In Vitro vs. In Vivo Transcriptomic Approach Revealed Core Pathways of Nitrogen Deficiency Response in Tea Plant (Camellia sinensis (L.) Kuntze)
Abstract
:1. Introduction
2. Results
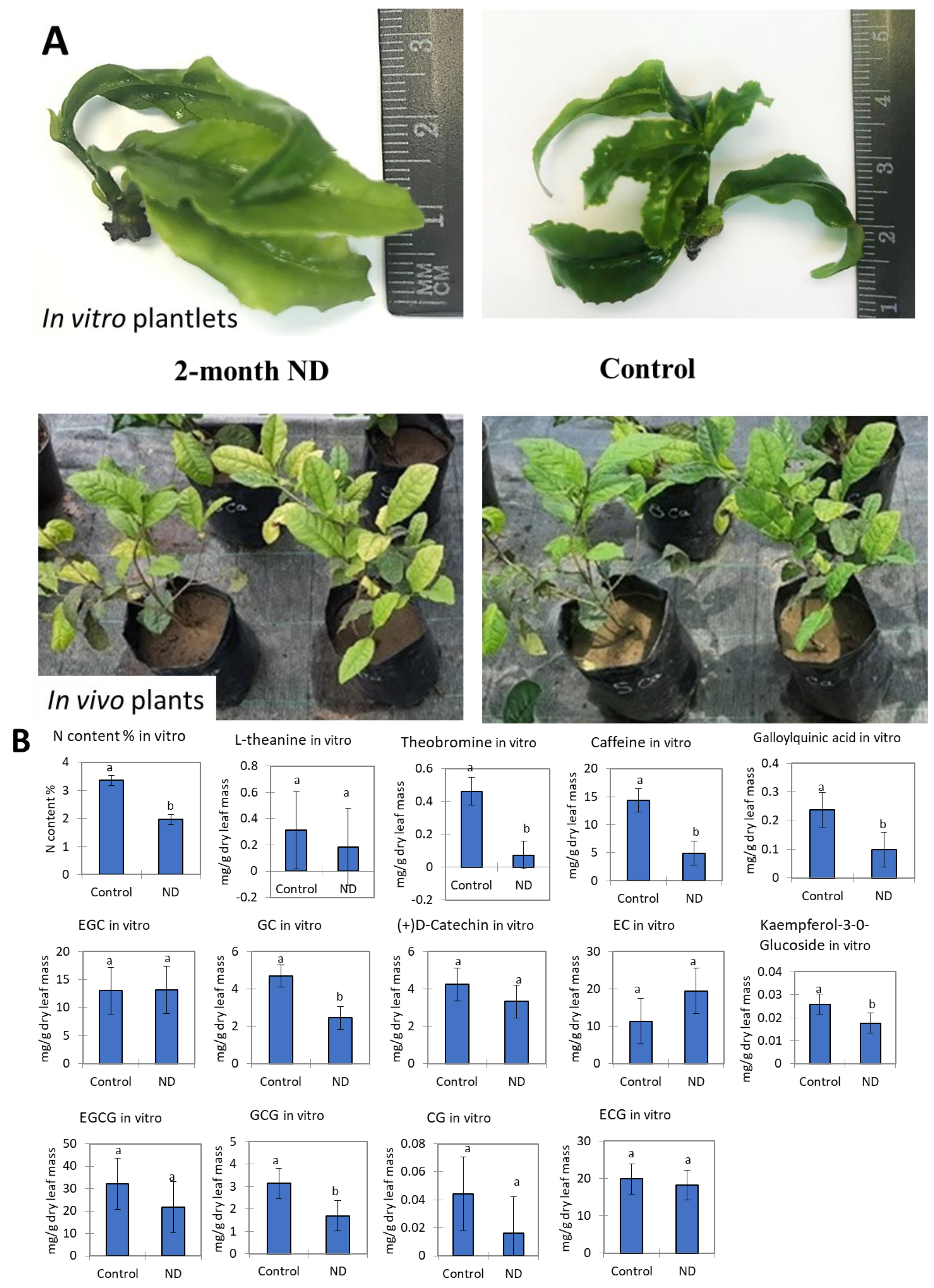
2.1. Leaf Quality Evaluation under ND In Vitro and In Vivo
