Epigenetic DNA Methylation and Protein Homocysteinylation: Key Players in Hypertensive Renovascular Damage
Abstract
1. Introduction
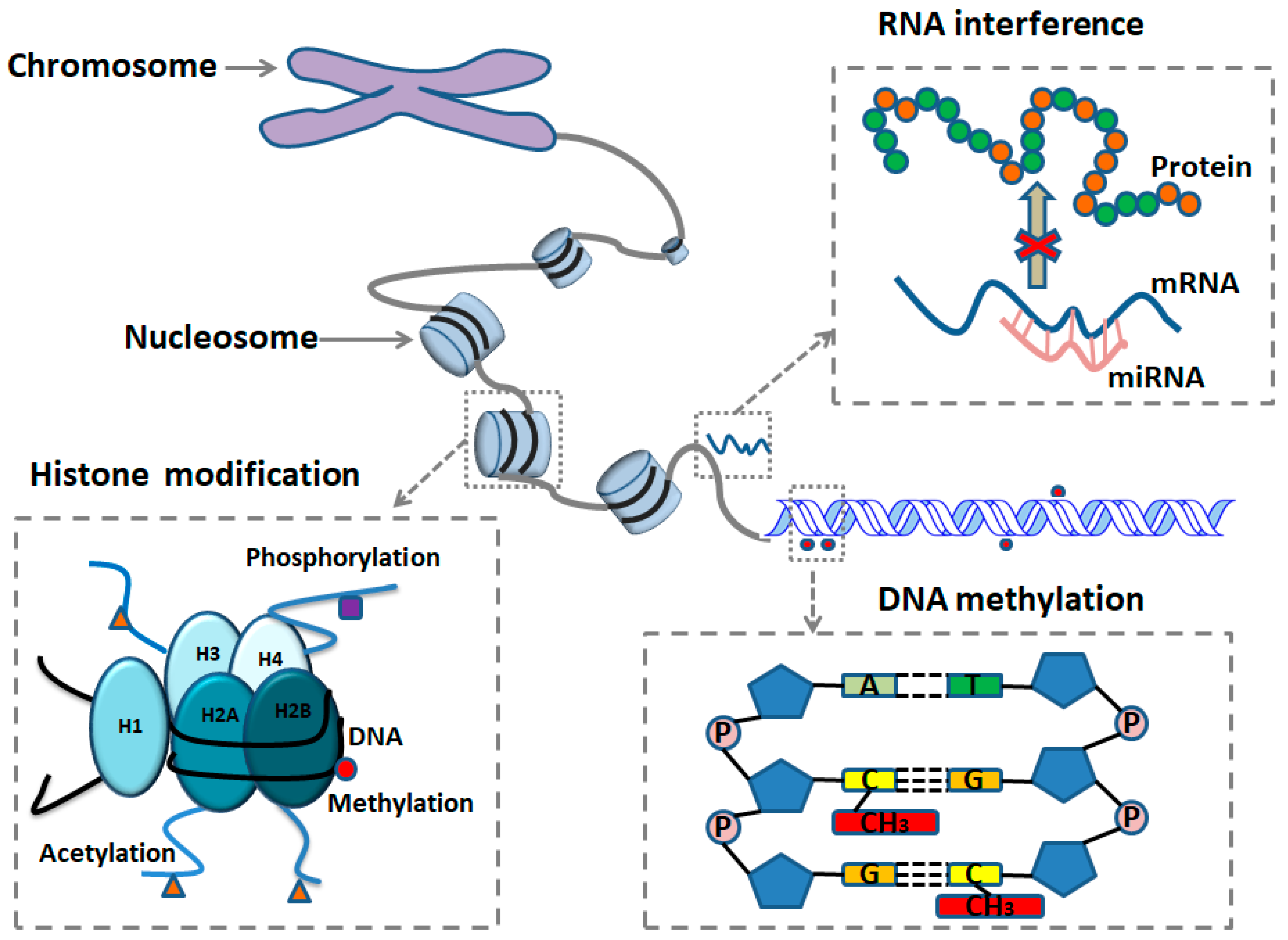
2. Epigenetics of Disease Mechanism
2.1. DNA Methylation
2.2. Histone Modification
2.3. Noncoding RNA
3. Epigenetic Cellular Modification and Hypertension
3.1. Endothelial Cells
3.1.1. Effect of DNA Methylation on Endothelial Cells
3.1.2. Effect of Histone Modifications on Endothelial Cells
3.1.3. Effect of Noncoding RNA Regulation on Endothelial Cells
3.2. Smooth Muscle Cells
3.2.1. Effect of DNA Methylation on SMCs
3.2.2. Effect of Histone Modifications on SMCs
3.2.3. Effect of Noncoding RNA Regulation on SMCs
3.3. Kidney
3.3.1. Effect of DNA Methylation on Podocyte, Mesangial, and Epithelial Cells
3.3.2. Effect of Histone Modifications on Podocyte, Mesangial, and Epithelial Cells
3.3.3. Effect of Noncoding RNA Regulation on Podocyte, Mesangial, and Epithelial Cells
4. Hypertension-Related Kidney Damages
4.1. Nephron Loss
4.2. Glomerular Sclerosis or Necrosis
4.3. Renal Fibrogenesis
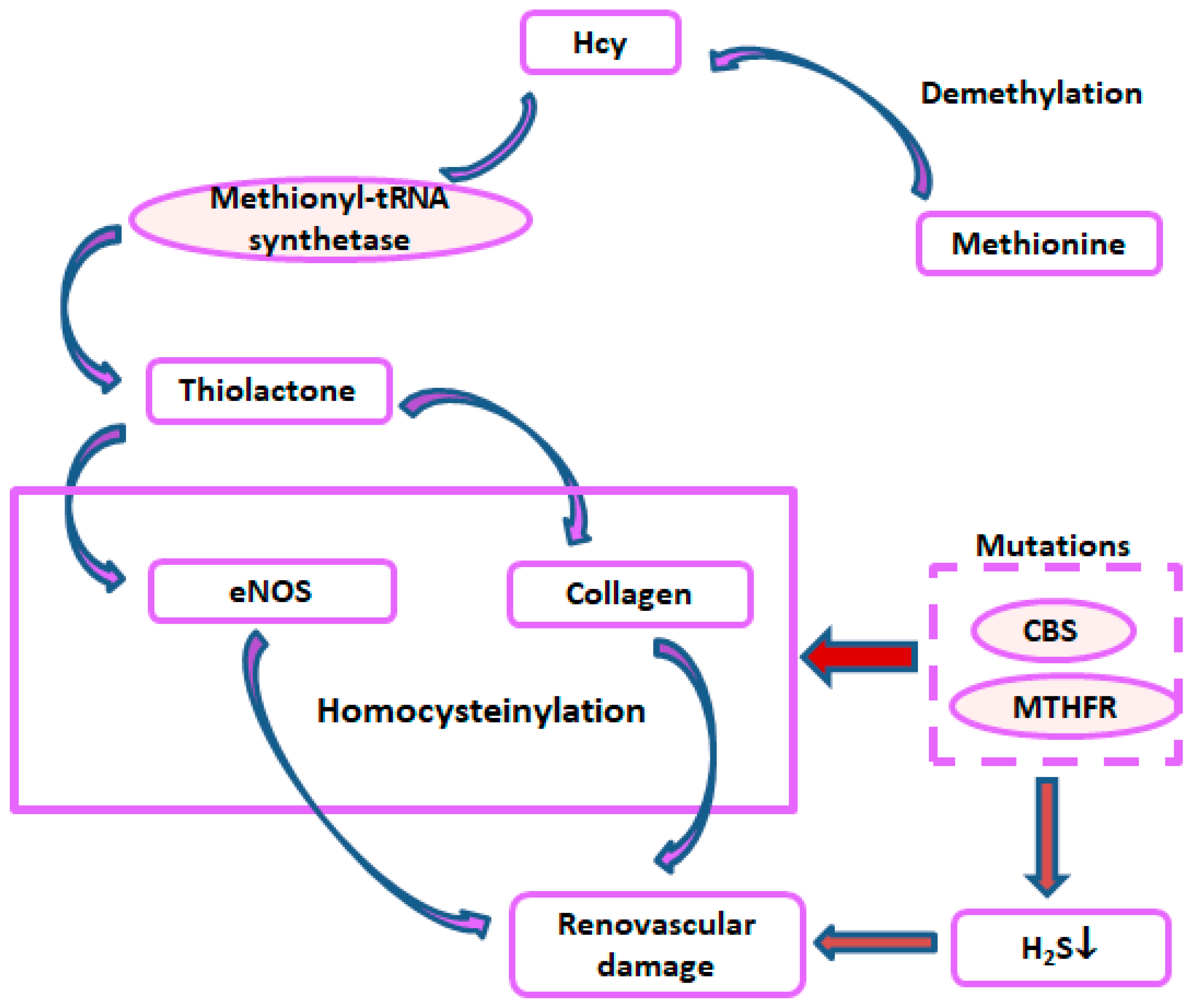
5. Metabolic Disorders, Hyperhomocysteinemia, and Hypertension
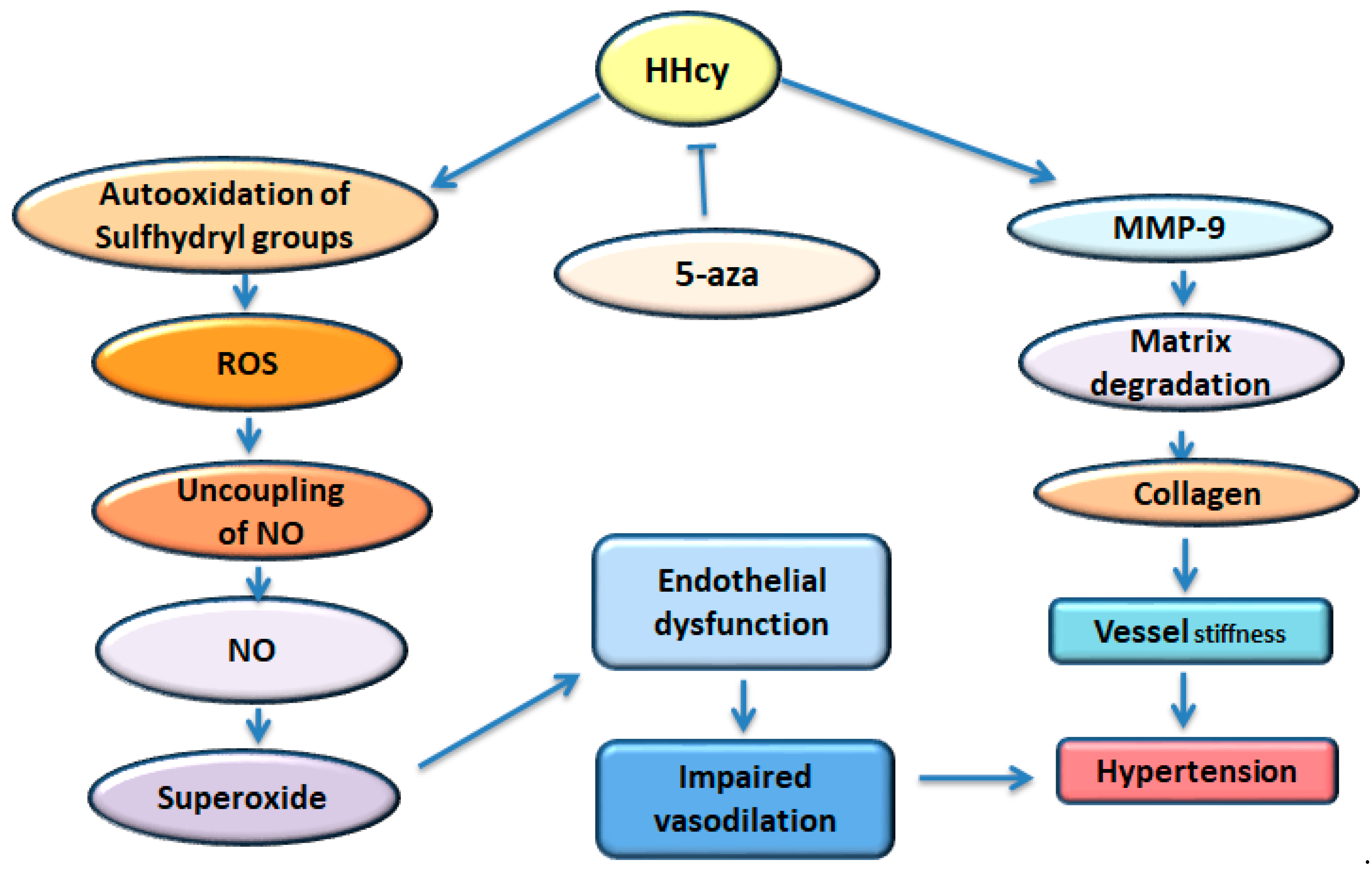
6. Protein Homocysteinylation and Hypertension
7. Oxidative Stress
8. Contribution of Biologically Active Gases in Epigenetic Hypertension
8.1. Hydrogen Sulfide (H2S)
8.2. Carbon Monoxide (CO)
8.3. Nitric Oxide (NO)
8.4. Interaction of Biological Gases
9. Other Mechanisms
9.1. Asymmetric Dimethylarginine
9.2. Glucocorticoid Level
9.3. Taurine
10. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E., Jr.; Collins, K.J.; Dennison Himmelfarb, C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018, 71, e13–e115. [Google Scholar] [CrossRef] [PubMed]
- Chobufo, M.D.; Gayam, V.; Soluny, J.; Rahman, E.U.; Enoru, S.; Foryoung, J.B.; Agbor, V.N.; Dufresne, A.; Nfor, T. Prevalence and control rates of hypertension in the USA: 2017–2018. Int. J. Cardiol. Hypertens. 2020, 6, 100044. [Google Scholar] [CrossRef] [PubMed]
- Raftopoulos, L.; Katsi, V.; Makris, T.; Tousoulis, D.; Stefanadis, C.; Kallikazaros, I. Epigenetics, the missing link in hypertension. Life Sci. 2014, 129, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.-Y.; Cowley, A.W.; Mattson, D.L.; Kotchen, T.A.; Liu, Y. Epigenomics of Hypertension. Semin. Nephrol. 2013, 33, 392–399. [Google Scholar] [CrossRef]
- Carriazo, S.; Vanessa Perez-Gomez, M.; Ortiz, A. Hypertensive nephropathy: A major roadblock hindering the advance of precision nephrology. Clin. Kidney J. 2020, 13, 504–509. [Google Scholar] [CrossRef]
- Freedman, B.I.; Cohen, A.H. Hypertension-attributed nephropathy: What’s in a name? Nat. Rev. Nephrol. 2016, 12, 27–36. [Google Scholar] [CrossRef]
- Xu, Y.; Ren, Y.; Zhang, J.; Niu, B.; Liu, M.; Xu, T.; Zhang, X.; Shen, J.; Wang, K.; Cao, Z. Discovery of pyridazinone derivatives bearing tetrahydroimidazo[1,2-a]pyrazine scaffold as potent inhibitors of transient receptor potential canonical 5 to ameliorate hypertension-induced renal injury in rats. Eur. J. Med. Chem. 2024, 275, 116565. [Google Scholar] [CrossRef]
- Mennuni, S.; Rubattu, S.; Pierelli, G.; Tocci, G.; Fofi, C.; Volpe, M. Hypertension and kidneys: Unraveling complex molecular mechanisms underlying hypertensive renal damage. J. Hum. Hypertens. 2014, 28, 74–79. [Google Scholar] [CrossRef]
- Irigoyen, M.C.; Fetter, C.; De Angelis, K. Advances on the Experimental Research in Resistant Hypertension. Curr. Hypertens. Rep. 2024, 26, 475–482. [Google Scholar] [CrossRef]
- Padmanabhan, S.; Dominiczak, A.F. Genomics of hypertension: The road to precision medicine. Nat. Rev. Cardiol. 2021, 18, 235–250. [Google Scholar] [CrossRef]
- Van Duijvenboden, S.; Ramirez, J.; Young, W.J.; Olczak, K.J.; Ahmed, F.; Alhammadi, M.; International Consortium of Blood, P.; Bell, C.G.; Morris, A.P.; Munroe, P.B. Integration of genetic fine-mapping and multi-omics data reveals candidate effector genes for hypertension. Am. J. Hum. Genet. 2023, 110, 1718–1734. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, S.; Delles, C.; Dominiczak, A.F. Beyond Genome-Wide Scans: Advancing Hypertension Genomics Into the Future. Hypertension 2024, 81, 1186–1189. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Snieder, H. Genome-wide association studies and beyond: What’s next in blood pressure genetics? Hypertension 2010, 56, 1035–1037. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Ko, Y.-A.; Suszak, K. Epigenomics: The science of no-longer-junk DNA. Why study it in chronic kidney disease? Semin. Nephrol. 2013, 33, 354–362. [Google Scholar] [CrossRef][Green Version]
- Kuneš, J.; Kadlecová, M.; Vaněčková, I.; Zicha, J. Critical developmental periods in the pathogenesis of hypertension. Physiol. Res. 2012, 61, S9–S17. [Google Scholar] [CrossRef]
- Miranda, T.B.; Jones, P.A. DNA methylation: The nuts and bolts of repression. J. Cell. Physiol. 2007, 213, 384–390. [Google Scholar] [CrossRef]
- Millis, R.M. Epigenetics and hypertension. Curr. Hypertens. Rep. 2011, 13, 21–28. [Google Scholar] [CrossRef]
- Cusack, M.; King, H.W.; Spingardi, P.; Kessler, B.M.; Klose, R.J.; Kriaucionis, S. Distinct contributions of DNA methylation and histone acetylation to the genomic occupancy of transcription factors. Genome Res. 2020, 30, 1393–1406. [Google Scholar] [CrossRef]
- Kaluscha, S.; Domcke, S.; Wirbelauer, C.; Stadler, M.B.; Durdu, S.; Burger, L.; Schubeler, D. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat. Genet. 2022, 54, 1895–1906. [Google Scholar] [CrossRef]
