Genetic Adaptations of the Tibetan Pig to High-Altitude Hypoxia on the Qinghai–Tibet Plateau
Abstract
:1. Introduction
2. Result
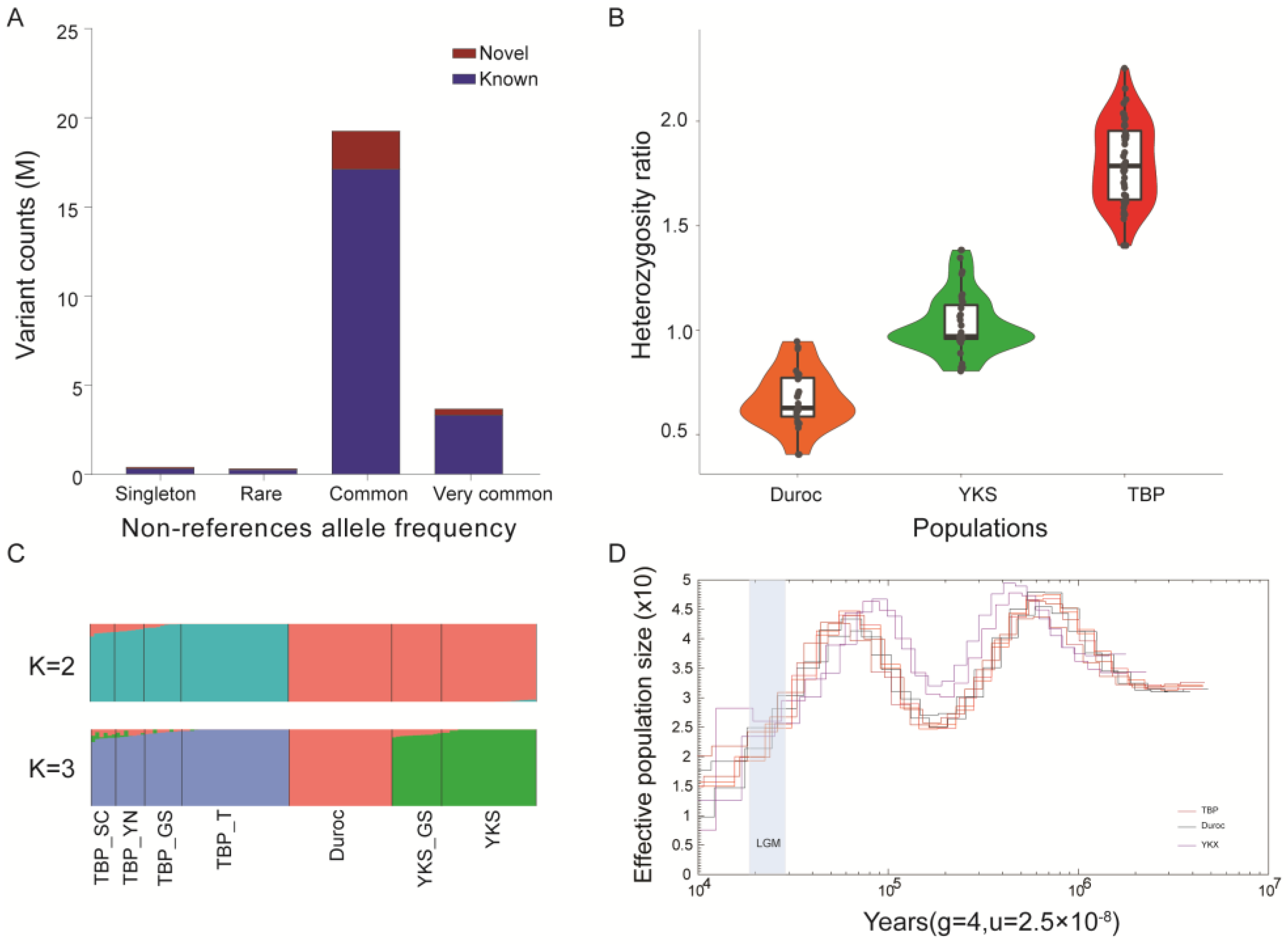
2.1. Genomic Dataset and Population Structure
2.2. Genome-Wide Scan for Darwin Positive Selection Signatures in TBP
2.3. The Newly Identified Top TBPSGs Explain the Adaption of Cardiorespiratory Function and Fat Metabolism
3. Discussion
4. Materials and Methods
4.1. Samples and Sequencing
4.2. Quality Control and Alignment
4.3. Variant Calling
4.4. Data Quality Control and Variant Annotations
4.4.1. Quality Control at Sample Level
4.4.2. Quality Control at SNVs Level
4.4.3. Variant Annotations
4.5. Detection of Genomic Signatures of Positive Selection
4.5.1. Population Structure Analysis
4.5.2. FST
4.5.3. Linkage Disequilibrium (LD) Decay
4.5.4. Population Branch Statistic (PBS)
4.5.5. XPEHH and iHS
4.5.6. CMS
4.5.7. Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, M.; Tian, S.; Jin, L.; Zhou, G.; Li, Y.; Zhang, Y.; Wang, T.; Yeung, C.K.; Chen, L.; Ma, J.; et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 2013, 45, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Groenen, M.A.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.J.; et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.J.; Yang, J.; Xie, X.L.; Lv, F.H.; Cao, Y.H.; Li, W.R.; Liu, M.J.; Wang, Y.T.; Li, J.Q.; Liu, Y.G.; et al. The Genome Landscape of Tibetan Sheep Reveals Adaptive Introgression from Argali and the History of Early Human Settlements on the Qinghai-Tibetan Plateau. Mol. Biol. Evol. 2019, 36, 283–303. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, S.; Wang, Y.F.; Li, S.; Wu, S.X.; Yan, R.G.; Zhang, Y.W.; Wan, R.D.; He, Z.; Song, R.D.; et al. Long read genome assemblies complemented by single cell RNA-sequencing reveal genetic and cellular mechanisms underlying the adaptive evolution of yak. Nat. Commun. 2022, 13, 4887. [Google Scholar] [CrossRef]
- Liu, L.; Li, R.G.; Martin, J.F. An improved method reveals the dual role of Tnni3k in promoting S-phase entry while suppressing cell division in cardiomyocytes. J. Mol. Cell Cardiol. 2023, 183, 100–101. [Google Scholar] [CrossRef]