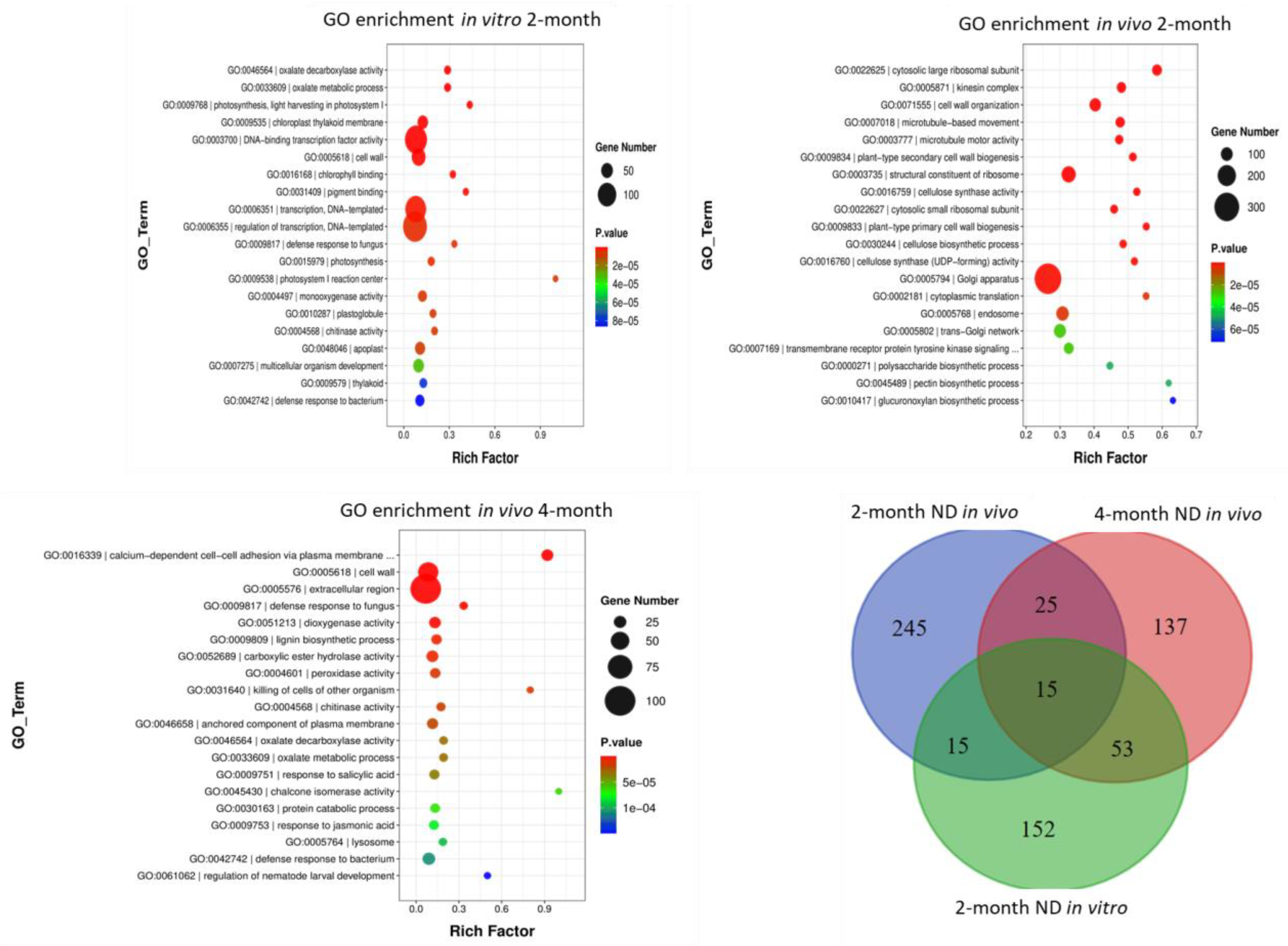
2.2. GO and KEGG Pathways Enrichment under ND In Vitro and In Vivo
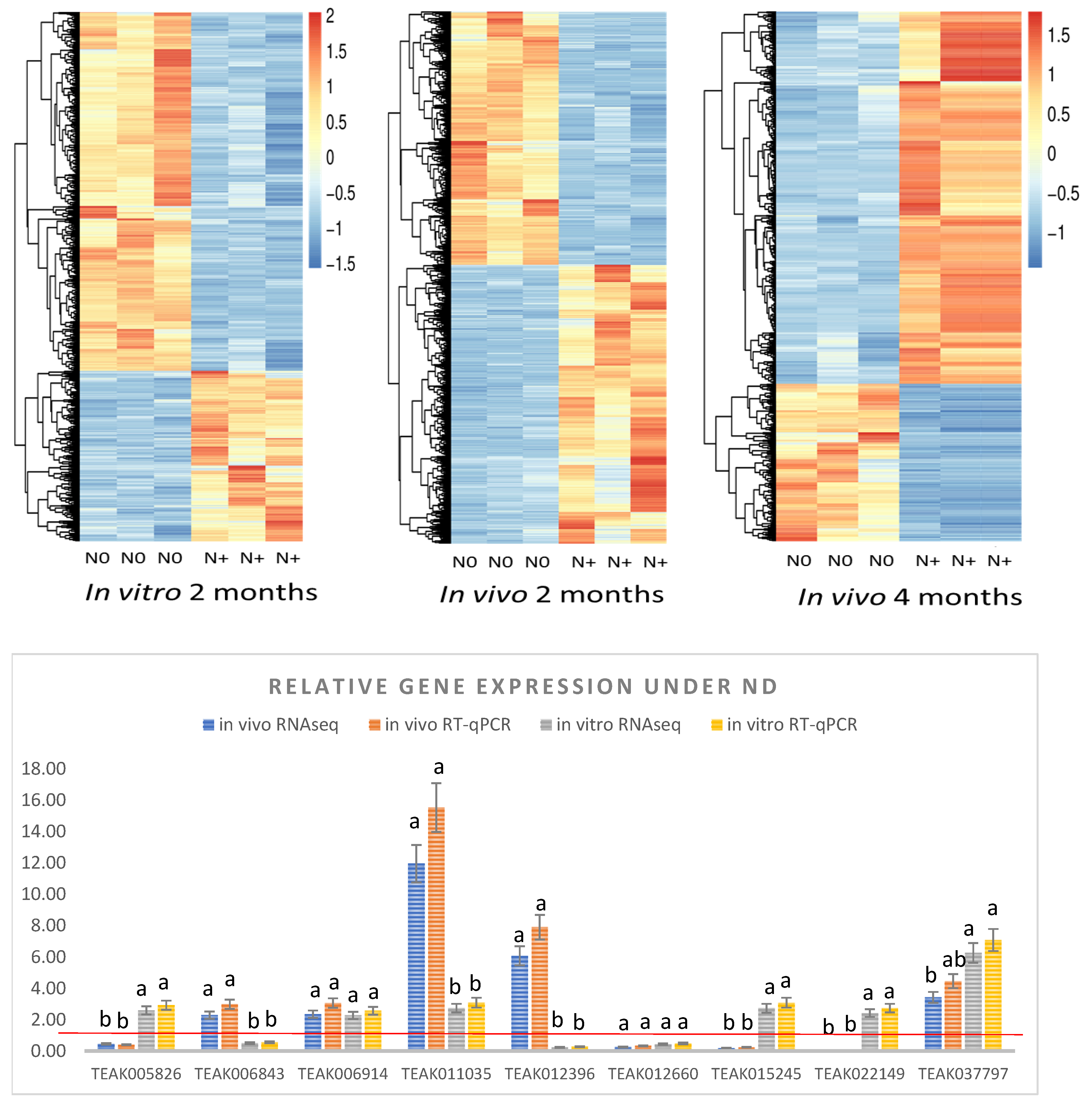
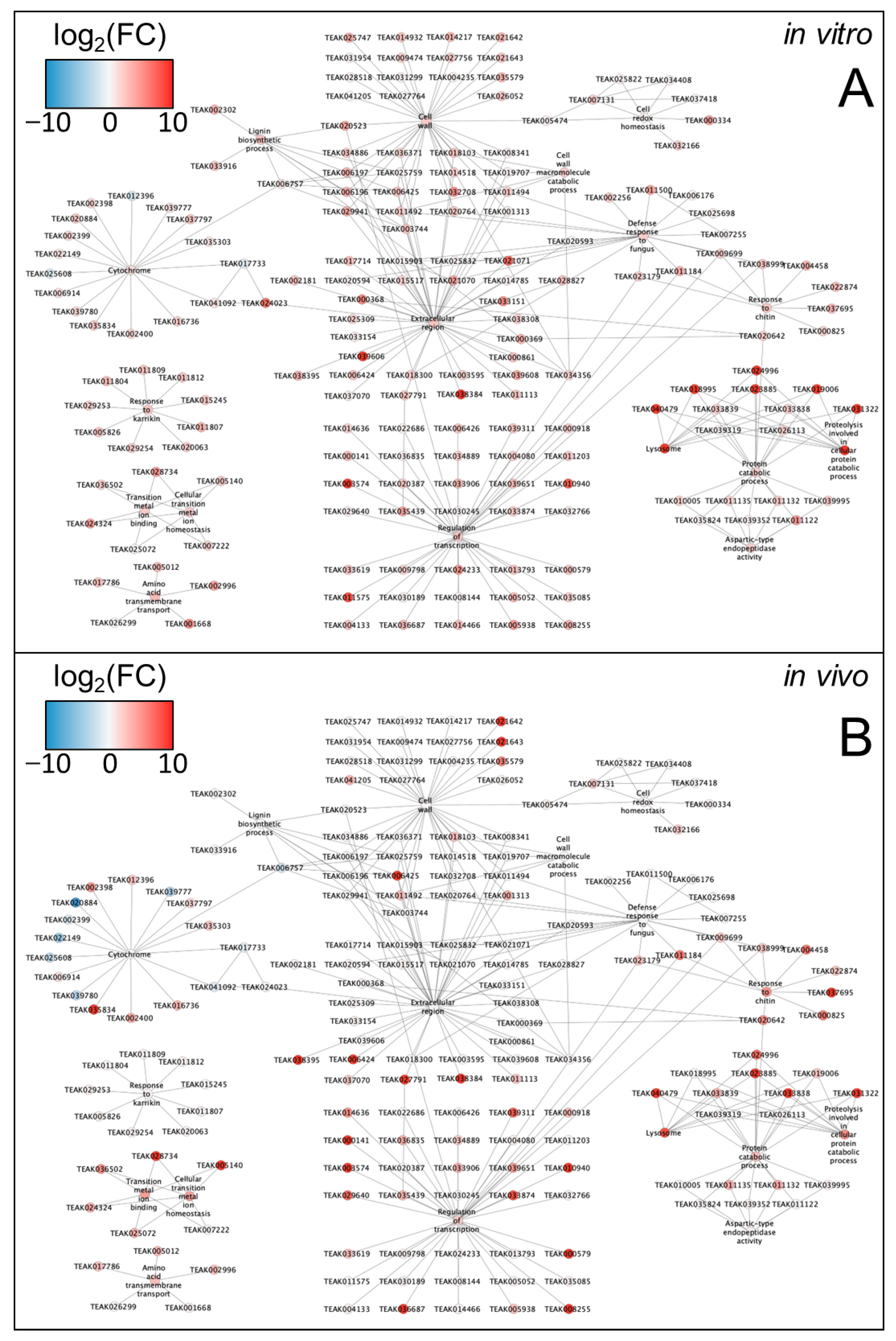
2.3. Key Common DEGs Significantly Affected by ND In Vitro and In Vivo
- (A)
- Calcium signaling included 818 DEGs with
- 40 DEGs directly related to MAPK signaling: Serine-threonine kinases, protein-tyrosine-phosphatases, histidine kinases, leucine-rich repeat receptors, and sucrose non-fermenting-1-related protein kinases.
