deficiency: The economic case. J. Nutr. 2002, 132, 794S–801S. [Google Scholar] [CrossRef] [PubMed]
- Minton, D.R.; Nam, M.; Mclaughlin, D.J.; Shin, J.; Bayraktar, E.C.; Alvarez, S.W.; Sviderskiy, V.O.; Papagiannakopoulos, T.; Sabatini, D.M.; Birsoyet, K.; et al. Serine catabolism by SHMT2 is required for proper mitochondrial translation initiation and maintenance of formylmethionyl-tRNAs. Mol. Cell 2018, 69, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Parsa, S.; Ortega-Molina, A.; Ying, H.-Y.; Jiang, M.; Teater, M.; Wang, J.; Zhao, C.; Reznik, E.; Pasion, J.P.; Kuo, D.; et al. The serine hydroxymethyltransferase-2 (SHMT2) initiates lymphoma development through epigenetic tumor suppressor silencing. Nat. Cancer 2020, 1, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Haley, K. Congenital Hemolytic Anemia. Med. Clin. N. Am. 2017, 101, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Moreira, M.; Brás, R.; Gonçalves, D.; Alencoão, I.; Inocêncio, G.; Rodrigues, M.; Braga, J. Fetal Splenomegaly: A Review. Ultrasound Q. 2018, 34, 32–33. [Google Scholar] [CrossRef]
- Takasaki, K.; Kacena, M.A.; Raskind, W.H.; Weiss, M.J.; Chou, T. GATA1-Related Cytopenia. In GeneReviews®; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Brady, C.A.; Jiang, D.; Mello, S.S.; Johnson, T.M.; Jarvis, L.A.; Kozak, M.M.; Broz, D.K.; Basak, S.; Park, E.J.; McLaughlin, M.E.; et al. Distinct p53 Transcriptional Programs Dictate Acute DNA-Damage Responses and Tumor Suppression. Cell 2011, 145, 571–583. [Google Scholar] [CrossRef]
- Le Goff, S.; Boussaid, I.; Floquet, C.; Raimbault, A.; Hatin, I.; Andrieu-Soler, C.; Salma, M.; Leduc, M.; Gautier, E.-F.; Guyot, B.; et al. p53 activation during ribosome biogenesis regulates normal erythroid differentiation. Blood 2021, 137, 89–102. [Google Scholar] [CrossRef]
- Lan, Y.; He, W.; Li, Z.; Wang, Y.; Wang, J.; Gao, J.; Wang, W.; Cheng, T.; Liu, B.; Yang, X. Endothelial Smad4 restrains the transition to hematopoietic progenitors via suppression of ERK activation. Blood 2014, 123, 2161–2171. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Zhang, B.; Li, Y.; Liu, C.; Tang, X.; Zhao, J.; Pei, X.; Li, Y. Serine Hydroxymethyltransferase 2 Deficiency in the Hematopoietic System Disrupts Erythropoiesis and Induces Anemia in Murine Models. Int. J. Mol. Sci. 2024, 25, 11072. https://doi.org/10.3390/ijms252011072
Li J, Zhang B, Li Y, Liu C, Tang X, Zhao J, Pei X, Li Y. Serine Hydroxymethyltransferase 2 Deficiency in the Hematopoietic System Disrupts Erythropoiesis and Induces Anemia in Murine Models. International Journal of Molecular Sciences. 2024; 25(20):11072. https://doi.org/10.3390/ijms252011072
Chicago/Turabian StyleLi, Jisheng, Bowen Zhang, Yunqiao Li, Chuanli Liu, Xuan Tang, Jiahui Zhao, Xuetao Pei, and Yanhua Li. 2024. "Serine Hydroxymethyltransferase 2 Deficiency in the Hematopoietic System Disrupts Erythropoiesis and Induces Anemia in Murine Models" International Journal of Molecular Sciences 25, no. 20: 11072. https://doi.org/10.3390/ijms252011072
APA StyleLi, J., Zhang, B., Li, Y., Liu, C., Tang, X., Zhao, J., Pei, X., & Li, Y. (2024). Serine Hydroxymethyltransferase 2 Deficiency in the Hematopoietic System Disrupts Erythropoiesis and Induces Anemia in Murine Models. International Journal of Molecular Sciences, 25(20), 11072. https://doi.org/10.3390/ijms252011072