- Du, Q.; Luu, P.L.; Stirzaker, C.; Clark, S.J. Methyl-CpG-binding domain proteins: Readers of the epigenome. Epigenomics 2015, 7, 1051–1073. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, L.; Nakielny, S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J. Biol. Chem. 2004, 279, 49479–49487. [Google Scholar] [CrossRef] [PubMed]
- Holoch, D.; Margueron, R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem. Sci. 2017, 42, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Veland, N.; Lu, Y.; Hardikar, S.; Gaddis, S.; Zeng, Y.; Liu, B.; Estecio, M.R.; Takata, Y.; Lin, K.; Tomida, M.W.; et al. DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 2019, 47, 152–167. [Google Scholar] [CrossRef]
- Aapola, U.; Kawasaki, K.; Scott, H.S.; Ollila, J.; Vihinen, M.; Heino, M.; Shintani, A.; Kawasaki, K.; Minoshima, S.; Krohn, K.; et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 2000, 65, 293–298. [Google Scholar] [CrossRef]
- Aapola, U.; Lyle, R.; Krohn, K.; Antonarakis, S.E.; Peterson, P. Isolation and initial characterization of the mouse Dnmt3l gene. Cytogenet. Cell Genet. 2001, 92, 122–126. [Google Scholar] [CrossRef]
- Hata, K.; Okano, M.; Lei, H.; Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002, 129, 1983–1993. [Google Scholar] [CrossRef]
- Chedin, F.; Lieber, M.R.; Hsieh, C.L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 2002, 99, 16916–16921. [Google Scholar] [CrossRef]
- Suetake, I.; Shinozaki, F.; Miyagawa, J.; Takeshima, H.; Tajima, S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004, 279, 27816–27823. [Google Scholar] [CrossRef]
- Chen, Z.X.; Mann, J.R.; Hsieh, C.L.; Riggs, A.D.; Chédin, F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 2005, 95, 902–917. [Google Scholar] [CrossRef]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Haws, S.A.; Leech, C.M.; Denu, J.M. Metabolism and the Epigenome: A Dynamic Relationship. Trends Biochem. Sci. 2020, 45, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Goffin, J.; Eisenhauer, E. DNA methyltransferase inhibitors-state of the art. Ann. Oncol. 2002, 13, 1699–1716. [Google Scholar] [CrossRef]
- De Oca, A.M.; Madueño, J.A.; Martinez-Moreno, J.M.; Guerrero, F.; Muñoz-Castañeda, J.; Rodriguez-Ortiz, M.E.; Mendoza, F.J.; Almaden, Y.; Lopez, I.; Rodriguez, M.; et al. High-Phosphate-Induced Calcification Is Related to α Promoter Methylation in Vascular Smooth Muscle Cells. J. Bone Miner. Res. 2010, 25, 1996–2005. [Google Scholar] [CrossRef]
- Lifton, R.P.; Gharavi, A.G.; Geller, D.S. Molecular mechanisms of human hypertension. Cell 2001, 104, 545–556. [Google Scholar] [CrossRef]
- Escher, G.; Vogt, B.; Beck, T.; Guntern, D.; Frey, B.M.; Frey, F.J. Reduced 11beta-hydroxysteroid dehydrogenase activity in the remaining kidney following nephrectomy. Endocrinology 1998, 139, 1533–1539. [Google Scholar] [CrossRef]
- Alikhani-Koopaei, R.; Fouladkou, F.; Frey, F.J.; Frey, B.M. Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J. Clin. Investig. 2004, 114, 1146–1157. [Google Scholar] [CrossRef]
- Funke-Kaiser, H.; Thomas, A.; Bremer, J.; Kovacevic, S.D.; Scheuch, K.; Bolbrinker, J.; Theis, S.; Lemmer, J.; Zimmermann, A.; Zollmann, F.S.; et al. Regulation of the major isoform of human endothelin-converting enzyme-1 by a strong housekeeping promoter modulated by polymorphic microsatellites. J. Hypertens. 2003, 21, 2111–2124. [Google Scholar] [CrossRef]
- Yan, M.S.; Matouk, C.C.; Marsden, P.A. Epigenetics of the vascular endothelium. J. Appl. Physiol. (1985) 2010, 109, 916–926. [Google Scholar] [CrossRef]
- Wang, G.G.; Allis, C.D.; Chi, P. Chromatin remodeling and cancer. Part I: Covalent histone modifications. Trends Mol. Med. 2007, 13, 363–372. [Google Scholar] [CrossRef]
- Samavat, S.; Ahmadpoor, P.; Samadian, F. Aldosterone, hypertension, and beyond. Iran. J. Kidney Dis. 2011, 5, 71–76. [Google Scholar] [PubMed]
- Tampe, D.; Zeisberg, M. A primer on the epigenetics of kidney fibrosis. Minerva Medica 2012, 103, 267–278. [Google Scholar] [PubMed]
- Lukauskas, S.; Tvardovskiy, A.; Nguyen, N.V.; Stadler, M.; Faull, P.; Ravnsborg, T.; Ozdemir Aygenli, B.; Dornauer, S.; Flynn, H.; Lindeboom, R.G.H.; et al. Decoding chromatin states by proteomic profiling of nucleosome readers. Nature 2024, 627, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Policarpi, C.; Munafo, M.; Tsagkris, S.; Carlini, V.; Hackett, J.A. Systematic epigenome editing captures the context-dependent instructive function of chromatin modifications. Nat. Genet. 2024, 56, 1168–1180. [Google Scholar] [CrossRef]
- Bush, E.W.; McKinsey, T.A. Protein acetylation in the cardiorenal axis: The promise of histone deacetylase inhibitors. Circ. Res. 2010, 106, 272–284. [Google Scholar] [CrossRef]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef]
- Chen, S.; Bellew, C.; Yao, X.; Stefkova, J.; Dipp, S.; Saifudeen, Z.; Bachvarov, D.; El-Dahr, S.S. Histone deacetylase (HDAC) activity is critical for embryonic kidney gene expression, growth, and differentiation. J. Biol. Chem. 2011, 286, 32775–32789. [Google Scholar] [CrossRef]
- Consalvi, S.; Saccone, V.; Giordani, L.; Minetti, G.; Mozzetta, C.; Puri, P.L. Histone deacetylase inhibitors in the treatment of muscular dystrophies: Epigenetic drugs for genetic diseases. Mol. Med. 2011, 17, 457–465. [Google Scholar] [CrossRef]
- Margariti, A.; Zampetaki, A.; Xiao, Q.; Zhou, B.; Karamariti, E.; Martin, D.; Yin, X.; Mayr, M.; Li, H.; Zhang, Z.; et al. Histone deacetylase 7 controls endothelial cell growth through modulation of betacatenin. Circ. Res. 2010, 106, 1202–1211. [Google Scholar] [CrossRef]
- Kook, H.; Lepore, J.J.; Gitler, A.D.; Lu, M.M.; Yung, W.W.-M.; Mackay, J.; Zhou, R.; Ferrari, V.; Gruber, P.; Epstein, J.A. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J. Clin. Investig. 2003, 112, 863–871. [Google Scholar] [CrossRef]