- Wang, M.S.; Li, Y.; Peng, M.S.; Zhong, L.; Wang, Z.J.; Li, Q.Y.; Tu, X.L.; Dong, Y.; Zhu, C.L.; Wang, L.; et al. Genomic Analyses Reveal Potential Independent Adaptation to High Altitude in Tibetan Chickens. Mol. Biol. Evol. 2015, 32, 1880–1889. [Google Scholar] [CrossRef]
- Speakman, J.R.; Chi, Q.; Ołdakowski, Ł.; Fu, H.; Fletcher, Q.E.; Hambly, C.; Togo, J.; Liu, X.; Piertney, S.B.; Wang, X.; et al. Surviving winter on the Qinghai-Tibetan Plateau: Pikas suppress energy demands and exploit yak feces to survive winter. Proc. Natl. Acad. Sci. USA 2021, 118, e2100707118. [Google Scholar] [CrossRef]
- Zheng, W.; He, Y.; Guo, Y.; Yue, T.; Zhang, H.; Li, J.; Zhou, B.; Zeng, X.; Li, L.; Wang, B.; et al. Large-scale genome sequencing redefines the genetic footprints of high-altitude adaptation in Tibetans. Genome Biol. 2023, 24, 73. [Google Scholar] [CrossRef]
- Wu, D.D.; Yang, C.P.; Wang, M.S.; Dong, K.Z.; Yan, D.W.; Hao, Z.Q.; Fan, S.Q.; Chu, S.Z.; Shen, Q.S.; Jiang, L.P.; et al. Convergent genomic signatures of high-altitude adaptation among domestic mammals. Natl. Sci. Rev. 2020, 7, 952–963. [Google Scholar] [CrossRef]
- Palomera-Sanchez, Z.; Zurita, M. Open, repair and close again: Chromatin dynamics and the response to UV-induced DNA damage. DNA Repair 2011, 10, 119–125. [Google Scholar] [CrossRef]
- Ma, Y.F.; Han, X.M.; Huang, C.P.; Zhong, L.; Adeola, A.C.; Irwin, D.M.; Xie, H.B.; Zhang, Y.P. Population Genomics Analysis Revealed Origin and High-altitude Adaptation of Tibetan Pigs. Sci. Rep. 2019, 9, 11463. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Li, Q.; Li, M.; Li, C. Transcriptome analysis reveals the long intergenic noncoding RNAs contributed to skeletal muscle differences between Yorkshire and Tibetan pig. Sci. Rep. 2021, 11, 2622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hu, S.; Cao, C.; Chen, C.; Liu, J.; Wang, Y.; Liu, J.; Zhao, J.; Tao, C.; Wang, Y. Functional and Genetic Characterization of Porcine Beige Adipocytes. Cells 2022, 11, 751. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Cao, C.; Liu, J.; Zhao, Y.; Pan, J.; Tao, C.; Wang, Y. Distinct Transcriptional Responses of Skeletal Muscle to Short-Term Cold Exposure in Tibetan Pigs and Bama Pigs. Int. J. Mol. Sci. 2023, 24, 7431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chamba, Y.; Shang, P.; Wang, Z.; Ma, J.; Wang, L.; Zhang, H. Comparative transcriptomic and proteomic analyses provide insights into the key genes involved in high-altitude adaptation in the Tibetan pig. Sci. Rep. 2017, 7, 3654. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, H.; Yang, T.; Li, Y.; Gao, C.; Jiao, T.; Cai, Y.; Zhao, S. The Expression Regulatory Network in the Lung Tissue of Tibetan Pigs Provides Insight into Hypoxia-Sensitive Pathways in High-Altitude Hypoxia. Front. Genet. 2021, 12, 691592. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, L.; Zhang, T.; Zhuang, D.; Wu, Q.; Yu, J.; Tian, C.; Zhang, Z. Gut microbiome signatures of extreme environment adaption in Tibetan pig. npj Biofilms Microbiomes 2023, 9, 27. [Google Scholar] [CrossRef]
- Kong, X.; Dong, X.; Yang, S.; Qian, J.; Yang, J.; Jiang, Q.; Li, X.; Wang, B.; Yan, D.; Lu, S.; et al. Natural selection on TMPRSS6 associated with the blunted erythropoiesis and improved blood viscosity in Tibetan pigs. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 233, 11–22. [Google Scholar] [CrossRef]
- Witt, K.E.; Huerta-Sánchez, E. Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos. Trans. R. Soc. B 2019, 374, 20180235. [Google Scholar] [CrossRef]
- Katz, A.E.; Gupte, T.; Ganesh, S.K. From Atherosclerosis to Spontaneous Coronary Artery Dissection: Defining a Clinical and Genetic Risk Spectrum for Myocardial Infarction. Curr. Atheroscler. Rep. 2024, 26, 331–340. [Google Scholar] [CrossRef]
- Ai, H.; Yang, B.; Li, J.; Xie, X.; Chen, H.; Ren, J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genom. 2014, 15, 834. [Google Scholar] [CrossRef] [PubMed]
- Grossman, S.R.; Shlyakhter, I.; Karlsson, E.K.; Byrne, E.H.; Morales, S.; Frieden, G.; Hostetter, E.; Angelino, E.; Garber, M.; Zuk, O.; et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 2010, 327, 883–886. