The Structural Role of RPN10 in the 26S Proteasome and an RPN2-Binding Residue on RPN13 Are Functionally Important in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. N-Terminal Portion (1–215) of Arabidopsis RPN10 (N215) Is Fully Functional In Vivo
2.2. The D11 Residue of RPN10 Plays a Critical Role In Vivo and for Assembly into the 26S Proteasome
2.3. Presence of Specific Paralogues for Several Core Subunits and Increased ECM29 and PA200 Protein Levels in Single-Capped 20S Proteasomes from rpn10-2
2.4. Expression of Genes Encoding Subunits and Biosynthesis Components of 26S Proteasomes Was Generally Significantly Increased in rpn10-2
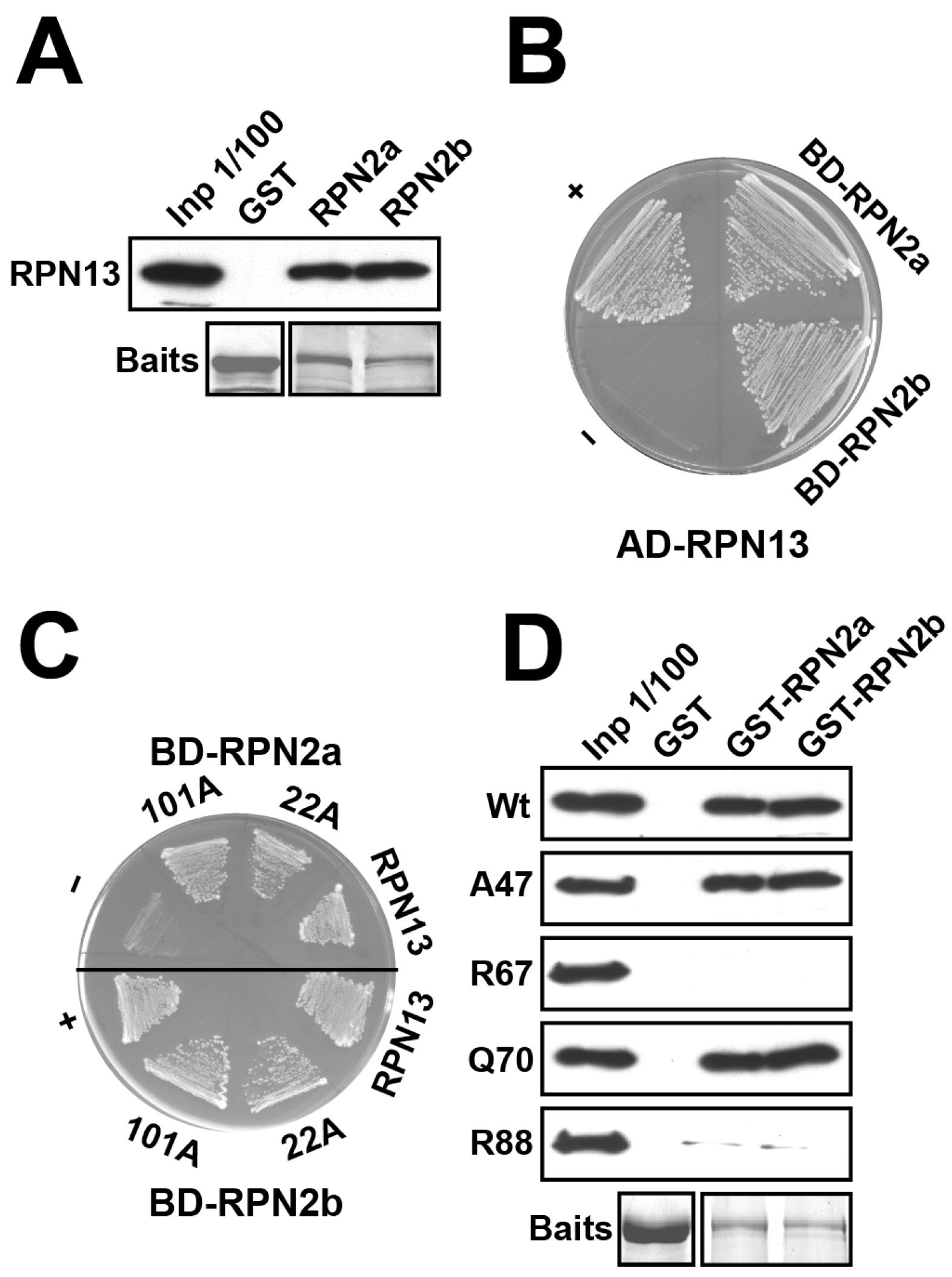
2.5. RPN13 Is Associated with RPN2 and UCH2 but Not UCH1
2.6. Domains and Residues on Arabidopsis RPN13 and UCH2 Critical for RPN13–RPN2 and RPN13–UCH2 Interactions
2.7. F67 Residue of RPN13, Critical for Both RPN2 and Ubiquitin Binding, Is Important In Vivo
3. Discussion
3.1. The Ubiquitin-Binding Motifs UIM1–3 of RPN10 Are Dispensable In Vivo
3.2. The N-Terminal Region of RPN10 Harboring the vWA Domain Is Functionally Important In Vivo
3.3. Conformational Changes or Structural Defects Are Likely Associated with 26S Proteasomes in rpn10-2
3.4. RPN13’s Role in UCH2 Recruitment to 26S Proteasomes by RPN2 Could Be Relevant In Vivo
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Complementation and Segregation Analyses of rpn10-2
4.3. Isolation and Analyses of Proteasome Complexes
4.4. In-Gel Trypsin Digestion and Mass Spectrometry Analysis for Protein Identification and Analysis
4.5. RNA-seq Analysis
4.6. GST Pull-Down Analyses
4.7. Yeast Two-Hybrid (Y2H) Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.H.; Shi, Y.; Zhang, N.; de Poot, S.A.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351, 831. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Lin, Y.-L.; Fatimababy, A.S. Proteasomal recognition of ubiquitylated substrates. Trends Plant Sci. 2010, 15, 375–386. [Google Scholar] [CrossRef]
- Fatimababy, A.S.; Lin, Y.-L.; Usharani, R.; Radjacommare, R.; Wang, H.-T.; Tsai, H.-L.; Lee, Y.; Fu, H. Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J. 2010, 277, 796–816. [Google Scholar] [CrossRef]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef]
- Sakata, E.; Bohn, S.; Mihalache, O.; Kiss, P.; Beck, F.; Nagy, I.; Nickell, S.; Tanaka, K.; Saeki, Y.; Förster, F.; et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 1479–1484. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, Q.; Ehlinger, A.; Randles, L.; Lary, J.W.; Kang, Y.; Haririnia, A.; Storaska, A.J.; Cole, J.L.; Fushman, D.; et al. Structure of the S5a:K48-linked diubiquitin complex and its interactions with Rpn13. Mol. Cell 2009, 35, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Peth, A.; Uchiki, T.; Goldberg, A.L. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol. Cell 2010, 40, 671–681. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Sung, S.-C.; Tsai, H.-L.; Yu, T.-T.; Radjacommare, R.; Usharani, R.; Fatimababy, A.S.; Lin, H.-Y.; Wang, Y.-Y.; Fu, H. The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell 2011, 23, 2754–2773. [Google Scholar] [CrossRef]
