Tail Tales: What We Have Learned About Regeneration from Xenopus Laevis Tadpoles
Abstract
:1. Introduction

2. Signaling in Regeneration
2.1. Proinflammatory Signaling from Regeneration Organizing Cells (ROCs)
2.2. Oxidative Eustress in Tail Regeneration
2.3. Mechanotransduction Signaling in Tail Regeneration
2.4. Metabolic Alterations to Meet the Demands of Regeneration
3. Epigenetic Control of Tail Regeneration
4. Innate Immune Responses in Tail Regeneration
4.1. The Contribution of the Microbiome to Regeneration
4.2. Amputation-Induced Recruitment of Immune Cells During Regeneration
5. Remodeling of the ECM During Regeneration
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Daponte, V.; Tylzanowski, P.; Forlino, A. Appendage Regeneration in Vertebrates: What Makes This Possible? Cells 2021, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Agata, K.; Saito, Y.; Nakajima, E. Unifying principles of regeneration I: Epimorphosis versus morphallaxis. Dev. Growth Differ. 2007, 49, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Pellettieri, J. Regenerative tissue remodeling in planarians—The mysteries of morphallaxis. Semin. Cell Dev. Biol. 2019, 87, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Stoick-Cooper, C.L.; Moon, R.T.; Weidinger, G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007, 21, 1292–1315. [Google Scholar] [CrossRef]
- Franco, C.; Soares, R.; Pires, E.; Koci, K.; Almeida, A.M.; Santos, R.; Coelho, A.V. Understanding regeneration through proteomics. Proteomics 2013, 13, 686–709. [Google Scholar] [CrossRef]
- Jin, Y.; Li, S.; Yu, Q.; Chen, T.; Liu, D. Application of stem cells in regeneration medicine. MedComm 2023, 4, e291. [Google Scholar] [CrossRef]
- Londono, R.; Sun, A.X.; Tuan, R.S.; Lozito, T.P. Tissue Repair and Epimorphic Regeneration: An Overview. Curr. Pathobiol. Rep. 2018, 6, 61–69. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Amaya, E. The cellular and molecular mechanisms of tissue repair and regeneration as revealed by studies in Xenopus. Regeneration 2016, 3, 198–208. [Google Scholar] [CrossRef]
- Reddy, P.C.; Gungi, A.; Unni, M. Cellular and Molecular Mechanisms of Hydra Regeneration. Results Probl. Cell Differ. 2019, 68, 259–290. [Google Scholar] [CrossRef]
- Reddien, P.W. The Cellular and Molecular Basis for Planarian Regeneration. Cell 2018, 175, 327–345. [Google Scholar] [CrossRef]
- Cordero-Espinoza, L.; Huch, M. The balancing act of the liver: Tissue regeneration versus fibrosis. J. Clin. Investig. 2018, 128, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Bizzaro, D.; Russo, F.P.; Burra, P. New Perspectives in Liver Transplantation: From Regeneration to Bioengineering. Bioengineering 2019, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Rees, W.D.; Tandun, R.; Yau, E.; Zachos, N.C.; Steiner, T.S. Regenerative Intestinal Stem Cells Induced by Acute and Chronic Injury: The Saving Grace of the Epithelium? Front. Cell Dev. Biol. 2020, 8, 583919. [Google Scholar] [CrossRef] [PubMed]
- Biermann, M.; Reya, T. Hematopoietic Stem Cells and Regeneration. Cold Spring Harb. Perspect. Biol. 2022, 14, a040774. [Google Scholar] [CrossRef]
- Schmidt, M.; Schuler, S.C.; Huttner, S.S.; von Eyss, B.; von Maltzahn, J. Adult stem cells at work: Regenerating skeletal muscle. Cell. Mol. Life Sci. 2019, 76, 2559–2570. [Google Scholar] [CrossRef]
- Jopling, C.; Boue, S.; Izpisua Belmonte, J.C. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef]