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Liang, Y.; Huerta-Sanchez, E.; Jin, X.; Cuo, Z.X.; Pool, J.E.; Xu, X.; Jiang, H.; Vinckenbosch, N.; Korneliussen, T.S.; et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 2010, 329, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Cui, C.; He, Y.; Ouzhuluobu; Zhang, H.; Yang, D.; Zhang, Q.; Bianbazhuoma; Yang, L.; He, Y.; et al. Down-Regulation of EPASTranscription and Genetic Adaptation of Tibetans to High-Altitude Hypoxia. Mol. Biol. Evol. 2017, 34, 818–830. [Google Scholar] [PubMed]
- Liang, X.; Duan, Q.; Li, B.; Wang, Y.; Bu, Y.; Zhang, Y.; Kuang, Z.; Mao, L.; An, X.; Wang, H.; et al. Genomic structural variation contributes to evolved changes in gene expression in high-altitude Tibetan sheep. Proc. Natl. Acad. Sci. USA 2024, 121, e2322291121. [Google Scholar] [CrossRef]
- Nawrocki, M.J.; Jopek, K.; Kaczmarek, M.; Zdun, M.; Mozdziak, P.; Jemielity, M.; Perek, B.; Bukowska, D.; Kempisty, B. Transcriptomic Profile of Genes Regulating the Structural Organization of Porcine Atrial Cardiomyocytes during Primary In Vitro Culture. Genes 2022, 13, 1205. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Lenstra, J.A.; Zheng, Z.; Wu, X.; Yang, J.; Li, B.; Yang, Y.; Qiu, Q.; Liu, H.; et al. Evolutionary origin of genomic structural variations in domestic yaks. Nat. Commun. 2023, 14, 5617. [Google Scholar] [CrossRef]
- Kodani, A.; Moyer, T.; Chen, A.; Holland, A.; Walsh, C.A.; Reiter, J.F. SFI1 promotes centriole duplication by recruiting USP9X to stabilize the microcephaly protein, STIL. J. Cell Biol. 2019, 218, 2185–2197. [Google Scholar] [CrossRef]
- Gu, Q.; Orgil, B.O.; Bajpai, A.K.; Chen, Y.; Ashbrook, D.G.; Starlard-Davenport, A.; Towbin, J.A.; Lebeche, D.; Purevjav, E.; Sheng, H.; et al. Expression Levels of the Tnni3k Gene in the Heart Are Highly Associated with Cardiac and Glucose Metabolism-Related Phenotypes and Functional Pathways. Int. J. Mol. Sci. 2023, 24, 12759. [Google Scholar] [CrossRef]
- Zheng, Z.; Hua, R.; Xu, G.; Yang, H.; Shi, P. Gene losses may contribute to subterranean adaptations in naked mole-rat and blind mole-rat. BMC Biol. 2022, 20, 44. [Google Scholar] [CrossRef]