- Schreiner, P.; Chen, X.; Husnjak, K.; Randles, L.; Zhang, N.; Elsasser, S.; Finley, D.; Dikic, I.; Walters, K.J.; Groll, M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 2008, 453, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Deveraux, Q.; Ustrell, V.; Pickart, C.; Rechsteiner, M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994, 269, 7059–7061. [Google Scholar] [CrossRef] [PubMed]
- van Nocker, S.; Deveraux, Q.; Rechsteiner, M.; Vierstra, R.D. Arabidopsis MBP1 gene encodes a conserved ubiquitin recognition component of the 26S proteasome. Proc. Natl. Acad. Sci. USA 1996, 93, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Reis, N.; Lee, Y.; Glickman, M.H.; Vierstra, R.D. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001, 20, 7096–7107. [Google Scholar] [CrossRef]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef]
- van Nocker, S.; Sadis, S.; Rubin, D.M.; Glickman, M.; Fu, H.; Coux, O.; Wefes, I.; Finley, D.; Vierstra, R.D. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 1996, 16, 6020–6028. [Google Scholar] [CrossRef]
- Hamazaki, J.; Sasaki, K.; Kawahara, H.; Hisanaga, S.-I.; Tanaka, K.; Murata, S. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol. Cell. Biol. 2007, 27, 6629–6638. [Google Scholar] [CrossRef]
- Szlanka, T.; Haracska, L.; Kiss, I.; Deák, P.; Kurucz, É.; Andó, I.; Viragh, E.; Udvardy, A. Deletion of proteasomal subunit S5a/Rpn10/p54 causes lethality, multiple mitotic defects and overexpression of proteasomal genes in Drosophila melanogaster. J. Cell Sci. 2003, 116, 1023–1033. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Fu, H. In vivo relevance of substrate recognition function of major Arabidopsis ubiquitin receptors. Plant Signal. Behav. 2012, 7, 722–727. [Google Scholar] [CrossRef]
- Fu, H.; Sadis, S.; Rubin, D.M.; Glickman, M.; van Nocker, S.; Finley, D.; Vierstra, R.D. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J. Biol. Chem. 1998, 273, 1970–1981. [Google Scholar] [CrossRef]
- Smalle, J.; Kurepa, J.; Yang, P.; Emborg, T.J.; Babiychuk, E.; Kushnir, S.; Vierstra, R.D. The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 2003, 15, 965–980. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Chen, S.; Feldman, R.; Schieltz, D.; Yates, J.; Dohmen, J.; Deshaies, R.J. Proteasomal proteomics: Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 2000, 11, 3425–3439. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Sakurai, N.; Kinoshita, T. Xoom is maternally stored and functions as a transmembrane protein for gastrulation movement in Xenopus embryos. Dev. Growth Differ. 2001, 43, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Hamazaki, J.; Hirayama, S.; Murata, S. Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis. PLoS Genet. 2015, 11, e1005401. [Google Scholar] [CrossRef]
- Hamazaki, J.; Iemura, S.; Natsume, T.; Yashiroda, H.; Tanaka, K.; Murata, S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006, 25, 4524–4536. [Google Scholar] [CrossRef]