- Miska, E.A.; Karlsson, C.; Langley, E.; Nielsen, S.J.; Pines, J.; T Kouzarides, T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999, 18, 5099–5107. [Google Scholar] [CrossRef]
- Morris, K.V. siRNA-mediated transcriptional gene silencing: The potential mechanism and a possible role in the histone code. Cell. Mol. Life Sci. 2005, 62, 3057–3066. [Google Scholar] [CrossRef] [PubMed]
- Kuehbacher, A.; Urbich, C.; Dimmeler, S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol. Sci. 2008, 29, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea, M.D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Saco, T.V.; Parthasarathy, P.T.; Cho, Y.; Lockey, R.F.; Kolliputi, N. Role of epigenetics in pulmonary hypertension. Am. J. Physiol. Cell Physiol. 2014, 306, C1101–C1105. [Google Scholar] [CrossRef]
- Friso, S.; Carvajal, C.A.; Fardella, C.E.; Olivieri, O. Epigenetics and arterial hypertension: The challenge of emerging evidence. Transl. Res. J. Lab. Clin. Med. 2015, 165, 154–165. [Google Scholar] [CrossRef]
- Stary, H.C.; Chandler, A.B.; Glagov, S.; Guyton, J.R.; Insull, W.J.; Rosenfeld, M.E.; Schaffer, S.A.; Schwartz, C.J.; Wagner, W.D.; Wissler, R.W. A definition of initial, fatty streak, and intermediate lesions of atherosclerosis. A report from the Committee on Vascular Lesions of the Council on Arteriosclerosis, American Heart Association. Circulation 1994, 89, 2462–2478. [Google Scholar] [CrossRef]
- Ross, R. The pathogenesis of atherosclerosis: A perspective for the 1990s. Nature 1993, 362, 801–809. [Google Scholar] [CrossRef]
- Wang, X.; Teng, X.; Luo, C.; Kong, L. Mechanisms and Advances of Epigenetic Regulation in Cardiovascular Disease. Front Biosci (Landmark Ed.) 2024, 29, 205. [Google Scholar] [CrossRef]
- Xu, X.F.; Ma, X.L.; Shen, Z.; Wu, X.L.; Cheng, F.; Du, L.Z. Epigenetic regulation of the endothelial nitric oxide synthase gene in persistent pulmonary hypertension of the newborn rat. J. Hypertens. 2010, 28, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Atkins, G.B.; Jain, M.K.; Hamik, A. Endothelial differentiation: Molecular mechanisms of specification and heterogeneity. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Sumpio, B.E.; Riley, J.T.; Dardik, A. Cells in focus: Endothelial cell. Int. J. Biochem. Cell Biol. 2002, 34, 1508–1512. [Google Scholar] [CrossRef] [PubMed]
- Teichert, A.M.; Miller, T.L.; Tai, S.C.; Wang, Y.; Bei, X.; Robb, G.B.; Phillips, M.J.; Marsden, P.A. In vivo expression profile of an endothelial nitric oxide synthase promoter-reporter transgene. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H1352–H1361. [Google Scholar] [CrossRef]
- Fish, J.E.; Matouk, C.C.; Rachlis, A.; Lin, S.; Tai, S.C.; D’Abreo, C.; Marsden, P.A. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J. Biol. Chem. 2005, 280, 24824–24838. [Google Scholar] [CrossRef]
- Matouk, C.C.; Marsden, P.A. Epigenetic regulation of vascular endothelial gene expression. Circ. Res. 2008, 102, 873–887. [Google Scholar] [CrossRef]
- Esper, R.J.; Nordaby, R.A.; Vilarino, J.O.; Paragano, A.; Cacharron, J.L.; Machado, R.A. Endothelial dysfunction: A comprehensive appraisal. Cardiovasc. Diabetol. 2006, 5, 4. [Google Scholar] [CrossRef]
- Kubis, N.; Levy, B.I. Understanding angiogenesis: A clue for understanding vascular malformations. J. Neuroradiol. J. De. Neuroradiol. 2004, 31, 365–368. [Google Scholar] [CrossRef]
- Hamik, A.; Wang, B.; Jain, M.K. Transcriptional regulators of angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1936–1947. [Google Scholar] [CrossRef]
- Chan, Y.; Fish, J.E.; D’Abreo, C.; Lin, S.; Robb, G.B.; Teichert, A.M.; Karantzoulis-Fegaras, F.; Keightley, A.; Steer, B.M.; Marsden, P.A. The cellspecific expression of endothelial nitric-oxide synthase: A role for DNA methylation. J. Biol. Chem. 2004, 279, 35087–35100. [Google Scholar] [CrossRef]
- Lagarkova, M.A.; Shutova, M.V.; Bogomazova, A.N.; Vassina, E.M.; Glazov, E.A.; Zhang, P.; Rizvanov, A.A.; Chestkov, I.V.; Kiselev, S.L. Induction of pluripotency in human endothelial cells resets epigenetic profile on genome scale. Cell Cycle 2010, 9, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Lagarkova, M.A.; Volchkov, P.Y.; Philonenko, E.S.; Kiselev, S.L. Efficient differentiation of hESCs into endothelial cells in vitro is secured by epigenetic changes. Cell Cycle 2008, 7, 2929–2935. [Google Scholar] [CrossRef][Green Version]
- Kim, J.; Kim, J.Y.; Song, K.S.; Lee, Y.H.; Seo, J.S.; Jelinek, J.; Goldschmidt-Clermont, P.J.; Issa, J.P. Epigenetic changes in estrogen receptor beta gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochim. Et. Biophys. Acta 2007, 1772, 72–80. [Google Scholar] [CrossRef]
- Pojoga, L.H.; Williams, J.S.; Yao, T.M.; Kumar, A.; Raffetto, J.D.; do Nascimento, G.R.; Reslan, O.M.; Adler, G.K.; Williams, G.H.; Shi, Y.; et al. Histone demethylase LSD1 deficiency during high-salt diet is associated with enhanced vascular contraction, altered NO-cGMP relaxation pathway, and hypertension. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H1862–H1871. [Google Scholar] [CrossRef] [PubMed]
- Bastiaansen, A.J.; Ewing, M.M.; de Boer, H.C.; van der Pouw Kraan, T.C.; de Vries, M.R.; Peters, E.A.; Welten, S.M.; Arens, R.; Moore, S.M.; Faber, J.E.; et al. Lysine acetyltransferase PCAF is a key regulator of arteriogenesis. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Kueh, A.J.; Dixon, M.P.; Voss, A.K.; Thomas, T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol. 2011, 31, 845–860. [Google Scholar] [CrossRef]