- Qu, H.; Zhang, Y.; Zhang, W.; Zhu, Y.; Xu, R. Knockout of cardiac troponin I-interacting kinase leads to cardiac dysfunction and remodelling. Clin. Exp. Pharmacol. Physiol. 2022, 49, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Zafra-Puerta, L.; Iglesias-Cabeza, N.; Burgos, D.F.; Sciaccaluga, M.; González-Fernández, J.; Bellingacci, L.; Canonichesi, J.; Sánchez-Martín, G.; Costa, C.; Sánchez, M.P.; et al. Gene therapy for Lafora disease in the Epm2a(−/−) mouse model. Mol. Ther. 2024, 32, 2130–2149. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, S.; Cai, Z.; Kuang, Z.; Wan, N.; Wang, Y.; Mao, L.; An, X.; Li, F.; Feng, T.; et al. Genomic insights into zokors’ phylogeny and speciation in China. Proc. Natl. Acad. Sci. USA 2022, 119, e2121819119. [Google Scholar] [CrossRef]
- Smith, J.L.; Wilson, M.L.; Nilson, S.M.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E.; Seabury, C.M. Genome-wide association and genotype by environment interactions for growth traits in U.S. Red Angus cattle. BMC Genom. 2022, 23, 517. [Google Scholar]
- Ding, R.; Zhuang, Z.; Qiu, Y.; Ruan, D.; Wu, J.; Ye, J.; Cao, L.; Zhou, S.; Zheng, E.; Huang, W.; et al. Identify known and novel candidate genes associated with backfat thickness in Duroc pigs by large-scale genome-wide association analysis. J. Anim. Sci. 2022, 100, skac012. [Google Scholar] [CrossRef]
- Chen, M.H.; Raffield, L.M.; Mousas, A.; Sakaue, S.; Huffman, J.E.; Moscati, A.; Trivedi, B.; Jiang, T.; Akbari, P.; Vuckovic, D.; et al. Trans-ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations. Cell 2020, 182, 1198–1213.e14. [Google Scholar] [CrossRef]
- Roberts, M.A.; Deol, K.K.; Mathiowetz, A.J.; Lange, M.; Leto, D.E.; Stevenson, J.; Hashemi, S.H.; Morgens, D.W.; Easter, E.; Heydari, K.; et al. Parallel CRISPR-Cas9 screens identify mechanisms of PLIN2 and lipid droplet regulation. Dev. Cell 2023, 58, 1782–1800.e10. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Hernandez, R.D. Selscan: An efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef]
- Deng, L.; Zhang, C.; Yuan, K.; Gao, Y.; Pan, Y.; Ge, X.; He, Y.; Yuan, Y.; Lu, Y.; Zhang, X.; et al. Prioritizing natural-selection signals from the deep-sequencing genomic data suggests multi-variant adaptation in Tibetan highlanders. Natl. Sci. Rev. 2019, 6, 1201–1222. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]