- Qiu, X.-B.; Ouyang, S.-Y.; Li, C.-J.; Miao, S.; Wang, L.; Goldberg, A. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006, 25, 5742–5753. [Google Scholar] [CrossRef]
- Yao, T.; Song, L.; Xu, W.; DeMartino, G.N.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Conaway, R.C.; Conaway, J.W.; Cohen, R.E. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006, 8, 994–1002. [Google Scholar] [CrossRef]
- Chen, X.; Lee, B.-H.; Finley, D.; Walters, K.J. Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol. Cell 2010, 38, 404–415. [Google Scholar] [CrossRef]
- Sahtoe, D.D.; van Dijk, W.J.; El Oualid, F.; Ekkebus, R.; Ovaa, H.; Sixma, T.K. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol. Cell 2015, 57, 887–900. [Google Scholar] [CrossRef]
- VanderLinden, R.T.; Hemmis, C.W.; Schmitt, B.; Ndoja, A.; Whitby, F.G.; Robinson, H.; Cohen, R.E.; Yao, T.; Hill, C.P. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol. Cell 2015, 57, 901–911. [Google Scholar] [CrossRef]
- Yang, P.; Smalle, J.; Lee, S.; Yan, N.; Emborg, T.J.; Vierstra, R.D. Ubiquitin C-terminal hydrolases 1 and 2 affect shoot architecture in Arabidopsis. Plant J. 2007, 51, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Yang, P.; Valverde, F.; Mizoguchi, T.; Furutani-Hayama, I.; Vierstra, R.D.; Coupland, G. Ubiquitin carboxyl-terminal hydrolases are required for period maintenance of the circadian clock at high temperature in Arabidopsis. Sci. Rep. 2019, 9, 17030. [Google Scholar] [CrossRef] [PubMed]
- Hölzl, H.; Kapelari, B.; Kellermann, J.; Seemüller, E.; Sümegi, M.; Udvardy, A.; Medalia, O.; Sperling, J.; Müller, S.A.; Engel, A.; et al. The regulatory complex of Drosophila melanogaster 26S proteasomes: Subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol. 2000, 150, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.A.; Xu, W.; DeMartino, G.N.; Cohen, R.E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 1997, 385, 737–740. [Google Scholar] [CrossRef]
- Emsley, J.; Cruz, M.; Handin, R.; Liddington, R. Crystal structure of the von Willebrand Factor A1 domain and implications for the binding of platelet glycoprotein Ib. J. Biol. Chem. 1998, 273, 10396–10401. [Google Scholar] [CrossRef]
- Yang, P.; Fu, H.; Walker, J.; Papa, C.M.; Smalle, J.; Ju, Y.-M.; Vierstra, R.D. Purification of the Arabidopsis 26 S proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J. Biol. Chem. 2004, 279, 6401–6413. [Google Scholar] [CrossRef]
- Book, A.J.; Gladman, N.P.; Lee, S.-S.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J. Biol. Chem. 2010, 285, 25554–25569. [Google Scholar] [CrossRef]
- Gemperline, D.C.; Marshall, R.S.; Lee, K.H.; Zhao, Q.; Hu, W.; McLoughlin, F.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Proteomic analysis of affinity-purified 26S proteasomes identifies a suite of assembly chaperones in Arabidopsis. J. Biol. Chem. 2019, 294, 17570–17592. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Dynamic regulation of the 26S proteasome: From synthesis to degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef]
