Overview on Current Selectable Marker Systems and Novel Marker Free Approaches in Fruit Tree Genetic Engineering
Abstract
:1. Introduction
2. Selectable Marker Systems Based on Toxic Compounds
2.1. Selection Based on Antibiotics
2.2. Selection Based on Herbicides
3. Selectable Marker Systems Based on Non-Metabolizable Compounds
4. Selectable Marker Systems Based on Morphogenic Regulators
5. Non-Selectable Marker Systems Based on Reporter Genes
6. Marker-Free Systems
6.1. SMG Removal by Co-Transformation
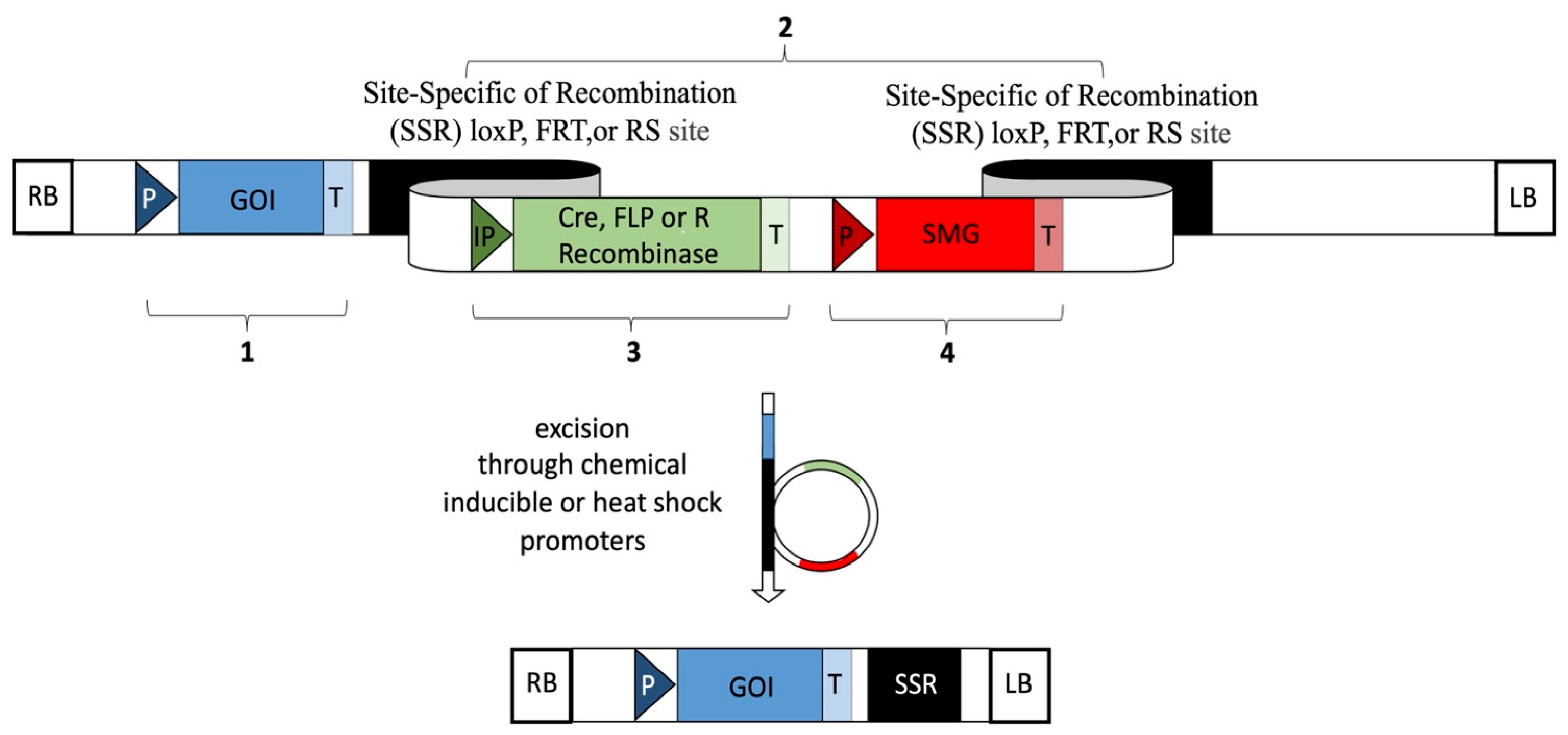
6.2. SMG Removal Using Site-Specific Recombinases
6.3. Production of DNA-Free Genetically Edited Fruits
7. Biosafety Considerations on the Use of SMGs in Fruit Crops
7.1. SMGs for Conditional Positive Selection Are Important for Fruit Crops, Can We Accept Them?
7.2. Are Other SMG Systems More Acceptable?
7.3. SMGs for New Breeding Techniques
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ricci, A.; Mezzetti, B.; Navacchi, O.; Burgos, L.; Sabbadini, S. In Vitro Regeneration, via Organogenesis, from Leaves of the Peach Rootstock GF677 (P. persica × P. amygdalus). Acta Hortic. 2023, 1359, 81–85. [Google Scholar] [CrossRef]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New Biotechnological Tools for the Genetic Improvement of Major Woody Fruit Species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef] [PubMed]
- Bevan, M.W.; Flavell, R.B.; Chilton, M.D. A Chimaeric Antibiotic Resistance Gene as a Selectable Marker for Plant Cell Transformation. Nature 1983, 304, 184–187. [Google Scholar] [CrossRef]
- Fraley, R.T.; Rogers, S.G.; Horsch, R.B.; Sanders, P.R.; Flick, J.S.; Adams, S.P.; Bittner, M.L.; Brand, L.A.; Fink, C.L.; Fry, J.S.; et al. Expression of Bacterial Genes in Plant Cells. Proc. Natl. Acad. Sci. USA 1983, 80, 4803–4807. [Google Scholar] [CrossRef] [PubMed]