- 28 DEGs of calcium-dependent protein kinases, calcium/calmodulin receptors, and CDPK-related kinases.
- 39 DEGs of hormonal signaling: histidine kinases, leucine-rich repeat receptor-like protein kinases, LRR receptor-like serine/threonine protein kinases, Serine/threonine protein kinases, and sucrose non-fermenting-1-related protein kinases.
- (B)
- Hormone signaling included 157 DEGs with
- 24 DEGs directly related to MAPK signaling: abscisic acid receptors, Dentin sialoproteins, EIN2-CEND, ethylene-responsive TFs, CUMW_237540, inducer of CBF expression 2, pathogenesis-related protein 1, LIGHT-DEPENDENT SHORT HYPOCOTYLS 10, bHLH14, bHLH18, bHLH25, protein phosphatase 2C 24, and protein phosphatase 2C 16,
- 44 DEGs of auxin signaling: auxin early response proteins GH3.1, GH3.9, SAUR23, SAUR41, and SAUR68, auxin responsive factors ARF2, ARF5, and ARF6, auxin transporter proteins, auxin-induced proteins, E3 ubiquitin proteins, EIN2-CEND, GH3 auxin-responsive promoters, indole-3-acetic acid-amido synthetase GH3.1, and transport inhibitor response 1.
- 34 DEGs of ABA signaling: abscisic acid receptors, ACT domain-containing proteins, bZIP1, bZIP8, Hb-ZIP, WCOR413, MFT1, general transcriptional corepressor trfA, bHLH14, MYB-related TF, DMP2, DMP6, IQ-DOMAIN-like, NRT1/PTR FAMILY 5.2.
- 14 DEGs of JA signaling: DELLA protein 3, DELLA4, EIN2-CEND, ethylene-responsive transcription factors, jasmonate-zim-domain proteins, Jasmonic acid-amido synthetase, transcription factor bHLH14, NRT1/ PTR FAMILY 5.2, TIFI, and bHLH.
3. Discussion
4. Materials and Methods
4.1. Plant Material and Stress Induction
4.2. Phenotyping of ND Response
4.3. mRNA-Sequencing, DEG Identification, and Gene Annotation
4.4. Verification of the RNAseq Results by qRT-PCR
4.5. Data Analysis and Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, J.; Sun, B.; Cheng, R.; Shi, Z.; Luo, D.; Liu, S.; Centritto, M. Effects of soil nitrogen (N) deficiency on photosynthetic N-use efficiency in N-fixing and non-N-fixing tree seedlings in subtropical China. Sci. Rep. 2019, 9, 4604. [Google Scholar] [CrossRef] [PubMed]
- Carranca, C.; Brunetto, G.; Tagliavini, M. Nitrogen Nutrition of Fruit Trees to Reconcile Productivity and Environmental Concerns. Plants 2018, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, H.; Pilon-Smits, E.; Huang, W.; Wang, P.; Wang, M.; Guo, F.; Wang, Y.; Li, R.; Zhao, H.; et al. Transcriptome-wide analysis of nitrogen-regulated genes in tea plant (Camellia sinensis LO Kuntze) and characterization of amino acid transporter CsCAT9.1. Plants 2020, 9, 1218. [Google Scholar] [CrossRef]
- Elrys, A.S.; Elnahal, A.S.; Abdo, A.I.; Desoky, E.S.M.; Selem, E.; Rady, M.M. Traditional, modern, and molecular strategies for improving the efficiency of nitrogen use in crops for sustainable agriculture: A fresh look at an old issue. J. Soil Sci. Plant Nutr. 2022, 22, 3130–3156. [Google Scholar] [CrossRef]
- Lin, Z.H.; Chen, C.S.; Zhong, Q.S.; Ruan, Q.C.; Chen, Z.H.; You, X.M.; Shan, R.Y.; Li, X.L. The GC-TOF/MS-based Metabolomic analysis reveals altered metabolic profiles in nitrogen-deficient leaves and roots of tea plants (Camellia sinensis). BMC Plant Biol. 2021, 21, 506. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.F.; Lin, J.Y. Estimating the gross budget of applied nitrogen and phosphorus in tea plantations. Sustain. Environ. Res. 2016, 26, 124–130. [Google Scholar] [CrossRef]
- Yan, P.; Shen, C.; Fan, L.; Li, X.; Zhang, L.; Zhang, L.; Han, W. Tea planting affects soil acidification and nitrogen and phosphorus distribution in soil. Agric. Ecosyst. Environ. 2018, 254, 20–25. [Google Scholar] [CrossRef]
- Yang, X.; Ma, L.; Ji, L.; Shi, Y.; Yi, X.; Yang, Q.; Ni, K.; Ruan, J. Long-term nitrogen fertilization indirectly affects soil fungi community structure by changing soil and pruned litter in a subtropical tea (Camellia sinensis L.) plantation in China. Plant Soil 2019, 444, 409–426. [Google Scholar] [CrossRef]