- Phipps, L.S.; Marshall, L.; Dorey, K.; Amaya, E. Model systems for regeneration: Xenopus. Development 2020, 147, dev180844. [Google Scholar] [CrossRef]
- Liu, Y.; Lou, W.P.; Fei, J.F. The engine initiating tissue regeneration: Does a common mechanism exist during evolution? Cell Regen. 2021, 10, 12. [Google Scholar] [CrossRef]
- Chowdhury, K.; Lin, S.; Lai, S.L. Comparative Study in Zebrafish and Medaka Unravels the Mechanisms of Tissue Regeneration. Front. Ecol. Evol. 2022, 10, 783818. [Google Scholar] [CrossRef]
- Kawakami, A. Stem cell system in tissue regeneration in fish. Dev. Growth Differ. 2010, 52, 77–87. [Google Scholar] [CrossRef]
- Barr, J.I.; Boisvert, C.A.; Bateman, P.W. At What Cost? Trade-Offs and Influences on Energetic Investment in Tail Regeneration in Lizards Following Autotomy. J. Dev. Biol. 2021, 9, 53. [Google Scholar] [CrossRef]
- Gilbert, E.A.; Delorme, S.L.; Vickaryous, M.K. The regeneration blastema of lizards: An amniote model for the study of appendage replacement. Regeneration 2015, 2, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Vonk, A.C.; Zhao, X.; Pan, Z.; Hudnall, M.L.; Oakes, C.G.; Lopez, G.A.; Hasel-Kolossa, S.C.; Kuncz, A.W.C.; Sengelmann, S.B.; Gamble, D.J.; et al. Single-cell analysis of lizard blastema fibroblasts reveals phagocyte-dependent activation of Hedgehog-responsive chondrogenesis. Nat. Commun. 2023, 14, 4489. [Google Scholar] [CrossRef] [PubMed]
- Vieira, W.A.; Wells, K.M.; McCusker, C.D. Advancements to the Axolotl Model for Regeneration and Aging. Gerontology 2020, 66, 212–222. [Google Scholar] [CrossRef]
- Chuong, C.M.; Randall, V.A.; Widelitz, R.B.; Wu, P.; Jiang, T.X. Physiological regeneration of skin appendages and implications for regenerative medicine. Physiology 2012, 27, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Zhu, Z.; Sun, X.; Fu, X. Functional hair follicle regeneration: An updated review. Signal Transduct. Target. Ther. 2021, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M.; Watt, F.M. Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature 2018, 557, 322–328. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Beck, C.W. Studying regeneration in Xenopus. Methods Mol. Biol. 2012, 917, 525–539. [Google Scholar] [CrossRef]
- Deuchar, E.M. Regeneration of the tail bud in Xenopus embryos. J. Exp. Zool. 1975, 192, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Tucker, A.S.; Slack, J.M.W. The Xenopus-Laevis Tail-Forming Region. Development 1995, 121, 249–262. [Google Scholar] [CrossRef]
- Beck, C.W. Development of the vertebrate tailbud. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.W.; Izpisua Belmonte, J.C.; Christen, B. Beyond early development: Xenopus as an emerging model for the study of regenerative mechanisms. Dev. Dyn. 2009, 238, 1226–1248. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.W.; Christen, B.; Slack, J.M. Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev. Cell 2003, 5, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Aztekin, C.; Hiscock, T.W.; Marioni, J.C.; Gurdon, J.B.; Simons, B.D.; Jullien, J. Identification of a regeneration-organizing cell in the Xenopus tail. Science 2019, 364, 653–658. [Google Scholar] [CrossRef]
- Aztekin, C.; Storer, M.A. To regenerate or not to regenerate: Vertebrate model organisms of regeneration-competency and-incompetency. Wound Repair Regen. 2022, 30, 623–635. [Google Scholar] [CrossRef]