- Potente, M.; Ghaeni, L.; Baldessari, D.; Mostoslavsky, R.; Rossig, L.; Dequiedt, F.; Haendeler, J.; Mione, M.; Dejana, E.; Alt, F.W.; et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes. Dev. 2007, 21, 2644–2658. [Google Scholar] [CrossRef]
- Kuehbacher, A.; Urbich, C.; Zeiher, A.M.; Dimmeler, S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 2007, 101, 59–68. [Google Scholar] [CrossRef]
- Otsuka, M.; Zheng, M.; Hayashi, M.; Lee, J.D.; Yoshino, O.; Lin, S.; Han, J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J. Clin. Investig. 2008, 118, 1944–1954. [Google Scholar] [CrossRef]
- Poliseno, L.; Tuccoli, A.; Mariani, L.; Evangelista, M.; Citti, L.; Woods, K.; Mercatanti, A.; Hammond, S.; Rainaldi, G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 2006, 108, 3068–3071. [Google Scholar] [CrossRef]
- Watkins, H.; Farrall, M. Genetic susceptibility to coronary artery disease: From promise to progress. Nat. Rev. Genet. 2006, 7, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Rzucidlo, E.M.; Martin, K.A.; Powell, R.J. Regulation of vascular smoothmuscle cell differentiation. J. Vasc. Surg. 2007, 45, A25–A32. [Google Scholar] [CrossRef] [PubMed]
- Laukkanen, M.O.; Mannermaa, S.; Hiltunen, M.O.; Aittomaki, S.; Airenne, K.; Janne, J.; Yla-Herttuala, S. Local hypomethylation in atherosclerosis found in rabbit ec-sod gene. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 2171–2178. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Lu, G.; Zhang, Q.; Ding, N.; Jie, Y.; Zhang, H.; Xu, L.; Xie, L.; Yang, X.; Zhang, H.; et al. Extracellular-superoxide dismutase DNA methylation promotes oxidative stress in homocysteine-induced atherosclerosis. Acta Biochim. Biophys. Sin. 2022, 54, 1222–1233. [Google Scholar] [CrossRef] [PubMed]
- Azechi, T.; Sato, F.; Sudo, R.; Wachi, H. 5-Aza-2-deoxycytidine, a DNA methyltransferase inhibitor, facilitates the inorganic phosphorus-induced mineralization of vascular smooth muscle cells. J. Atheroscler. Thromb. 2014, 21, 463–476. [Google Scholar] [CrossRef]
- Shirodkar, A.V.; Marsden, P.A. Epigenetics in cardiovascular disease. Curr. Opin. Cardiol. 2011, 26, 209–215. [Google Scholar] [CrossRef]
- Salmon, M.; Gomez, D.; Greene, E.; Shankman, L.; Owens, G.K. Cooperative binding of KLF4, pELK-1, and HDAC2 to a G/C repressor element in the SM22alpha promoter mediates transcriptional silencing during SMC phenotypic switching in vivo. Circ. Res. 2012, 111, 685–696. [Google Scholar] [CrossRef]
- Xu, X.; Ha, C.H.; Wong, C.; Wang, W.; Hausser, A.; Pfizenmaier, K.; Olson, E.N.; McKinsey, T.A.; Jin, Z.G. Angiotensin II stimulates protein kinase D-dependent histone deacetylase 5 phosphorylation and nuclear export leading to vascular smooth muscle cell hypertrophy. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 2355–2362. [Google Scholar] [CrossRef]
- Li, H.; Li, W.; Gupta, A.K.; Mohler, P.J.; Anderson, M.E.; Grumbach, I.M. Calmodulin kinase II is required for angiotensin II-mediated vascular smooth muscle hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 2010, 298, H688–H698. [Google Scholar] [CrossRef]
- Usui, T.; Okada, M.; Mizuno, W.; Oda, M.; Ide, N.; Morita, T.; Hara, Y.; Yamawaki, H. HDAC4 mediates development of hypertension via vascular inflammation in spontaneous hypertensive rats. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H1894–H1904. [Google Scholar] [CrossRef]
- Caruso, P.; Dempsie, Y.; Stevens, H.C.; McDonald, R.A.; Long, L.; Lu, R.; White, K.; Mair, K.M.; McClure, J.D.; Southwood, M.; et al. A role for miR-145 in pulmonary arterial hypertension: Evidence from mouse models and patient samples. Circ. Res. 2012, 111, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Courboulin, A.; Paulin, R.; Giguère, N.J.; Saksouk, N.; Perreault, T.; Meloche, J.; Paquet, E.R.; Biardel, S.; Provencher, S.; Côté, J.; et al. Role for miR-204 in human pulmonary arterial hypertension. J. Exp. Med. 2011, 208, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kang, Y.; Kojima, Y.; Lighthouse, J.K.; Hu, X.; Aldred, M.A.; McLean, D.L.; Park, H.; Comhair, S.A.; Greif, D.M.; et al. An endothelial apelin-FGF link mediated by miR-424 and miR-503 is disrupted in pulmonary arterial hypertension. Nat. Med. 2013, 19, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kontaraki, J.E.; Marketou, M.E.; Zacharis, E.A.; Parthenakis, F.I.; Vardas, P.E. Differential expression of vascular smooth musclemodulating microRNAs in human peripheral blood mononuclear cells: Novel targets in essential hypertension. J. Hum. Hypertens. 2014, 28, 510–516. [Google Scholar] [CrossRef]
- Sapienza, C.; Lee, J.; Powell, J.; Erinle, O.; Yafai, F.; Reichert, J.; Siraj, E.S.; Madaio, M. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics 2011, 6, 20–28. [Google Scholar] [CrossRef]
- Lake, B.B.; Menon, R.; Winfree, S.; Hu, Q.; Melo Ferreira, R.; Kalhor, K.; Barwinska, D.; Otto, E.A.; Ferkowicz, M.; Diep, D.; et al. An atlas of healthy and injured cell states and niches in the human kidney. Nature 2023, 619, 585–594. [Google Scholar] [CrossRef]
- Ko, Y.A.; Mohtat, D.; Suzuki, M.; Park, A.S.; Izquierdo, M.C.; Han, S.Y.; Kang, H.M.; Si, H.; Hostetter, T.; Pullman, J.M.; et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 2013, 14, R108. [Google Scholar] [CrossRef]
- Wyrwoll, C.S.; Mark, P.J.; Waddell, B.J. Developmental programming of renal glucocorticoid sensitivity and the renin-angiotensin system. Hypertension 2007, 50, 579–584. [Google Scholar] [CrossRef]
- Bogdarina, I.; Welham, S.; King, P.J.; Burns, S.P.; Clark, A.J. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ. Res. 2007, 100, 520–526. [Google Scholar] [CrossRef]