- Cochetel, N.; Escudié, F.; Cookson, S.J.; Dai, Z.; Vivin, P.; Bert, P.F.; Muñoz, M.S.; Delrot, S.; Klopp, C.; Ollat, N.; et al. Root transcriptomic responses of grafted grapevines to heterogeneous nitrogen availability depend on rootstock genotype. J. Exp. Bot. 2017, 68, 4339–4355. [Google Scholar] [CrossRef]
- Pan, S.Y.; Nie, Q.; Tai, H.C.; Song, X.L.; Tong, Y.F.; Zhang, L.J.F.; Wu, X.W.; Lin, Z.H.; Zhang, Y.Y.; Ye, D.Y.; et al. Tea and tea drinking: China’s outstanding contributions to the mankind. Chin. Med. 2022, 17, 27. [Google Scholar] [CrossRef]
- Zhang, W.; Ni, K.; Long, L.; Ruan, J. Nitrogen transport and assimilation in tea plant (Camellia sinensis): A review. Front. Plant Sci. 2023, 14, 1249202. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.H.; Tong, W.; Wu, Q.; Wei, S.; Zhao, J.; Zhang, Z.Z.; Wei, C.L.; Wan, X.C. Tea plant genomics: Achievements, challenges and perspectives. Hortic. Res. 2020, 7, 7. [Google Scholar] [CrossRef]
- Fang, K.; Xia, Z.; Li, H.; Jiang, X.; Qin, D.; Wang, Q.; Pan, C.; Li, B.; Wu, H. Genome-wide association analysis identified molecular markers associated with important tea flavor-related metabolites. Hortic. Res. 2021, 8, 42. [Google Scholar] [CrossRef]
- Li, F.; Dong, C.; Yang, T.; Bao, S.; Fang, W.; Lucas, W.J.; Zhang, Z. The tea plant CsLHT1 and CsLHT6 transporters take up amino acids, as a nitrogen source, from the soil of organic tea plantations. Hortic. Res. 2021, 8, 178. [Google Scholar] [CrossRef] [PubMed]
- Samynathan, R.; Thiruvengadam, M.; Nile, S.H.; Shariati, M.A.; Rebezov, M.; Mishra, R.K.; Venkidasamy, B.; Periyasamy, S.; Chung, I.M.; Pateiro, M.; et al. Recent insights on tea metabolites, their biosynthesis and chemo-preventing effects: A review. Crit. Rev. Food Sci. Nutr. 2021, 63, 3130–3149. [Google Scholar] [CrossRef]
- Zhu, J.; Xu, Q.; Zhao, S.; Xia, X.; Ya, X.; An, Y.; Mi, X.; Guo, L.; Samarina, L.S.; Wei, C. Comprehensive co-expression analysis provides novel insights into temporal variation of flavonoids in fresh leaves of the tea plant (Camellia sinensis). Plant Sci. 2020, 290, 110306. [Google Scholar] [CrossRef]
- Fan, K.; Zhang, Q.; Tang, D.; Shi, Y.; Ma, L.; Liu, M.; Ruan, J. Dynamics of nitrogen translocation from mature leaves to new shoots and related gene expression during spring shoots development in tea plants (Camellia sinensis L.). J. Plant Nutr. Soil Sci. 2020, 183, 180–191. [Google Scholar] [CrossRef]
- Huang, H.; Yao, Q.; Xia, E.; Gao, L. Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and amino acids in young shoots of tea plant (Camellia sinensis L.) associated with tea flavor. J. Agric. Food Chem. 2018, 66, 9828–9838. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Yang, T.; Su, Y.; Lin, S.; Zhang, S.; Zhang, Z. Nitrogen-regulated theanine and flavonoid biosynthesis in tea plant roots: Protein-level regulation revealed by multiomics analyses. J. Agric. Food Chem. 2021, 69, 10002–10016. [Google Scholar] [CrossRef]
- Zhang, F.; He, W.; Yuan, Q.; Wei, K.; Ruan, L.; Wang, L.; Cheng, H. Transcriptome analysis identifies CsNRT genes involved in nitrogen uptake in tea plants, with a major role of CsNRT2.4. Plant Physiol. Biochem. 2021, 167, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.-H.; Chen, C.-S.; Zhao, S.-Q.; Liu, Y.; Zhong, Q.-S.; Ruan, Q.-C.; Chen, Z.-H.; You, X.-M.; Shan, R.-Y.; Li, X.-L.; et al. Molecular and physiological mechanisms of tea (Camellia sinensis (L.) O. Kuntze) leaf and root in response to nitrogen deficiency. BMC Genom. 2023, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Sun, Z.; Zhang, X.; Han, X. Novel aspects of regulation of nitrogen responses in the tea plant (Camellia sinensis (L.)). Agronomy 2023, 13, 144. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Xu, H.; Jiang, S.; Fang, H.; Zhang, T.; Su, M.; Xu, L.; Zhang, Z.; Chen, X. Nitrogen affects anthocyanin biosynthesis by regulating MdLOB52 downstream of MdARF19 in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana). J. Plant Growth Regul. 2018, 37, 719–729. [Google Scholar] [CrossRef]