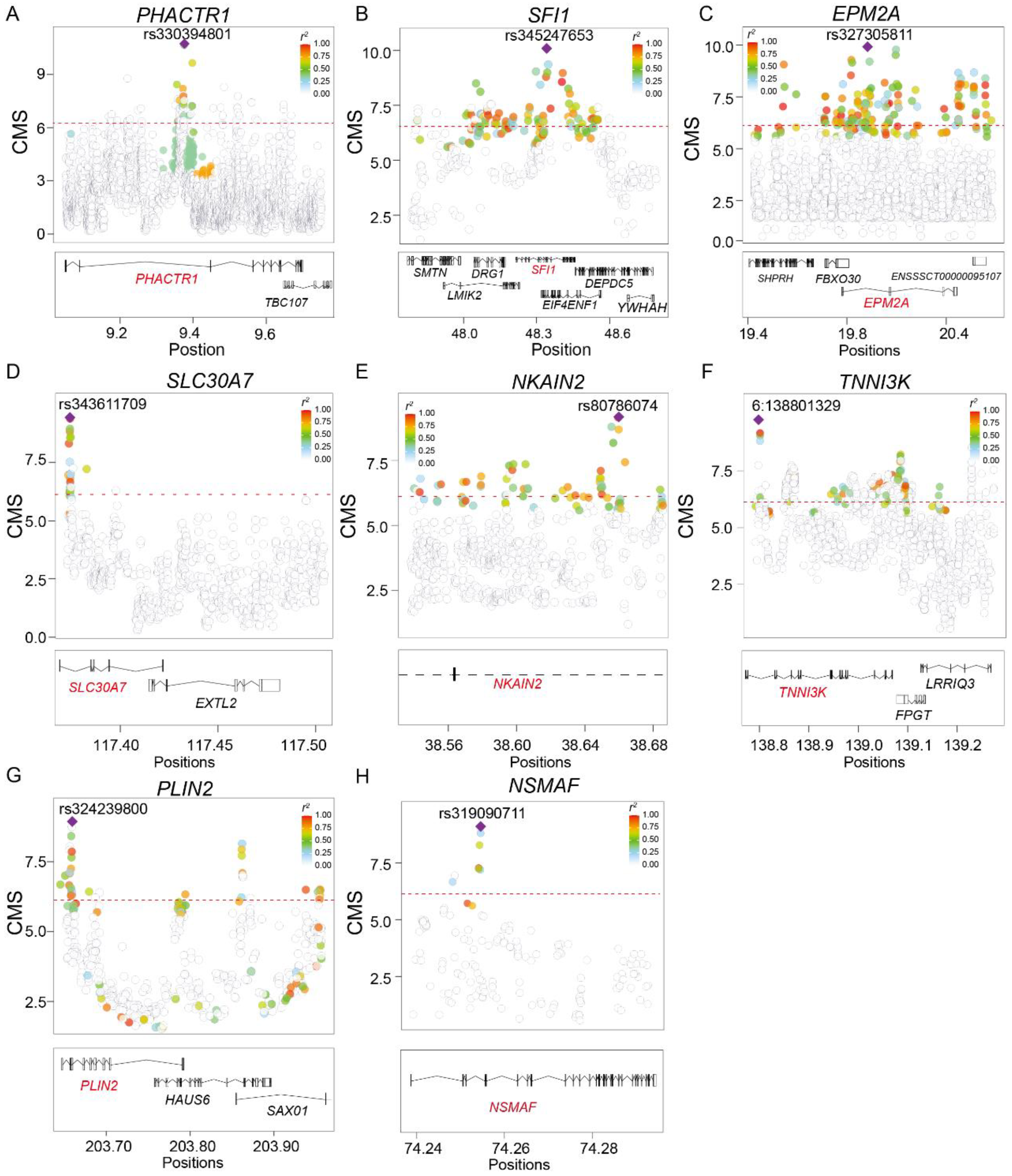
| Chromosome | rsID | Position (Sus scrofa 11.2) | CMS | PBS | Genes | Enriched Allele Frequency | ||
|---|---|---|---|---|---|---|---|---|
| TBPs (n = 58) | DU (n = 25) | YKS (n = 35) | ||||||
| 7 | rs330394801 | 9366281 | 11.14 | 0.63 | PHACTR1 | 0.93 | 0.33 | 0.06 |
| 14 | rs345247653 | 48268185 | 10.45 | 0.53 | SFI1 | 0.92 | 0.27 | 0.3 |
| 1 | rs327305811 | 19895804 | 9.92 | 0.43 | EPM2A | 0.88 | 0.45 | 0.43 |
| 4 | rs345409819 | 49294816 | 9.72 | 0.59 | BCR | 0.93 | 0.25 | 0.36 |
| 4 | rs343611709 | 117365692 | 9.42 | 0.55 | SLC30A7 | 0.77 | 0.17 | 0.12 |
| 1 | rs80786074 | 38655025 | 9.21 | 0.59 | NKAIN2 | 0.81 | 0.08 | 0.2 |
| 6 | 6:138801329 | 138801329 | 9.17 | 0.38 | TNNI3K | 0.76 | 0.01 | 0.31 |
| 1 | rs344915163 | 203690850 | 9.11 | 0.67 | PLIN2 | 0.75 | 0.02 | 0.03 |
| 18 | rs333217490 | 40016420 | 9.09 | 0.34 | BBS9 | 0.68 | 0.25 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Yuan, H.; Yao, B.; Zhao, S.; Wang, X.; Xu, L.; Zhang, L. Genetic Adaptations of the Tibetan Pig to High-Altitude Hypoxia on the Qinghai–Tibet Plateau. Int. J. Mol. Sci. 2024, 25, 11303. https://doi.org/10.3390/ijms252011303
Yang Y, Yuan H, Yao B, Zhao S, Wang X, Xu L, Zhang L. Genetic Adaptations of the Tibetan Pig to High-Altitude Hypoxia on the Qinghai–Tibet Plateau. International Journal of Molecular Sciences. 2024; 25(20):11303. https://doi.org/10.3390/ijms252011303
Chicago/Turabian StyleYang, Yanan, Haonan Yuan, Boyuan Yao, Shengguo Zhao, Xinrong Wang, Linna Xu, and Lingyun Zhang. 2024. "Genetic Adaptations of the Tibetan Pig to High-Altitude Hypoxia on the Qinghai–Tibet Plateau" International Journal of Molecular Sciences 25, no. 20: 11303. https://doi.org/10.3390/ijms252011303
APA StyleYang, Y., Yuan, H., Yao, B., Zhao, S., Wang, X., Xu, L., & Zhang, L. (2024). Genetic Adaptations of the Tibetan Pig to High-Altitude Hypoxia on the Qinghai–Tibet Plateau. International Journal of Molecular Sciences, 25(20), 11303. https://doi.org/10.3390/ijms252011303