- Nieuwkoop, P.D. Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis, 1st ed.; Garland Pub: New York, NY, USA, 1994. [Google Scholar]
- Wang, S.; Shi, Y.B. Correction to: Evolutionary divergence in tail regeneration between Xenopus laevis and Xenopus tropicalis. Cell Biosci. 2021, 11, 104. [Google Scholar] [CrossRef]
- Williams, M.C.; Patel, J.H.; Kakebeen, A.D.; Wills, A.E. Nutrient availability contributes to a graded refractory period for regeneration in Xenopus tropicalis. Dev. Biol. 2021, 473, 59–70. [Google Scholar] [CrossRef]
- Zahn, N.; James-Zorn, C.; Ponferrada, V.G.; Adams, D.S.; Grzymkowski, J.; Buchholz, D.R.; Nascone-Yoder, N.M.; Horb, M.; Moody, S.A.; Vize, P.D.; et al. Normal Table of Xenopus development: A new graphical resource. Development 2022, 149, dev200356. [Google Scholar] [CrossRef]
- Gargioli, C.; Slack, J.M. Cell lineage tracing during Xenopus tail regeneration. Development 2004, 131, 2669–2679. [Google Scholar] [CrossRef] [PubMed]
- Akhurst, R.J.; Hata, A. Targeting the TGFbeta signalling pathway in disease. Nat. Rev. Drug Discov. 2012, 11, 790–811. [Google Scholar] [CrossRef]
- Ho, D.M.; Whitman, M. TGF-beta signaling is required for multiple processes during Xenopus tail regeneration. Dev. Biol. 2008, 315, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Yoshida, H.; Moriyama, Y.; Kawakita, I.; Wlizla, M.; Takebayashi-Suzuki, K.; Horb, M.E.; Suzuki, A. TGF-beta1 signaling is essential for tissue regeneration in the Xenopus tadpole tail. Biochem. Biophys. Res. Commun. 2021, 565, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Yoshida, H.; Takahashi, E.; Wlizla, M.; Takebayashi-Suzuki, K.; Horb, M.E.; Suzuki, A. The AP-1 transcription factor JunB functions in Xenopus tail regeneration by positively regulating cell proliferation. Biochem. Biophys. Res. Commun. 2020, 522, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Okada, M.; Takebayashi-Suzuki, K.; Ueno, N.; Suzuki, A. Involvement of JunB Proto-Oncogene in Tail Formation During Early Xenopus Embryogenesis. Zool. Sci. 2016, 33, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Kyoda, T.; Yoshida, H.; Takebayashi-Suzuki, K.; Koike, R.; Takahashi, E.; Moriyama, Y.; Wlizla, M.; Horb, M.E.; Suzuki, A. Injury-induced cooperation of InhibinbetaA and JunB is essential for cell proliferation in Xenopus tadpole tail regeneration. Sci. Rep. 2024, 14, 3679. [Google Scholar] [CrossRef]
- Okumura, A.; Hayashi, T.; Ebisawa, M.; Yoshimura, M.; Sasagawa, Y.; Nikaido, I.; Umesono, Y.; Mochii, M. Cell type-specific transcriptome analysis unveils secreted signaling molecule genes expressed in apical epithelial cap during appendage regeneration. Dev. Growth Differ. 2019, 61, 447–456. [Google Scholar] [CrossRef]
- Lin, G.; Slack, J.M. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev. Biol. 2008, 316, 323–335. [Google Scholar] [CrossRef]
- Lin, G.; Chen, Y.; Slack, J.M. Transgenic analysis of signaling pathways required for Xenopus tadpole spinal cord and muscle regeneration. Anat. Rec. 2012, 295, 1532–1540. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Love, N.R.; Chen, Y.; Ishibashi, S.; Kritsiligkou, P.; Lea, R.; Koh, Y.; Gallop, J.L.; Dorey, K.; Amaya, E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat. Cell Biol. 2013, 15, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.; Raghunathan, V.; Luxardi, G.; Zhu, K.; Zhao, M. Early redox activities modulate Xenopus tail regeneration. Nat. Commun. 