- Fernandes, J.C.; Goulao, L.F.; Amâncio, S. Regulation of cell wall remodeling in grapevine (Vitis vinifera L.) callus under individual mineral stress deficiency. J. Plant Physiol. 2016, 190, 95–105. [Google Scholar] [CrossRef]
- Saad, K.R.; Kumar, G.; Giridhar, P. Differential expression of anthocyanin biosynthesis genes in Daucus carota callus culture in response to ammonium and potassium nitrate ratio in the culture medium. 3 Biotech 2018, 8, 431. [Google Scholar] [CrossRef]
- Oleszkiewicz, T.; Kruczek, M.; Baranski, R. Repression of Carotenoid Accumulation by Nitrogen and NH4+ Supply in Carrot Callus Cells In Vitro . Plants 2021, 10, 1813. [Google Scholar] [CrossRef]
- Xiang, P.; Zhu, Q.; Tukhvatshin, M.; Cheng, B.; Tan, M.; Liu, J.; Wang, X.; Huang, J.; Gao, S.; Lin, D.; et al. Light control of catechin accumulation is mediated by photosynthetic capacity in tea plant (Camellia sinensis). BMC Plant Biol. 2021, 21, 478. [Google Scholar] [CrossRef]
- Gong, A.D.; Lian, S.B.; Wu, N.N.; Zhou, Y.J.; Zhao, S.Q.; Zhang, L.M.; Cheng, L.; Yuan, H.Y. Integrated transcriptomics and metabolomics analysis of catechins, caffeine and theanine biosynthesis in tea plant (Camellia sinensis) over the course of seasons. BMC Plant Biol. 2020, 20, 294. [Google Scholar] [CrossRef]
- Zeng, L.; Zhou, X.; Liao, Y.; Yang, Z. Roles of specialized metabolites in biological function and environmental adaptability of tea plant (Camellia sinensis) as a metabolite studying model. J. Adv. Res. 2020, 34, 159–171. [Google Scholar] [CrossRef]
- Perchlik, M.; Tegeder, M. Improving plant nitrogen use efficiency through alteration of amino acid transport processes. Plant Physiol. 2017, 175, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Melzer, M. Regulation of root development and architecture by strigolactones under optimal and nutrient deficiency conditions. Int. J. Mol. Sci. 2018, 19, 1887. [Google Scholar] [CrossRef]
- Franciosini, A.; Rymen, B.; Shibata, M.; Favero, D.S.; Sugimoto, K. Molecular networks orchestrating plant cell growth. Curr. Opin. Plant Biol. 2017, 35, 98–104. [Google Scholar] [CrossRef]
- Wang, Y.; Ouyang, J.X.; Fan, D.M.; Wang, S.M.; Xuan, Y.M.; Wang, X.C.; Zheng, X.Q. Transcriptome analysis of tea (Camellia sinensis) leaves in response to ammonium starvation and recovery. Front. Plant Sci. 2022, 13, 963269. [Google Scholar] [CrossRef] [PubMed]
- Samarina, L.; Malyukova, L.; Wang, S.; Li, Y.; Doroshkov, A.; Bobrovskikh, A.; Shkhalakhova, R.; Koninskaya, N.; Matskiv, A.; Velikiy, A.; et al. Nitrogen deficiency differentially affects lignin biosynthesis genes and flavanols accumulation in tolerant and susceptible tea genotypes (Camellia sinensis (L.) Kuntze). Plant Stress 2024, 14, 100581. [Google Scholar] [CrossRef]
- Mu, X.; Chen, Y. The physiological response of photosynthesis to nitrogen deficiency. Plant Physiol. Biochem. 2021, 158, 76–82. [Google Scholar] [CrossRef]
- Kumari, M.; Naidu, S.; Kumari, B.; Singh, I.K.; Singh, A. Comparative transcriptome analysis of Zea mays upon mechanical wounding. Mol. Biol. Rep. 2023, 50, 5319–5343. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, X.; Yu, H.; Jiang, W.; Sun, N.; Liu, X.; Liu, X.; Zhang, X.; Wang, Y.; Gu, X. RNA-Seq-based transcriptome profiling of early nitrogen deficiency response in cucumber seedlings provides new insight into the putative nitrogen regulatory network. Plant Cell Physiol. 2015, 56, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.; Jugulam, M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Bigeard, J.; Hirt, H. Nuclear signaling of plant MAPKs. Front. Plant Sci. 2018, 9, 469. [Google Scholar] [CrossRef]