- Pushpakumar, S.; Ren, L.; Juin, S.K.; Majumder, S.; Kulkarni, R.; Sen, U. Methylation-dependent antioxidant-redox imbalance regulates hypertensive kidney injury in aging. Redox Biol. 2020, 37, 101754. [Google Scholar] [CrossRef]
- Hayashi, K.; Hishikawa, A.; Hashiguchi, A.; Azegami, T.; Yoshimoto, N.; Nakamichi, R.; Tokuyama, H.; Itoh, H. Association of glomerular DNA damage and DNA methylation with one-year eGFR decline in IgA nephropathy. Sci. Rep. 2020, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Hishikawa, A.; Hayashi, K.; Yoshimoto, N.; Nakamichi, R.; Homma, K.; Itoh, H. DNA damage and expression of DNA methylation modulators in urine-derived cells of patients with hypertension and diabetes. Sci. Rep. 2020, 10, 3377. [Google Scholar] [CrossRef]
- Yoshimoto, N.; Hayashi, K.; Hishikawa, A.; Hashiguchi, A.; Nakamichi, R.; Sugita-Nishimura, E.; Yoshida-Hama, E.; Azegami, T.; Nakayama, T.; Itoh, H. Significance of podocyte DNA damage and glomerular DNA methylation in CKD patients with proteinuria. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2023, 46, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Q.M.; Liu, S.X.; Chen, Y.H.; Li, R.Z.; Lin, T.; Yu, C.P.; Zhang, H.; Huang, Z.S.; Zhao, X.C.; et al. DNA methyltransferase 1 may be a therapy target for attenuating diabetic nephropathy and podocyte injury. Kidney Int. 2017, 92, 140–153. [Google Scholar] [CrossRef]
- Hayashi, K. Targeting DNA Methylation in Podocytes to Overcome Chronic Kidney Disease. Keio J. Med. 2023, 72, 67–76. [Google Scholar] [CrossRef]
- Zhang, D.; Yu, Z.Y.; Cruz, P.; Kong, Q.; Li, S.; Kone, B.C. Epigenetics and the control of epithelial sodium channel expression in collecting duct. Kidney Int. 2009, 75, 260–267. [Google Scholar] [CrossRef]
- Bhalla, V.; Hallows, K.R. Mechanisms of ENaC regulation and clinical implications. J. Am. Soc. Nephrol. 2008, 19, 1845–1854. [Google Scholar] [CrossRef]
- Wang, J.; Gong, L.; Tan, Y.; Hui, R.; Wang, Y. Hypertensive epigenetics: From DNA methylation to microRNAs. J. Hum. Hypertens. 2015, 29, 575–582. [Google Scholar] [CrossRef]
- Yu, Z.; Kong, Q.; Kone, B.C. Aldosterone reprograms promoter methylation to regulate alphaENaC transcription in the collecting duct. Am. J. Physiol. Ren. Physiol. 2013, 305, F1006–F1013. [Google Scholar] [CrossRef][Green Version]
- Wu, T.H.; Kuo, H.C.; Lin, I.C.; Chien, S.J.; Huang, L.T.; Tain, Y.L. Melatonin prevents neonatal dexamethasone induced programmed hypertension: Histone deacetylase inhibition. J. Steroid Biochem. Mol. Biol. 2014, 144 Pt B, 253–259. [Google Scholar] [CrossRef]
- Lee, H.A.; Lee, D.Y.; Lee, H.J.; Han, H.S.; Kim, I. Enrichment of (pro)renin receptor promoter with activating histone codes in the kidneys of spontaneously hypertensive rats. J. Renin-Angiotensin-Aldosterone Syst. JRAAS 2012, 13, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.; Fenning, A.; Lim, J.; Le, G.T.; Reid, R.C.; Halili, M.A.; David P Fairlie, D.P.; Brown, L. Antifibrotic activity of an inhibitor of histone deacetylases in DOCA-salt hypertensive rats. Br. J. Pharmacol. 2010, 159, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Medina Rangel, P.X.; Cross, E.; Liu, C.; Pedigo, C.E.; Tian, X.; Gutierrez-Calabres, E.; Nagata, S.; Priyadarshini, A.; Lerner, G.; Bunda, P.; et al. Cell Cycle and Senescence Regulation by Podocyte Histone Deacetylase 1 and 2. J. Am. Soc. Nephrol. 2023, 34, 433–450. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Gan, G.; Ciarleglio, M.; Zhang, Y.; Tian, X.; Pedigo, C.E.; Cavanaugh, C.; Tate, J.; Wang, Y.; Cross, E.; et al. Podocyte histone deacetylase activity regulates murine and human glomerular diseases. J. Clin. Investig. 2019, 129, 1295–1313. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Hasebe, N. Alteration of histone H3K4 methylation in glomerular podocytes associated with proteinuria in patients with membranous nephropathy. BMC Nephrol. 2016, 17, 179. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Y.; Zhan, P.; Sun, W.; Dong, C.; Liu, X.; Yang, Y.; Wang, X.; Xie, Y.; Gao, C.; et al. Histone deacetylase 9 exacerbates podocyte injury in hyperhomocysteinemia through epigenetic repression of Klotho. Pharmacol. Res. 2023, 198, 107009. [Google Scholar] [CrossRef]
- Liu, M.; Qiao, Z.; Zhang, Y.; Zhan, P.; Yi, F. Histone Deacetylases Take Center Stage on Regulation of Podocyte Function. Kidney Dis. 2020, 6, 236–246. [Google Scholar] [CrossRef]
- Reddy, M.A.; Sumanth, P.; Lanting, L.; Yuan, H.; Wang, M.; Mar, D.; Alpers, C.E.; Bomsztyk, K.; Natarajan, R. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int. 2014, 85, 362–373. [Google Scholar] [CrossRef]
- Hyndman, K.A.; Speed, J.S.; Mendoza, L.D.; Allan, J.M.; Colson, J.; Sedaka, R.; Jin, C.; Jung, H.J.; El-Dahr, S.; Pollock, D.M.; et al. Fluid-electrolyte homeostasis requires histone deacetylase function. JCI Insight 2020, 5, e137792. [Google Scholar] [CrossRef]
- Romero, D.G.; Plonczynski, M.W.; Carvajal, C.A.; Gomez-Sanchez, E.P.; Gomez-Sanchez, C.E. Microribonucleic acid-21 increases aldosterone secretion and proliferation in H295R human adrenocortical cells. Endocrinology 2008, 149, 2477–2483. [Google Scholar] [CrossRef]
- Ling, S.; Nanhwan, M.; Qian, J.; Kodakandla, M.; Castillo, A.C.; Thomas, B.; Liu, H.; Ye, Y. Modulation of microRNAs in hypertension-induced arterial remodeling through the β1 and β3-adrenoreceptor pathways. J. Mol. Cell. Cardiol. 2013, 65, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Campain, A.E.; Tomaszewski, M.; Zukowska-Szczechowska, E.; Yang, Y.H.; Charchar, F.J.; Morris, B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 2011, 58, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Eskildsen, T.V.; Jeppesen, P.L.; Schneider, M.; Nossent, A.Y.; Sandberg, M.B.; Hansen, P.B.; Jensen, C.H.; Hansen, M.L.; Marcussen, N.; Rasmussen, L.M.; et al. Angiotensin II regulates microRNA-132/-212 in hypertensive rats and humans. Int. J. Mol. Sci. 2013, 14, 11190–11207. [Google Scholar] [CrossRef] [PubMed]