2018, 9, 4296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, Y.; Xu, H.; Yang, L.; Yuan, F.; Li, L.; Xu, Y.; Chen, Y.; Zhang, C.; Lin, G. Melanocortin Receptor 4 Signaling Regulates Vertebrate Limb Regeneration. Dev. Cell 2018, 46, 397–409.e395. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Ouyang, Y.; Lu, N.; Li, N. The NF-kappaB Signaling Pathway, the Microbiota, and Gastrointestinal Tumorigenesis: Recent Advances. Front. Immunol. 2020, 11, 1387. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Bishop, T.F.; Beck, C.W. Bacterial lipopolysaccharides can initiate regeneration of the Xenopus tadpole tail. iScience 2021, 24, 103281. [Google Scholar] [CrossRef]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-kappaB in biology and targeted therapy: New insights and translational implications. Signal Transduct. Target. Ther. 2024, 9, 53. [Google Scholar] [CrossRef]
- Sugiura, T.; Tazaki, A.; Ueno, N.; Watanabe, K.; Mochii, M. Xenopus Wnt-5a induces an ectopic larval tail at injured site, suggesting a crucial role for noncanonical Wnt signal in tail regeneration. Mech. Dev. 2009, 126, 56–67. [Google Scholar] [CrossRef]
- Yin, C.; Ye, Z.; Wu, J.; Huang, C.; Pan, L.; Ding, H.; Zhong, L.; Guo, L.; Zou, Y.; Wang, X.; et al. Elevated Wnt2 and Wnt4 activate NF-kappaB signaling to promote cardiac fibrosis by cooperation of Fzd4/2 and LRP6 following myocardial infarction. EBioMedicine 2021, 74, 103745. [Google Scholar] [CrossRef]
- Shin, H.M.; Minter, L.M.; Cho, O.H.; Gottipati, S.; Fauq, A.H.; Golde, T.E.; Sonenshein, G.E.; Osborne, B.A. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 2006, 25, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.K.; Borodinsky, L.N. Spontaneous calcium transients manifest in the regenerating muscle and are necessary for skeletal muscle replenishment. Cell Calcium 2014, 56, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.B.; Borodinsky, L.N. Injury-induced Erk1/2 signaling tissue-specifically interacts with Ca2+ activity and is necessary for regeneration of spinal cord and skeletal muscle. Cell Calcium 2022, 102, 102540. [Google Scholar] [CrossRef] [PubMed]
- Korotkova, D.D.; Lyubetsky, V.A.; Ivanova, A.S.; Rubanov, L.I.; Seliverstov, A.V.; Zverkov, O.A.; Martynova, N.Y.; Nesterenko, A.M.; Tereshina, M.B.; Peshkin, L.; et al. Bioinformatics Screening of Genes Specific for Well-Regenerating Vertebrates Reveals c-answer, a Regulator of Brain Development and Regeneration. Cell Rep. 2019, 29, 1027–1040.e1026. [Google Scholar] [CrossRef] [PubMed]
- Woods, L.T.; Forti, K.M.; Shanbhag, V.C.; Camden, J.M.; Weisman, G.A. P2Y receptors for extracellular nucleotides: Contributions to cancer progression and therapeutic implications. Biochem. Pharmacol. 2021, 187, 114406. [Google Scholar] [CrossRef]
- Pannekoek, W.J.; de Rooij, J.; Gloerich, M. Force transduction by cadherin adhesions in morphogenesis. F1000Research 2019, 8, 1–14. [Google Scholar] [CrossRef]
- Moya, I.M.; Halder, G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019, 20, 211–226. [Google Scholar] [CrossRef]
- Panciera, T.; Azzolin, L.; Cordenonsi, M.; Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 758–770. [Google Scholar] [CrossRef]
- Hayashi, S.; Ochi, H.; Ogino, H.; Kawasumi, A.; Kamei, Y.; Tamura, K.; Yokoyama, H. Transcriptional regulators in the Hippo signaling pathway control organ growth in Xenopus tadpole tail regeneration. Dev. Biol. 2014, 396, 31–41. [Google Scholar] [CrossRef]