- Kumar, K.; Raina, S.K.; Sultan, S.M. Arabidopsis MAPK signaling pathways and their cross talks in abiotic stress response. J. Plant Biochem. Biotechnol. 2020, 29, 700–714. [Google Scholar] [CrossRef]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 2006, 97, 883–893. [Google Scholar] [CrossRef]
- Jia, Z.; Giehl, R.F.H.; Hartmann, A.; Estevez, J.M.; Bennett, M.J.; von Wiren, N. A spatially-concerted epidermal auxin signaling framework steers the root hair foraging response under low nitrogen. Curr. Biol. 2023, 33, 3926–3941. [Google Scholar] [CrossRef]
- Jung, J.Y.; Ahn, J.H.; Schachtman, D.P. CC-type glutaredoxins mediate plant response and signaling under nitrate starvation in Arabidopsis. BMC Plant Biol. 2018, 18, 281. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, Y.; Liu, Y.; Li, X.; Hao, G.; Han, Q.; Dirk, L.M.; Downie, A.B.; Ruan, Y.L.; Wang, J.; et al. Raffinose synthase enhances drought tolerance through raffinose synthesis or galactinol hydrolysis in maize and Arabidopsis plants. J. Biol. Chem. 2020, 295, 8064–8077. [Google Scholar] [CrossRef]
- Peng, M.Y.; Ren, Q.Q.; Lai, Y.H.; Zhang, J.; Chen, H.H.; Guo, J.; Yang, L.T.; Chen, L.S. Integration of physiology, metabolome and transcriptome for understanding of the adaptive strategies to long-term nitrogen deficiency in Citrus sinensis leaves. Sci. Hortic. 2023, 317, 112079. [Google Scholar] [CrossRef]
- Yang, H.; Duan, Y.; Wu, Y.; Zhang, C.; Wu, W.; Lyu, L.; Li, W. Physiological and transcriptional responses of carbohydrate and nitrogen metabolism and ion balance in blueberry plants under nitrogen deficiency. Plant Growth Regul. 2023, 101, 519–535. [Google Scholar] [CrossRef]
- Pask, A.J.D.; Sylvester-Bradley, R.; Jamieson, P.D.; Foulkes, M.J. Quantifying how winter wheat crops accumulate and use nitrogen reserves during growth. Field Crops Res. 2012, 126, 104–118. [Google Scholar] [CrossRef]
- Tzvetkova-Chevolleau, T.; Franck, F.; Alawady, A.E.; Dall’Osto, L.; Carrière, F.; Bassi, R.; Grimm, B.; Nussaume, L.; Havaux, M. The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J. 2007, 50, 795–809. [Google Scholar] [CrossRef]
- Li, X.W.; Liu, H.J.; Xie, S.X.; Yuan, H.Y. Isolation and characterization of two genes of the early light-induced proteins of Camellia sinensis. Photosynthetica 2013, 51, 305–311. [Google Scholar] [CrossRef]
- Samarina, L.S.; Wang, S.; Malyukova, L.; Bobrovskikh, A.; Doroshkov, A.; Koninskaya, N.; Shkhalakhova, R.; Matskiv, A.; Fedorina, J.; Fizikova, A.; et al. Long-term cold, freezing and drought: Overlapping and specific regulatory mechanisms and signal transduction in tea plant (Camellia sinensis (L.) Kuntze). Front. Plant Sci. 2023, 14, 1145793. [Google Scholar] [CrossRef] [PubMed]
- Curci, P.L.; Aiese Cigliano, R.; Zuluaga, D.L.; Janni, M.; Sanseverino, W.; Sonnante, G. Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 2017, 7, 1176. [Google Scholar] [CrossRef]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Q.; Yan, C.; Sun, Q.; Wang, J.; Li, C.; Yuan, C.; Mou, Y.; Shan, S. The bHLH transcription factor AhbHLH121 improves salt tolerance in peanut. Int. J. Biol. Macromol. 2024, 256, 128492. [Google Scholar] [CrossRef]
- Bhat, B.A.; Mir, R.A.; Mir, W.R.; Hamdani, S.S.; Mir, M.A. Transcription factors-golden keys to modulate the plant metabolism to develop salinity tolerance. Plant Stress 2024, 11, 100409. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Y.X.; Liu, Z.W.; Wang, W.L.; Li, H.; Zhuang, J. Transcriptome-wide identification and expression profile analysis of the bHLH family genes in Camellia sinensis. Funct. Integr. Genom. 2018, 18, 489–503. [Google Scholar] [CrossRef]
- Ma, Q.; Zhou, Q.; Chen, C.; Cui, Q.; Zhao, Y.; Wang, K.; Arkorful, E.; Chen, X.; Sun, K.; Li, X. Isolation and expression analysis of CsCML genes in response to abiotic stresses in the tea plant (Camellia sinensis). Sci. Rep. 2019, 9, 8211. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yan, X.; Sun, T.; Shen, Y.; Shi, Q.; Wang, W.; Bao, M.; Luo, H.; Nian, F.; Ning, G. Homeostatic regulation of flavonoid and lignin biosynthesis in phenylpropanoid pathway of transgenic tobacco. Gene 2022, 809, 146017. [Google Scholar] [CrossRef]