- Ursu, R.; Sopel, N.; Ohs, A.; Tati, R.; Buvall, L.; Nystrom, J.; Schiffer, M.; Muller-Deile, J. Glomerular Endothelial Cell-Derived miR-200c Impairs Glomerular Homeostasis by Targeting Podocyte VEGF-A. Int. J. Mol. Sci. 2022, 23, 15070. [Google Scholar] [CrossRef]
- Perez-Hernandez, J.; Riffo-Campos, A.L.; Ortega, A.; Martinez-Arroyo, O.; Perez-Gil, D.; Olivares, D.; Solaz, E.; Martinez, F.; Martinez-Hervas, S.; Chaves, F.J.; et al. Urinary- and Plasma-Derived Exosomes Reveal a Distinct MicroRNA Signature Associated With Albuminuria in Hypertension. Hypertension 2021, 77, 960–971. [Google Scholar] [CrossRef]
- Li, H.; Chen, Z.; Chen, W.; Li, J.; Liu, Y.; Ma, H.; Shi, M.; Sun, X.; Yao, X.; Li, Z.; et al. MicroRNA-23b-3p Deletion Induces an IgA Nephropathy-like Disease Associated with Dysregulated Mucosal IgA Synthesis. J. Am. Soc. Nephrol. 2021, 32, 2561–2578. [Google Scholar] [CrossRef]
- Denby, L.; Ramdas, V.; McBride, M.W.; Wang, J.; Robinson, H.; McClure, J.; Crawford, W.; Lu, R.; Hillyard, D.Z.; Khanin, R.; et al. miR-21 and miR-214 are consistently modulated during renal injury in rodent models. Am. J. Pathol. 2011, 179, 661–672. [Google Scholar] [CrossRef]
- Zandi-Nejad, K.; Luyckx, V.A.; Brenner, B.M. Adult hypertension and kidney disease: The role of fetal programming. Hypertension 2006, 47, 502–508. [Google Scholar] [CrossRef]
- Liakopoulos, V.; Georgianos, P.I.; Eleftheriadis, T.; Sarafidis, P.A. Epigenetic Mechanisms and Kidney Diseases. Curr. Med. Chem. 2011, 18, 1733–1739. [Google Scholar] [CrossRef]
- Saleem, M.A. Biology of the human podocyte. Nephron. Exp. Nephrol. 2003, 95, e87–e92. [Google Scholar] [CrossRef]
- Levidiotis, V.; Power, D.A. New insights into the molecular biology of the glomerular filtration barrier and associated disease. Nephrology 2005, 10, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Kriz, W.; LeHir, M. Pathways to nephron loss starting from glomerular diseases-insights from animal models. Kidney Int. 2005, 67, 404–419. [Google Scholar] [CrossRef] [PubMed]
- Marfella, C.G.A.; Henninger, N.; LeBlanc, S.E.; Krishnan, N.; Garlick, D.S.; Holzman, L.B.; Imbalzanoa, A.N. A mutation in the mouse Chd2 chromatin remodeling enzyme results in a complex renal phenotype. Kidney Blood Press. Res. 2009, 31, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Zeisberg, M.; Strutz, F.; Muller, G.A. Role of fibroblast activation in inducing interstitial fibrosis. J. Nephrol. 2000, 13, S111–S120. [Google Scholar]
- Neilson, E.G. Mechanisms of disease: Fibroblasts—A new look at an old problem. Nat. Clin. Pract. Nephrol. 2006, 2, 101–108. [Google Scholar] [CrossRef]
- Bechtel, W.; McGoohan, S.; Zeisberg, E.M.; Muller, G.A.; Kalbacher, H.; Salant, D.J.; Müller, C.A.; Kalluri, R.; Zeisberg, M. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med. 2010, 16, 544–550. [Google Scholar] [CrossRef]
- Tonna, S.; El-Osta, A.; Cooper, M.E.; Tikellis, C. Metabolic memory and diabetic nephropathy: Potential role for epigenetic mechanisms. Nat. Rev. Nephrol. 2010, 6, 332–342. [Google Scholar] [CrossRef]
- El-Osta, A.; Brasacchio, D.; Yao, D.; Pocai, A.; Jones, P.L.; Roeder, R.G.; Cooper, M.E.; Brownlee, M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 2008, 205, 2409–2417. [Google Scholar] [CrossRef]
- Macconi, D.; Tomasoni, S.; Romagnani, P.; Trionfini, P.; Sangalli, F.; Mazzinghi, B.; Rizzo, P.; Lazzeri, E.; Abbate, M.; Remuzzi, G.; et al. MicroRNA-324-3p promotes renal fibrosis and is a target of ACE inhibition. J. Am. Soc. Nephrol. JASN 2012, 23, 1496–1505. [Google Scholar] [CrossRef]
- Perna, A.F.; Ingrosso, D.; Lombardi, C.; Cesare, C.M.; Acantora, F.; Satta, E.; De Santo, N.G. Homocysteine in uremia. Am. J. Kidney Dis. 2003, 41, S123–S126. [Google Scholar] [CrossRef]
- Ekstrom, T.J.; Stenvinkel, P. The epigenetic conductor: A genomic orchestrator in chronic kidney disease complications? J. Nephrol. 2009, 22, 442–449. [Google Scholar] [PubMed]
- Jamaluddin, M.S.; Yang, X.; Wang, H. Hyperhomocysteinemia, DNA methylation and vascular disease. Clin. Chem. Lab. Med. 2007, 45, 1660–1666. [Google Scholar] [CrossRef] [PubMed]
- Topal, G.; Brunet, A.; Millanvoye, E.; Boucher, J.L.; Rendu, F.; Devynck, M.A.; David-Dufilho, M. Homocysteine induces oxidative stress by uncoupling of NO synthase activity through reduction of tetrahydrobiopterin. Free Radic. Biol. Med. 2004, 36, 1532–1541. [Google Scholar] [CrossRef] [PubMed]
- Juonala, M.; Viikari, J.S.; Ronnemaa, T.; Helenius, H.; Taittonen, L.; Raitakari, O.T. Elevated blood pressure in adolescent boys predicts endothelial dysfunction: The cardiovascular risk in young Finns study. Hypertension 2006, 48, 424–430. [Google Scholar] [CrossRef]
- Sundstrom, J.; Sullivan, L.; D’Agostino, R.B.; Jacques, P.F.; Selhub, J.; Rosenberg, I.H.; Wilson, P.W.; Levy, D.; Vasan, R.S. Plasma homocysteine, hypertension incidence, and blood pressure tracking: The Framingham Heart Study. Hypertension 2003, 42, 1100–1105. [Google Scholar] [CrossRef]
- Tyagi, S.C.; Matsubara, L.; Weber, K.T. Direct extraction and estimation of collagenase(s) activity by zymography in microquantities of rat myocardium and uterus. Clin. Biochem. 1993, 26, 191–198. [Google Scholar] [CrossRef]
- Sen, U.; Mishra, P.K.; Tyagi, N.; Tyagi, S.C. Homocysteine to hydrogen sulfide or hypertension. Cell Biochem. Biophys. 2010, 57, 49–58. [Google Scholar] [CrossRef]