- Hayashi, S.; Tamura, K.; Yokoyama, H. Yap1, transcription regulator in the Hippo signaling pathway, is required for Xenopus limb bud regeneration. Dev. Biol. 2014, 388, 57–67. [Google Scholar] [CrossRef]
- Bay, S.; Ozturk, G.; Emekli, N.; Demircan, T. Downregulation of Yap1 during limb regeneration results in defective bone formation in axolotl. Dev. Biol. 2023, 500, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, X.; Zhou, Q.; Tian, Y.; Zuo, J.; Yuan, Z.; Liu, Y.; Li, J.; Sun, J. Hippo Signaling Regulates Blastema Formation During Limb Regeneration in Chinese Mitten Crab (Eriocheir sinensis). Mar. Biotechnol. 2023, 25, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Nikitovic, D.; Corsini, E.; Kouretas, D.; Tsatsakis, A.; Tzanakakis, G. ROS-major mediators of extracellular matrix remodeling during tumor progression. Food Chem. Toxicol. 2013, 61, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, S.; Heallen, T.; Martin, J.F. The Hippo pathway in the heart: Pivotal roles in development, disease, and regeneration. Nat. Rev. Cardiol. 2018, 15, 672–684. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Afzal, S.; Abdul Manap, A.S.; Attiq, A.; Albokhadaim, I.; Kandeel, M.; Alhojaily, S.M. From imbalance to impairment: The central role of reactive oxygen species in oxidative stress-induced disorders and therapeutic exploration. Front. Pharmacol. 2023, 14, 1269581. [Google Scholar] [CrossRef]
- Hong, Y.; Boiti, A.; Vallone, D.; Foulkes, N.S. Reactive Oxygen Species Signaling and Oxidative Stress: Transcriptional Regulation and Evolution. Antioxidants 2024, 13, 312. [Google Scholar] [CrossRef]
- Love, N.R.; Ziegler, M.; Chen, Y.; Amaya, E. Carbohydrate metabolism during vertebrate appendage regeneration: What is its role? How is it regulated?: A postulation that regenerating vertebrate appendages facilitate glycolytic and pentose phosphate pathways to fuel macromolecule biosynthesis. Bioessays 2014, 36, 27–33. [Google Scholar] [CrossRef]
- Denko, N.C. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer 2008, 8, 705–713. [Google Scholar] [CrossRef]
- Anastasiou, D.; Poulogiannis, G.; Asara, J.M.; Boxer, M.B.; Jiang, J.K.; Shen, M.; Bellinger, G.; Sasaki, A.T.; Locasale, J.W.; Auld, D.S.; et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011, 334, 1278–1283. [Google Scholar] [CrossRef]
- Min, S.; Whited, J.L. Limb blastema formation: How much do we know at a genetic and epigenetic level? J. Biol. Chem. 2023, 299, 102858. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Thakur, J.K.; Prasad, M. Histone acetylation dynamics regulating plant development and stress responses. Cell. Mol. Life Sci. 2021, 78, 4467–4486. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Takagi, C.; Miura, S.; Sakane, Y.; Suzuki, M.; Sakuma, T.; Sakamoto, N.; Endo, T.; Kamei, Y.; Sato, Y.; et al. In vivo tracking of histone H3 lysine 9 acetylation in Xenopus laevis during tail regeneration. Genes Cells 2016, 21, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Tseng, A.S.; Carneiro, K.; Lemire, J.M.; Levin, M. HDAC activity is required during Xenopus tail regeneration. PLoS ONE 2011, 6, e26382. [Google Scholar] [CrossRef]
- Taylor, A.J.; Beck, C.W. Histone deacetylases are required for amphibian tail and limb regeneration but not development. Mech. Dev. 2012, 129, 208–218. [Google Scholar] [CrossRef]