- Zentgraf, U.; Doll, J. Arabidopsis WRKY53, a node of multi-layer regulation in the network of senescence. Plants 2019, 8, 578. [Google Scholar] [CrossRef]
- Xiang, P.; Zhu, Q.; Xu, P.; Liu, L.; Li, Y.; Cheng, B.; Wang, X.; Liu, J.; Shi, Y.; Wu, L.; et al. Integrative analyses of transcriptome and metabolome reveal comprehensive mechanisms of Epigallocatechin-3-gallate (EGCG) biosynthesis in response to ecological factors in tea plant (Camellia sinensis). Food Res. Int. 2023, 166, 112591. [Google Scholar] [CrossRef]
- Huang, D.; Gong, Z.; Chen, X.; Wang, H.; Tan, R.; Mao, Y. Transcriptomic responses to aluminum stress in tea plant leaves. Sci. Rep. 2021, 11, 5800. [Google Scholar] [CrossRef] [PubMed]
- Mehari, T.G.; Hou, Y.; Xu, Y.; Umer, M.J.; Shiraku, M.L.; Wang, Y.; Wang, H.; Peng, R.; Wei, Y.; Cai, X.; et al. Overexpression of cotton GhNAC072 gene enhances drought and salt stress tolerance in transgenic Arabidopsis. BMC Genom. 2022, 23, 648. [Google Scholar] [CrossRef]
- Lin, Z.H.; Zhong, Q.S.; Chen, C.S.; Ruan, Q.C.; Chen, Z.H.; You, X.M. Carbon dioxide assimilation and photosynthetic electron transport of tea leaves under nitrogen deficiency. Bot. Stud. 2016, 57, 37. [Google Scholar] [CrossRef] [PubMed]
- Samarina, L.; Fedorina, J.; Kuzmina, D.; Malyukova, L.; Manakhova, K.; Kovalenko, T.; Matskiv, A.; Xia, E.; Tong, W.; Zhang, Z.; et al. Analysis of Functional Single-Nucleotide Polymorphisms (SNPs) and Leaf Quality in Tea Collection under Nitrogen-Deficient Conditions. Int. J. Mol. Sci. 2023, 24, 14538. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Raivo, K. Package ‘Pheatmap’. 2019. Available online: https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf (accessed on 1 December 2023).
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fan, W.; Tian, G.; Zhu, H.; He, L.; Cai, J.; Huang, Q.; Cai, Q.; Li, B.; Bai, Y.; et al. The sequence and de novo assembly of the giant panda genome. Nature 2010, 463, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Korf, I.; Robb, S.M.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Alvarado, A.S.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarina, L.; Malyukova, L.; Wang, S.; Bobrovskikh, A.; Doroshkov, A.; Shkhalakhova, R.; Manakhova, K.; Koninskaya, N.; Matskiv, A.; Ryndin, A.; et al. In Vitro vs. In Vivo Transcriptomic Approach Revealed Core Pathways of Nitrogen Deficiency Response in Tea Plant (Camellia sinensis (L.) Kuntze). Int. J. Mol. Sci. 2024, 25, 11726. https://doi.org/10.3390/ijms252111726
Samarina L, Malyukova L, Wang S, Bobrovskikh A, Doroshkov A, Shkhalakhova R, Manakhova K, Koninskaya N, Matskiv A, Ryndin A, et al. In Vitro vs. In Vivo Transcriptomic Approach Revealed Core Pathways of Nitrogen Deficiency Response in Tea Plant (Camellia sinensis (L.) Kuntze). International Journal of Molecular Sciences. 2024; 25(21):11726. https://doi.org/10.3390/ijms252111726
Chicago/Turabian StyleSamarina, Lidiia, Lyudmila Malyukova, Songbo Wang, Aleksandr Bobrovskikh, Alexey Doroshkov, Ruset Shkhalakhova, Karina Manakhova, Natalia Koninskaya, Alexandra Matskiv, Alexey Ryndin, and et al. 2024. "In Vitro vs. In Vivo Transcriptomic Approach Revealed Core Pathways of Nitrogen Deficiency Response in Tea Plant (Camellia sinensis (L.) Kuntze)" International Journal of Molecular Sciences 25, no. 21: 11726. https://doi.org/10.3390/ijms252111726
APA StyleSamarina, L., Malyukova, L., Wang, S., Bobrovskikh, A., Doroshkov, A., Shkhalakhova, R., Manakhova, K., Koninskaya, N., Matskiv, A., Ryndin, A., Khlestkina, E., & Orlov, Y. (2024). In Vitro vs. In Vivo Transcriptomic Approach Revealed Core Pathways of Nitrogen Deficiency Response in Tea Plant (Camellia sinensis (L.) Kuntze). International Journal of Molecular Sciences, 25(21), 11726. https://doi.org/10.3390/ijms252111726