- Narayanan, N.; Pushpakumar, S.B.; Givvimani, S.; Kundu, S.; Metreveli, N.; James, D.; Bratcher, A.P.; Tyagi, S.C. Epigenetic regulation of aortic remodeling in hyperhomocysteinemia. FASEB J. 2014, 28, 3411–3422. [Google Scholar] [CrossRef]
- Weng, X.; Cheng, X.; Wu, X.; Xu, H.; Fang, M.; Xu, Y. Sin3B mediates collagen type I gene repression by interferon gamma in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2014, 447, 263–270. [Google Scholar] [CrossRef]
- Selhub, J. Homocysteine metabolism. Annu. Rev. Nutr. 1999, 19, 217–246. [Google Scholar] [CrossRef]
- Pushpakumar, S.; Kundu, S.; Sen, U. Hydrogen Sulfide Protects Hyperhomocysteinemia-Induced Renal Damage by Modulation of Caveolin and eNOS Interaction. Sci. Rep. 2019, 9, 2223. [Google Scholar] [CrossRef] [PubMed]
- Perla-Kajan, J.; Jakubowski, H. Dysregulation of Epigenetic Mechanisms of Gene Expression in the Pathologies of Hyperhomocysteinemia. Int. J. Mol. Sci. 2019, 20, 3140. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H. Protein homocysteinylation: Possible mechanism underlying pathological consequences of elevated homocysteine levels. FASEB J. 1999, 13, 2277–2283. [Google Scholar] [CrossRef] [PubMed]
- Perla-Kajan, J.; Marczak, L.; Kajan, L.; Skowronek, P.; Twardowski, T.; Jakubowski, H. Modification by homocysteine thiolactone affects redox status of cytochrome C. Biochemistry 2007, 46, 6225–6231. [Google Scholar] [CrossRef]
- Jakubowski, H. Calcium-dependent human serum homocysteine thiolactone hydrolase. A protective mechanism against protein N-homocysteinylation. J. Biol. Chem. 2000, 275, 3957–3962. [Google Scholar] [CrossRef]
- Jakubowski, H.; Zhang, L.; Bardeguez, A.; Aviv, A. Homocysteine thiolactone and protein homocysteinylation in human endothelial cells: Implications for atherosclerosis. Circ. Res. 2000, 87, 45–51. [Google Scholar] [CrossRef]
- Glowacki, R.; Jakubowski, H. Cross-talk between Cys34 and lysine residues in human serum albumin revealed by N-homocysteinylation. J. Biol. Chem. 2004, 279, 10864–10871. [Google Scholar] [CrossRef]
- Undas, A.; Jankowski, M.; Twardowska, M.; Padjas, A.; Jakubowski, H.; Szczeklik, A. Antibodies to N-homocysteinylated albumin as a marker for early-onset coronary artery disease in men. Thromb. Haemost. 2005, 93, 346–350. [Google Scholar] [CrossRef]
- Undas, A.; Stepien, E.; Glowacki, R.; Tisonczyk, J.; Tracz, W.; Jakubowski, H. Folic acid administration and antibodies against homocysteinylated proteins in subjects with hyperhomocysteinemia. Thromb. Haemost. 2006, 96, 342–347. [Google Scholar] [CrossRef]
- Lonn, E.; Yusuf, S.; Arnold, M.J.; Sheridan, P.; Pogue, J.; Micks, M.; McQueen, M.J.; Probstfield, J.; Fodor, G.; Held, C.; et al. Homocysteine lowering with folic acid and B vitamins in vascular disease. N. Engl. J. Med. 2006, 354, 1567–1577. [Google Scholar] [CrossRef]
- Bonaa, K.H.; Njolstad, I.; Ueland, P.M.; Schirmer, H.; Tverdal, A.; Steigen, T.; Wang, H.; Nordrehaug, J.E.; Arnesen, E.; Rasmussen, K.; et al. Homocysteine lowering and cardiovascular events after acute myocardial infarction. N. Engl. J. Med. 2006, 354, 1578–1588. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H.; Boers, G.H.; Strauss, K.A. Mutations in cystathionine beta-synthase or methylenetetrahydrofolate reductase gene increase N-homocysteinylated protein levels in humans. FASEB J. 2008, 22, 4071–4076. [Google Scholar] [CrossRef] [PubMed]
- Perla-Kajan, J.; Utyro, O.; Rusek, M.; Malinowska, A.; Sitkiewicz, E.; Jakubowski, H. N-Homocysteinylation impairs collagen cross-linking in cystathionine beta-synthase-deficient mice: A novel mechanism of connective tissue abnormalities. FASEB J. 2016, 30, 3810–3821. [Google Scholar] [CrossRef] [PubMed]
- Borowczyk, K.; Wroblewski, J.; Suliburska, J.; Akahoshi, N.; Ishii, I.; Jakubowski, H. Mutations in Homocysteine Metabolism Genes Increase Keratin N-Homocysteinylation and Damage in Mice. Int. J. Genom. 2018, 2018, 7570850. [Google Scholar] [CrossRef]
- Zhang, L.; Zeng, X.X.; Li, Y.M.; Chen, S.K.; Tang, L.Y.; Wang, N.; Yang, X.; Lin, M.J. Keratin 1 attenuates hypoxic pulmonary artery hypertension by suppressing pulmonary artery media smooth muscle expansion. Acta Physiol. 2021, 231, e13558. [Google Scholar] [CrossRef]
- Wilcox, C.S. Oxidative stress and nitric oxide deficiency in the kidney: A critical link to hypertension? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 289, R913–R935. [Google Scholar] [CrossRef]
- Pratt, J.R.; Parker, M.D.; Affleck, L.J.; Corps, C.; Hostert, L.; Michalak, E.; Lodge, J.P. Ischemic epigenetics and the transplanted kidney. Transplant. Proc. 2006, 38, 3344–3346. [Google Scholar] [CrossRef]
- Li, L.; Hsu, A.; Moore, P.K. Actions and interactions of nitric oxide, carbon monoxide and hydrogen sulphide in the cardiovascular system and in inflammation—A tale of three gases! Pharmacol. Ther. 2009, 123, 386–400. [Google Scholar] [CrossRef]
- Tang, G.; Yang, G.; Jiang, B.; Ju, Y.; Wu, L.; Wang, R. H2S is an endothelium-derived hyperpolarizing factor. Antioxid. Redox Signal. 2013, 19, 1634–1646. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, R. H2S-induced vasorelaxation and underlying cellular and molecular mechanisms. Am. J. Physiol. Heart Circ. Physiol. 2002, 283, H474–H480. [Google Scholar] [CrossRef]
- Tang, G.; Wu, L.; Liang, W.; Wang, R. Direct stimulation of K(ATP) channels by exogenous and endogenous hydrogen sulfide in vascular smooth muscle cells. Mol. Pharmacol. 2005, 68, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Bełtowski, J. Hypoxia in the renal medulla: Implications for hydrogen sulfide signaling. J. Pharmacol. Exp. Ther. 2010, 334, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Maines, M.D. The heme oxygenase system: A regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 1997, 37, 517–554. [Google Scholar] [CrossRef]