- Kakebeen, A.D.; Chitsazan, A.D.; Williams, M.C.; Saunders, L.M.; Wills, A.E. Chromatin accessibility dynamics and single cell RNA-Seq reveal new regulators of regeneration in neural progenitors. eLife 2020, 9, e52648. [Google Scholar] [CrossRef]
- Lee, J.S.; Smith, E.; Shilatifard, A. The language of histone crosstalk. Cell 2010, 142, 682–685. [Google Scholar] [CrossRef]
- Chapman, P.A.; Gilbert, C.B.; Devine, T.J.; Hudson, D.T.; Ward, J.; Morgan, X.C.; Beck, C.W. Manipulating the microbiome alters regenerative outcomes in Xenopus laevis tadpoles via lipopolysaccharide signalling. Wound Repair. Regen. 2022, 30, 636–651. [Google Scholar] [CrossRef]
- Piccinni, M.Z.; Watts, J.E.M.; Fourny, M.; Guille, M.; Robson, S.C. The skin microbiome of Xenopus laevis and the effects of husbandry conditions. Anim. Microbiome 2021, 3, 17. [Google Scholar] [CrossRef]
- Tsujioka, H.; Kunieda, T.; Katou, Y.; Shirahige, K.; Kubo, T. Unique gene expression profile of the proliferating Xenopus tadpole tail blastema cells deciphered by RNA-sequencing analysis. PLoS ONE 2015, 10, e0111655. [Google Scholar] [CrossRef]
- Tsujioka, H.; Kunieda, T.; Katou, Y.; Shirahige, K.; Fukazawa, T.; Kubo, T. interleukin-11 induces and maintains progenitors of different cell lineages during Xenopus tadpole tail regeneration. Nat. Commun. 2017, 8, 495. [Google Scholar] [CrossRef]
- Suzuki, S.; Sasaki, K.; Fukazawa, T.; Kubo, T. Xenopus laevis il11ra.L is an experimentally proven interleukin-11 receptor component that is required for tadpole tail regeneration. Sci. Rep. 2022, 12, 1903. [Google Scholar] [CrossRef]
- Cook, S.A. The Pathobiology of Interleukin 11 in Mammalian Disease is Likely Explained by its Essential Evolutionary Role for Fin Regeneration. J. Cardiovasc. Transl. Res. 2023, 16, 755–757. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.; Cook, S.A.; Schafer, S. Interleukin-11 signaling underlies fibrosis, parenchymal dysfunction, and chronic inflammation of the airway. Exp. Mol. Med. 2020, 52, 1871–1878. [Google Scholar] [CrossRef] [PubMed]
- Paredes, S.D.; Rancan, L.; Kireev, R.; Gonzalez, A.; Louzao, P.; Gonzalez, P.; Rodriguez-Bobada, C.; Garcia, C.; Vara, E.; Tresguerres, J.A. Melatonin Counteracts at a Transcriptional Level the Inflammatory and Apoptotic Response Secondary to Ischemic Brain Injury Induced by Middle Cerebral Artery Blockade in Aging Rats. BioRes. Open Access 2015, 4, 407–416. [Google Scholar] [CrossRef]
- Lai, S.L.; Marin-Juez, R.; Moura, P.L.; Kuenne, C.; Lai, J.K.H.; Tsedeke, A.T.; Guenther, S.; Looso, M.; Stainier, D.Y. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. eLife 2017, 6, e25605. [Google Scholar] [CrossRef] [PubMed]
- Godwin, J.W.; Pinto, A.R.; Rosenthal, N.A. Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 9415–9420. [Google Scholar] [CrossRef]
- Simkin, J.; Gawriluk, T.R.; Gensel, J.C.; Seifert, A.W. Macrophages are necessary for epimorphic regeneration in African spiny mice. eLife 2017, 6, e24623. [Google Scholar] [CrossRef]
- Simkin, J.; Sammarco, M.C.; Marrero, L.; Dawson, L.A.; Yan, M.; Tucker, C.; Cammack, A.; Muneoka, K. Macrophages are required to coordinate mouse digit tip regeneration. Development 2017, 144, 3907–3916. [Google Scholar] [CrossRef]
- Kakebeen, A.D.; Wills, A.E. More Than Just a Bandage: Closing the Gap Between Injury and Appendage Regeneration. Front. Physiol. 2019, 10, 81. [Google Scholar] [CrossRef]