- Gladman, N.P.; Marshall, R.S.; Lee, K.-H.; Vierstra, R.D. The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell 2016, 28, 1279–1296. [Google Scholar] [CrossRef]
- Park, S.; Kim, W.; Tian, G.; Gygi, S.P.; Finley, D. Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J. Biol. Chem. 2011, 286, 36652–36666. [Google Scholar] [CrossRef] [PubMed]
- Nickell, S.; Beck, F.; Scheres, S.H.W.; Korinek, A.; Förster, F.; Lasker, K.; Mihalache, O.; Sun, N.; Nagy, I.; Sali, A.; et al. Insights into the molecular architecture of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2009, 106, 11943–11947. [Google Scholar] [CrossRef] [PubMed]
- Bohn, S.; Beck, F.; Sakata, E.; Walzthoeni, T.; Beck, M.; Aebersold, R.; Förster, F.; Baumeister, W.; Nickell, S. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc. Natl. Acad. Sci. USA 2010, 107, 20992–20997. [Google Scholar] [CrossRef] [PubMed]
- Lander, G.C.; Estrin, E.; Matyskiela, M.E.; Bashore, C.; Nogales, E.; Martin, A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012, 482, 186–191. [Google Scholar] [CrossRef]
- Pathare, G.R.; Nagy, I.; Bohn, S.; Unverdorben, P.; Hubert, A.; Körner, R.; Nickell, S.; Lasker, K.; Sali, A.; Tamura, T.; et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc. Natl. Acad. Sci. USA 2012, 109, 149–154. [Google Scholar] [CrossRef]
- Fu, H.; Doelling, J.H.; Arendt, C.S.; Hochstrasser, M.; Vierstra, R.D. Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics 1998, 149, 677–692. [Google Scholar] [CrossRef]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26S Proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Welk, V.; Coux, O.; Kleene, V.; Abeza, C.; Trümbach, D.; Eickelberg, O.; Meiners, S. Inhibition of proteasome activity induces formation of alternative proteasome complexes. J. Biol. Chem. 2016, 291, 13147–13159. [Google Scholar] [CrossRef]
- Marques, A.J.; Glanemann, C.; Ramos, P.C.; Dohmen, R.J. The C-terminal extension of the β7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation. J. Biol. Chem. 2007, 282, 34869–34876. [Google Scholar] [CrossRef]
- Li, X.; Kusmierczyk, A.R.; Wong, P.; Emili, A.; Hochstrasser, M. β-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007, 26, 2339–2349. [Google Scholar] [CrossRef]
- Lehmann, A.; Jechow, K.; Enenkel, C. Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Rep. 2008, 9, 1237–1243. [Google Scholar] [CrossRef] [PubMed]
- Weberruss, M.H.; Savulescu, A.F.; Jando, J.; Bissinger, T.; Harel, A.; Glickman, M.H.; Enenkel, C. Blm10 facilitates nuclear import of proteasome core particles. EMBO J. 2013, 32, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Leggett, D.S.; Hanna, J.; Borodovsky, A.; Crosas, B.; Schmidt, M.; Baker, R.T.; Walz, T.; Ploegh, H.; Finley, D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 2002, 10, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Gorbea, C.; Goellner, G.M.; Teter, K.; Holmes, R.K.; Rechsteiner, M. Characterization of mammalian Ecm29, a 26 S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J. Biol. Chem. 2004, 279, 54849–54861. [Google Scholar] [CrossRef]