- Prabhakar, N.R.; Dinerman, J.L.; Agani, F.H.; Snyder, S.H. Carbon monoxide: A role in carotid body chemoreception. Proc. Natl. Acad. Sci. USA 1995, 92, 1994–1997. [Google Scholar] [CrossRef]
- Prabhakar, N.R. Sensing hypoxia: Physiology, genetics and epigenetics. J. Physiol. 2013, 591, 2245–2257. [Google Scholar] [CrossRef]
- Montecucco, F.; Pende, A.; Quercioli, A.; Mach, F. Inflammation in the pathophysiology of essential hypertension. J. Nephrol. 2011, 24, 23–34. [Google Scholar] [CrossRef]
- Martin, C.; Cameron, J.; McGrath, B. Mechanical and circulating biomarkers in isolated clinic hypertension. Clin. Exp. Pharmacol. Physiol. 2008, 35, 402–408. [Google Scholar] [CrossRef]
- Chander, V.; Chopra, K. Possible role of nitric oxide in the protective effect of resveratrol in 5/6th nephrectomized rats. J. Surg. Res. 2006, 133, 129–135. [Google Scholar] [CrossRef]
- Baylis, C. Nitric oxide deficiency in chronic kidney disease. Am. J. Physiol. Ren. Physiol. 2008, 294, F1–F9. [Google Scholar] [CrossRef]
- Kondo, K.; Bhushan, S.; King, A.L.; Prabhu, S.D.; Hamid, T.; Koenig, S.; Murohara, T.; Predmore, B.L.; Gojon, G., Sr.; Gojon, G., Jr.; et al. H2S Protects Against Pressure Overload Induced Heart Failure via Upregulation of Endothelial Nitric Oxide Synthase (eNOS). Circulation 2013, 127, 1116–1127. [Google Scholar] [CrossRef]
- Jin, H.F.; Du, J.B.; Li, X.H.; Wang, Y.F.; Liang, Y.F.; Tang, C.S. Interaction between hydrogen sulfide/cystathionine gamma-lyase and carbon monoxide/heme oxygenase pathways in aortic smooth muscle cells. Acta Pharmacol. Sin. 2006, 27, 1561–1566. [Google Scholar] [CrossRef] [PubMed]
- Thorup, C.; Jones, C.L.; Gross, S.S.; Moore, L.C.; Goligorsky, M.S. Carbon monoxide induces vasodilation and nitric oxide release but suppresses endothelial NOS. Am. J. Physiol. 1999, 277, F882–F889. [Google Scholar] [CrossRef] [PubMed]
- Coletta, C.; Papapetropoulos, A.; Erdelyi, K.; Olah, G.; Módis, K.; Panopoulos, P.; Asimakopoulou, A.; Gerö, D.; Sharina, I.; Martin, E.; et al. Hydrogen sulfide and nitric oxide are mutually dependent in the regulation of angiogenesis and endothelium-dependent vasorelaxation. Proc. Natl. Acad. Sci. USA 2012, 109, 9161–9166. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, S.; Fledderus, J.O.; Verhaar, M.C.; Joles, J.A. Beneficial effects of diminished production of hydrogen sulfide or carbon monoxide on hypertension and renal injury induced by NO withdrawal. Br. J. Pharmacol. 2015, 172, 1607–1619. [Google Scholar] [CrossRef]
- Tain, Y.L.; Lee, W.C.; Hsu, C.N.; Lee, W.C.; Huang, L.T.; Lee, C.T.; Lin, C.Y. Asymmetric dimethylarginine is associated with developmental programming of adult kidney disease and hypertension in offspring of streptozotocintreated mothers. PLoS ONE 2013, 8, e55420. [Google Scholar] [CrossRef]
- Quaia, M.; Zancai, P.; Cariati, R.; Rizzo, S.; Boiocchi, M.; Dolcetti, R. Glucocorticoids promote the proliferation and antagonize the retinoic acidmediated growth suppression of Epstein–Barr virus-immortalized B lymphocytes. Blood 2000, 96, 711–718. [Google Scholar] [CrossRef]
- Fish, J.; Marsden, P. Endothelial nitric oxide synthase: Insight into cellspecific gene regulation in the vascular endothelium. Cell. Mol. Life Sci. CMLS 2006, 63, 144–162. [Google Scholar] [CrossRef]
- Gan, Y.; Shen, Y.H.; Wang, J.; Wang, X.; Utama, B.; Wang, J.; Wang, X.L. Role of histone deacetylation in cell-specific expression of endothelial nitric-oxide synthase. J. Biol. Chem. 2005, 280, 16467–16475. [Google Scholar] [CrossRef]
- Bouckenooghe, T.; Remacle, C.; Reusens, B. Is taurine a functional nutrient? Curr. Opin. Clin. Nutr. Metab. Care 2006, 9, 728–733. [Google Scholar] [CrossRef]
- Militante, J.D.; Lombardini, J.B. Treatment of hypertension with oral taurine: Experimental and clinical studies. Amino Acids 2002, 23, 381–393. [Google Scholar] [CrossRef]
- Yamori, Y.; Taguchi, T.; Hamada, A.; Kunimasa, K.; Mori, H.; Mori, M. Taurine in health and diseases: Consistent evidence from experimental and epidemiological studies. J. Biomed. Sci. 2010, 17, S6. [Google Scholar] [CrossRef]
- Schaffer, S.W.; Jong, C.J.; Ramila, K.C.; Azuma, J. Physiological roles of taurine in heart and muscle. J. Biomed. Sci. 2010, 17, S2. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, L.; Pushpakumar, S.; Almarshood, H.; Das, S.K.; Sen, U. Epigenetic DNA Methylation and Protein Homocysteinylation: Key Players in Hypertensive Renovascular Damage. Int. J. Mol. Sci. 2024, 25, 11599. https://doi.org/10.3390/ijms252111599
Ren L, Pushpakumar S, Almarshood H, Das SK, Sen U. Epigenetic DNA Methylation and Protein Homocysteinylation: Key Players in Hypertensive Renovascular Damage. International Journal of Molecular Sciences. 2024; 25(21):11599. https://doi.org/10.3390/ijms252111599
Chicago/Turabian StyleRen, Lu, Sathnur Pushpakumar, Hebah Almarshood, Swapan K. Das, and Utpal Sen. 2024. "Epigenetic DNA Methylation and Protein Homocysteinylation: Key Players in Hypertensive Renovascular Damage" International Journal of Molecular Sciences 25, no. 21: 11599. https://doi.org/10.3390/ijms252111599
APA StyleRen, L., Pushpakumar, S., Almarshood, H., Das, S. K., & Sen, U. (2024). Epigenetic DNA Methylation and Protein Homocysteinylation: Key Players in Hypertensive Renovascular Damage. International Journal of Molecular Sciences, 25(21), 11599. https://doi.org/10.3390/ijms252111599