- Aztekin, C.; Hiscock, T.W.; Butler, R.; De Jesus Andino, F.; Robert, J.; Gurdon, J.B.; Jullien, J. The myeloid lineage is required for the emergence of a regeneration-permissive environment following Xenopus tail amputation. Development 2020, 147, dev185496. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, T.; Naora, Y.; Kunieda, T.; Kubo, T. Suppression of the immune response potentiates tadpole tail regeneration during the refractory period. Development 2009, 136, 2323–2327. [Google Scholar] [CrossRef] [PubMed]
- Huffer, A.; Mao, M.; Ballard, K.; Ozdemir, T. Biomimetic Hyaluronan Binding Biomaterials to Capture the Complex Regulation of Hyaluronan in Tissue Development and Function. Biomimetics 2024, 9, 499. [Google Scholar] [CrossRef] [PubMed]
- Contreras, E.G.; Gaete, M.; Sanchez, N.; Carrasco, H.; Larrain, J. Early requirement of Hyaluronan for tail regeneration in Xenopus tadpoles. Development 2009, 136, 2987–2996. [Google Scholar] [CrossRef]
- Sindelka, R.; Naraine, R.; Abaffy, P.; Zucha, D.; Kraus, D.; Netusil, J.; Smetana, K., Jr.; Lacina, L.; Endaya, B.B.; Neuzil, J.; et al. Characterization of regeneration initiating cells during Xenopus laevis tail regeneration. Genome Biol. 2024, 25, 251. [Google Scholar] [CrossRef]
- Murugan, N.J.; Vigran, H.J.; Miller, K.A.; Golding, A.; Pham, Q.L.; Sperry, M.M.; Rasmussen-Ivey, C.; Kane, A.W.; Kaplan, D.L.; Levin, M. Acute multidrug delivery via a wearable bioreactor facilitates long-term limb regeneration and functional recovery in adult Xenopus laevis. Sci. Adv. 2022, 8, eabj2164. [Google Scholar] [CrossRef]
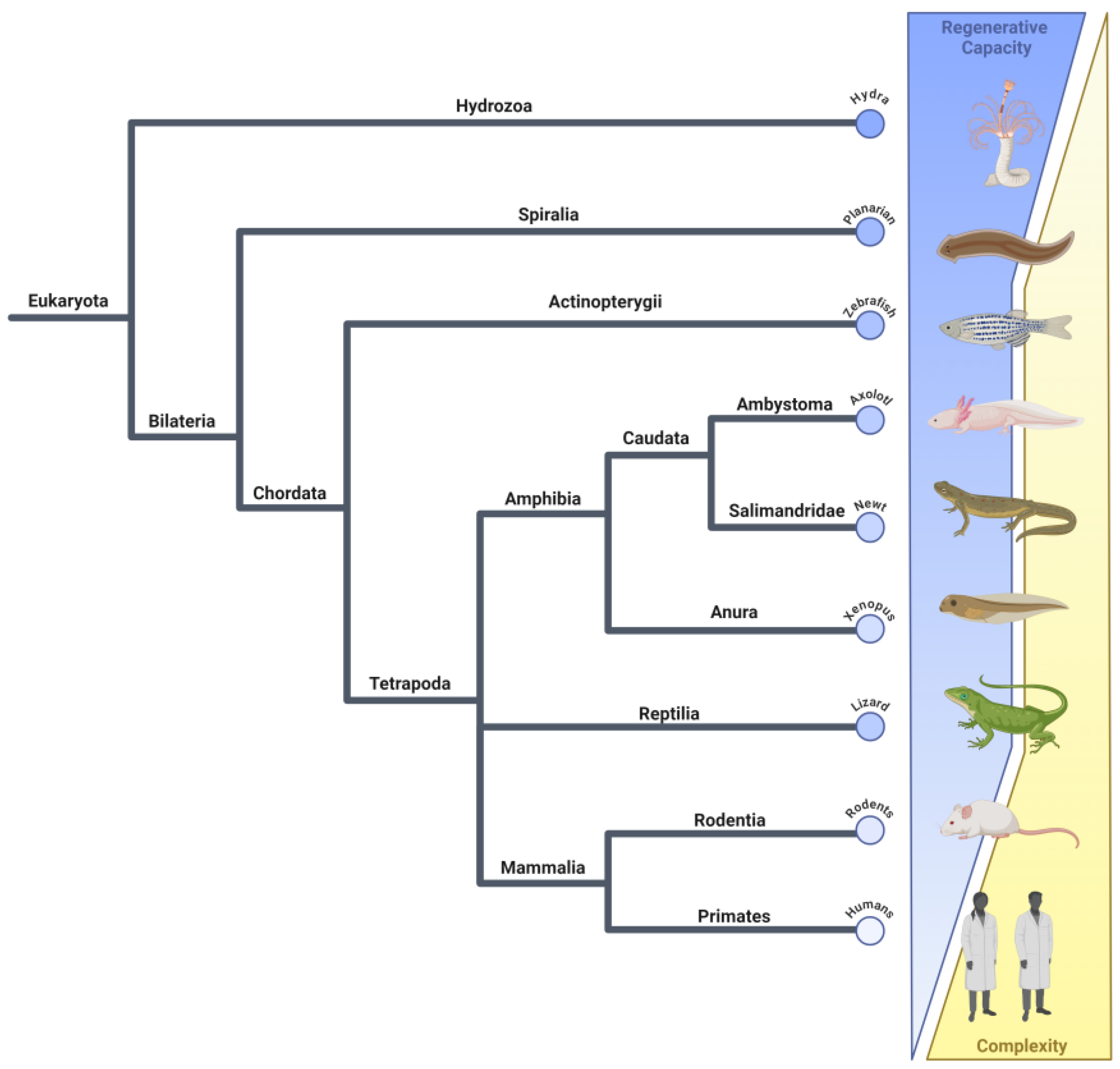

| Type of Regeneration | Definition | Examples |
|---|---|---|
| Epimorphosis | Involves the formation of a mass of undifferentiated cells, known as a blastema, at the site of injury. These cells differentiate into the various cell types needed to regrow the lost structure [2,7]. | Amphibians can regenerate entire limbs, including bones, muscles, nerves, and skin [2,7]. Xenopus tadpoles can regenerate their tails through the formation of a blastema that differentiates into the various tissues of the tail [8]. |
| Morphallaxis | Involves the reorganization of existing tissues without significant cell proliferation. This process typically results in the direct transformation of existing cells into a new structure [2,3]. | Hydra can regenerate its entire body from a small fragment by reorganizing its existing cells to form a complete organism [2,9]. Planarians can regenerate from small body fragments through a combination of morphallaxis and epimorphosis, involving both reorganization and proliferation of cells [2,3,10]. |
| Compensatory | Occurs when differentiated cells divide but maintain their original function. There is no formation of a blastema, and the regeneration typically restores function rather than form [4]. | The mammalian liver can regenerate lost tissue through compensatory hypertrophy and hyperplasia. Hepatocytes grow and divide to restore the liver’s mass and function without forming a blastema [11,12]. |
| Stem cell-mediated | This involves the activation and differentiation of stem cells for regeneration, either through undifferentiated stem cells or tissue-specific progenitors, without the formation of a blastema [6]. | The mammalian intestinal epithelium is continuously regenerated by stem cells located in the crypts of the intestinal lining [13,14]. In the hematopoietic system, blood cells are regenerated from hematopoietic stem cells in the bone marrow, which continuously produce new blood cells throughout an organism’s life [15]. Human skeletal muscle can regenerate through the activation of satellite cells, which are muscle-specific stem cells that proliferate and differentiate to repair muscle fibers [16]. Newts can regenerate the lens of their eyes when the cells from the iris dedifferentiate and proliferate to form a new lens [17]. |