- Lehmann, A.; Niewienda, A.; Jechow, K.; Janek, K.; Enenkel, C. Ecm29 fulfils quality control functions in proteasome assembly. Mol. Cell 2010, 38, 879–888. [Google Scholar] [CrossRef]
- Lee, S.Y.-C.; De La Mota-Peynado, A.; Roelofs, J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. J. Biol. Chem. 2011, 286, 36641–36651. [Google Scholar] [CrossRef]
- De La Mota-Peynado, A.; Lee, S.Y.-C.; Pierce, B.M.; Wani, P.; Singh, C.R.; Roelofs, J. The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. J. Biol. Chem. 2013, 288, 29467–29481. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chemmama, I.E.; Yu, C.; Huszagh, A.; Xu, Y.; Viner, R.; Block, S.A.; Cimermancic, P.; Rychnovsky, S.D.; Ye, Y.; et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Biol. Chem. 2017, 292, 16310–16320. [Google Scholar] [CrossRef]
- Choi, W.H.; Yun, Y.; Byun, I.; Kim, S.; Lee, S.; Sim, J.; Levi, S.; Park, S.H.; Jun, J.; Kleifeld, O.; et al. ECPAS/Ecm29-mediated 26S proteasome disassembly is an adaptive response to glucose starvation. Cell Rep. 2023, 42, 112701. [Google Scholar] [CrossRef]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Arike, L.; Peil, L. Spectral counting label-free proteomics. In Shotgun Proteomics: Methods and Protocols; Martins-de-Souza, D., Ed.; Springer: New York, NY, USA, 2014; pp. 213–222. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]





| Subunit | Locus | Ratio of Total Number of PSMs rpn10-2/Col-0 (% ± SD) 2 | Subunit | Locus | Ratio of Total Number of PSMs rpn10-2/Col-0 (% ± SD) |
|---|---|---|---|---|---|
| CP | Base | ||||
| PAA1 | At5g35590 | 118.3/100.8 (117.4 ± 8.6) 3 | RPT1a | At1g53750 | 141.3/106.6 (132.6 ± 11.8) |
| PAA2 | At2g05840 | 123.0/89.9 (136.8 ± 6.3) | RPT1b | At1g53780 | ND/ND (NA) |
| PAB1 | At1g16470 | 32.7/43.4 (75.3 ± 80.1) | RPT2a | At4g29040 | ND/ND (NA) |
| PAB2 | At1g79210 | 17.3/ND (NA) | RPT2b | At2g20140 | 93.3/98.7 (94.5 ± 19.0) |
| PAC1 | At3g22110 | 102.9/101.3 (101.5 ± 27.4) | RPT3 | At5g58290 | 139.4/118.4 (117.7 ± 16.2) |
| PAC2 | At4g15160 | ND/ND (NA) | RPT4a | At5g43010 | 17.5/16.8 (104.2 ± 180.6) |
| PAD1 | At3g51260 | 124.0/107.3 (115.6 ± 3.7) | RPT4b | At1g45000 | 111.8/120.1 (93.1 ± 35.8) |
| PAD2 | At5g66140 | 32.7/10.5 (312.5 ± 270.9) | RPT5a | At3g05530 | 145.2/138.2 (105.1 ± 30.4) |
| PAE1 | At1g53850 | 18.8/11.8 (158.6 ± 141.7) | RPT5b | At1g09100 | 11.54/ND (NA) |
| PAE2 | At3g14290 | 26.9/31.6 (85.2 ± 45.1) | RPT6a | At5g19990 | 44.2/84.2 (52.5 ± 91.0) |
| PAF1 | At5g42790 | 96.1/107.9 (89.1 ± 13.8) | RPT6b | At5g20000 | 73.1/ND (NA) |
| PAF2 | At1g47250 | 10.13/ND (NA) | RPN1a | At2g20580 | 330.8/272.8 (121.3 ± 20.0) |
| PAG1 | At2g27020 | 85.6/65.8 (130.1 ± 34.1) | RPN1b | At4g28470 | 34.6/91.7 (37.7 ± 37.8) |
| PBA1 | At4g31300 | 90.4/77.6 (116.4 ± 24.2) | RPN2a | At2g32730 | 157.8/105.0 (150.3 ± 36.6) |
| PBB1 | At3g27430 | 25.1/39.5 (63.7 ± 65.1) | RPN2b | At1g04810 | 71.1/44.3 (160.2 ± 66.6) |
| PBB2 | At5g40580 | 11.9/ND (NA) | RPN10 | At4g38630 | ND/29.0 (NA) * |
| PBC1 | At1g21720 | 72.7/89.5 (81.3 ± 11.4) | RPN13 | At2g26590 | ND/ND (NA) |
| PBC2 | At1g77440 | 10.9/ND (NA) | RPN15 | At1g64750 | ND/ND (NA) |
| PBD1 | At3g22630 | 74.0/31.5 (234.7 ± 64.7) | Lid | ||
| PBD2 | At4g14800 | 6.8/29.7 (23.0 ± 39.8) | RPN3a | At1g20200 | 157.9/181.6 (86.9 ± 12.5) |
| PBE1 | At1g13060 | 56.8/34.2 (166.0 ± 31.8) | RPN3b | At1g75990 | 19.5/48.7 (40.1 ± 40.6) |
| PBE2 | At3g26340 | 5.21/ND (NA) | RPN5a | At5g09900 | 51.1/40.5 (126.3 ± 17.5) |
| PBF1 | At3g60820 | 100.0/100.0 (100.0 ± 25.9) | RPN5b | At5g64760 | 41.2/21.4 (192.7 ± 172.1) |
| PBG1 | At1g56450 | 106.7/90.8 (117.6 ± 17.7) | RPN6 | At1g29150 | 158.7/148.7 (106.7 ± 20.4) |
| 26S proteasome-associated factors | RPN7 | At4g24820 | 131.7/117.1 (112.5 ± 23.7) | ||
| ECM29 | At2g26780 | 180.8/63.2 (286.2 ± 90.1) * | RPN8a | At5g05780 | 60.1/89.5 (67.1 ± 19.9) |
| PA200 | At3G13330 | 89.4/53.9 (165.8 ± 29.8) | RPN8b | At3g11270 | 39.5/ND (NA) |
| TPP-II | At4g20850 | 97.1/152.6 (63.6 ± 20.9) | RPN9a | At5g45620 | ND/73.5 (NA) * |
| RPN9b | At4g19006 | 157.7/67.3 (234.4 ± 20.3) * | |||
| RPN11 | At5g23540 | 74.0/59.2 (125.0 ± 63.4) | |||
| RPN12a | At1g64520 | 97.1/114.5 (84.8 ± 15.4) | |||
| RPN12b | At5g42040 | ND/ND (NA) | |||
| Genotype | Total Number of Seeds Examined (Germination Rate) | Homozygous rpn10-2 Number (%) | χ2 |
|---|---|---|---|
| Col-0 1 | 605 (99.00%) | NA 2 | |
| N10n10 N13N13 | 3610 (99.30%) | 69 (1.93) | |
| N10n10 n13n13 | 1348 (99.50%) | 0 (0.00) | |
| N10 n10 n13n13 [N13-1] | 2825 (98.80%) | 57 (2.04) | 0.17 3 |
| N10n10 n13n13 [N13-2] | 3330 (99.73%) | 59 (1.78) | 0.39 |
| N10 n10 n13n13 [R67-1] | 3155 (98.00%) | 34 (1.10) | 11.49 * |
| N10 n10 n13n13 [R67-2] | 3111 (99.10%) | 26 (0.84) | 18.82 * |
| N10 n10 n13n13 [Q70-1] | 3124 (99.60%) | 77 (2.48) | 4.91 |
| N10 n10 n13n13 [Q70-2] | 2955 (99.22%) | 66 (2.25) | 1.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.-Y.; Lin, Y.-L.; Usharani, R.; Radjacommare, R.; Fu, H. The Structural Role of RPN10 in the 26S Proteasome and an RPN2-Binding Residue on RPN13 Are Functionally Important in Arabidopsis. Int. J. Mol. Sci. 2024, 25, 11650. https://doi.org/10.3390/ijms252111650
Lin S-Y, Lin Y-L, Usharani R, Radjacommare R, Fu H. The Structural Role of RPN10 in the 26S Proteasome and an RPN2-Binding Residue on RPN13 Are Functionally Important in Arabidopsis. International Journal of Molecular Sciences. 2024; 25(21):11650. https://doi.org/10.3390/ijms252111650
Chicago/Turabian StyleLin, Shih-Yun, Ya-Ling Lin, Raju Usharani, Ramalingam Radjacommare, and Hongyong Fu. 2024. "The Structural Role of RPN10 in the 26S Proteasome and an RPN2-Binding Residue on RPN13 Are Functionally Important in Arabidopsis" International Journal of Molecular Sciences 25, no. 21: 11650. https://doi.org/10.3390/ijms252111650
APA StyleLin, S.-Y., Lin, Y.-L., Usharani, R., Radjacommare, R., & Fu, H. (2024). The Structural Role of RPN10 in the 26S Proteasome and an RPN2-Binding Residue on RPN13 Are Functionally Important in Arabidopsis. International Journal of Molecular Sciences, 25(21), 11650. https://doi.org/10.3390/ijms252111650