| Regeneration Stage (Time After Amputation) | Dominant Regulatory Pathways |
|---|---|
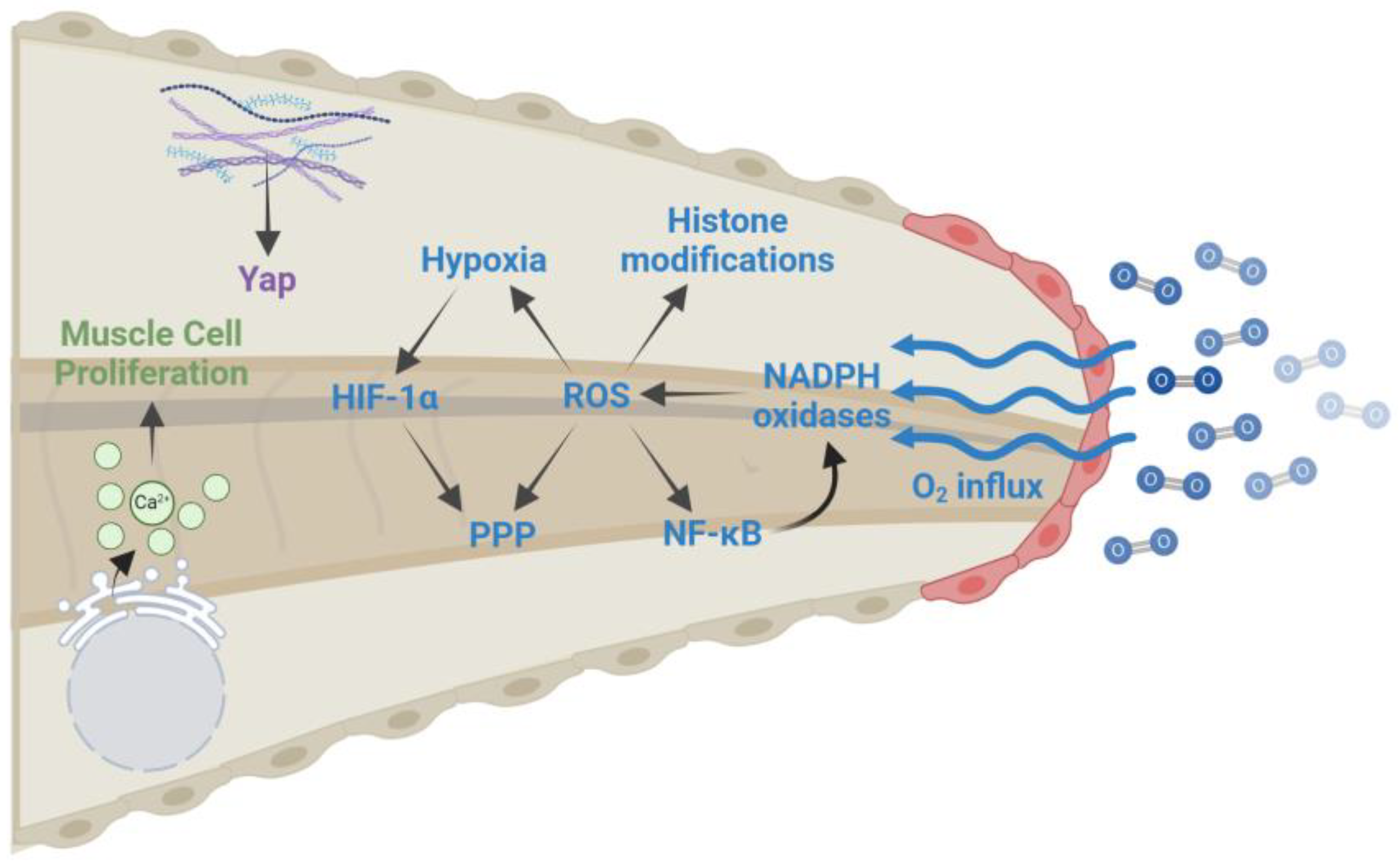
| Wound healing (0–6 hpa) | ROS production: Oxygen influx at the damage site leads to increased ROS levels that are essential for regeneration. ROS levels remain elevated for several days after amputation and are required for activating NF-κB, creating a hypoxic environment, altering histone modifications, and diverting glucose to the pentose phosphate pathway. |
| Calcium signaling: Calcium is released from the ER following injury and induces the activation of muscle satellite cells and the proliferation of muscle progenitor cells. | |
| Inflammatory response: The recruitment of innate immune cells, including macrophages, neutrophils, and myeloid cells, to the amputation site is required for regeneration. | |
| ECM remodeling: ECM remodeling genes are expressed in the RICs that emerge hours after amputation. RICs combined with upregulation of the HA pathway promote the migration of ROCs to the wound edge. | |
| TGFβ signaling: This signaling pathway is required for the proper formation of the wound epidermis following amputation. | |
| Blastema formation (6–48 hpa) | Growth factor signaling: Additional growth factors, like FGFs, BMPs, and Wnts, are secreted by ROCs to promote the proliferation of progenitor cells in the blastema. |
| Anti-inflammatory response: The inflammatory response must be dampened for regeneration to occur. Chronic inflammation leads to a loss of regenerative capabilities. | |
| Epigenetic changes: H3K9Ac peaks one day after amputation and likely facilitates the expression of genes required for cell proliferation and regenerative outgrowth. However, there are a variety of epigenetic changes in which the dynamics are not well known and likely regulate other epigenetic modifications. | |
| Regenerative outgrowth (2–7 dpa) | Hippo/YAP signaling: Yap is expressed several days after amputation and maintains the survival of neural progenitors during the growth of the regenerating tail. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lara, J.; Mastela, C.; Abd, M.; Pitstick, L.; Ventrella, R. Tail Tales: What We Have Learned About Regeneration from Xenopus Laevis Tadpoles. Int. J. Mol. Sci. 2024, 25, 11597. https://doi.org/10.3390/ijms252111597
Lara J, Mastela C, Abd M, Pitstick L, Ventrella R. Tail Tales: What We Have Learned About Regeneration from Xenopus Laevis Tadpoles. International Journal of Molecular Sciences. 2024; 25(21):11597. https://doi.org/10.3390/ijms252111597
Chicago/Turabian StyleLara, Jessica, Camilla Mastela, Magda Abd, Lenore Pitstick, and Rosa Ventrella. 2024. "Tail Tales: What We Have Learned About Regeneration from Xenopus Laevis Tadpoles" International Journal of Molecular Sciences 25, no. 21: 11597. https://doi.org/10.3390/ijms252111597
APA StyleLara, J., Mastela, C., Abd, M., Pitstick, L., & Ventrella, R. (2024). Tail Tales: What We Have Learned About Regeneration from Xenopus Laevis Tadpoles. International Journal of Molecular Sciences, 25(21), 11597. https://doi.org/10.3390/ijms252111597