- Sundar, I.K.; Sakthivel, N. Advances in Selectable Marker Genes for Plant Transformation. J. Plant Physiol. 2008, 165, 1698–1716. [Google Scholar] [CrossRef] [PubMed]
- Ramessar, K.; Peremarti, A.; Gómez-Galera, S.; Naqvi, S.; Moralejo, M.; Muñoz, P.; Capell, T.; Christou, P. Biosafety and Risk Assessment Framework for Selectable Marker Genes in Transgenic Crop Plants: A Case of the Science Not Supporting the Politics. Transgenic Res. 2007, 16, 261–280. [Google Scholar] [CrossRef]
- Sabbadini, S.; Capriotti, L.; Molesini, B.; Pandolfini, T.; Navacchi, O.; Limera, C.; Ricci, A.; Mezzetti, B. Comparison of Regeneration Capacity and Agrobacterium-Mediated Cell Transformation Efficiency of Different Cultivars and Rootstocks of Vitis spp. via Organogenesis. Sci. Rep. 2019, 9, 582. [Google Scholar] [CrossRef]
- Capriotti, L.; Limera, C.; Mezzetti, B.; Ricci, A.; Sabbadini, S. From Induction to Embryo Proliferation: Improved Somatic Embryogenesis Protocol in Grapevine for Italian Cultivars and Hybrid Vitis Rootstocks. Plant Cell. Tissue Organ Cult. 2022, 151, 221–233. [Google Scholar] [CrossRef]
- EFSA. Opinion of the Scientific Panel on Genetically Modified Organisms on the Use of Antibiotic Resistance Genes as Marker Genes in Genetically Modified Plants. EFSA J. 2004, 2, 48. [Google Scholar] [CrossRef]
- Manimaran, P.; Ramkumar, G.; Sakthivel, K.; Sundaram, R.M.; Madhav, M.S.; Balachandran, S.M. Suitability of Non-Lethal Marker and Marker-Free Systems for Development of Transgenic Crop Plants: Present Status and Future Prospects. Biotechnol. Adv. 2011, 29, 703–714. [Google Scholar] [CrossRef]
- Yau, Y.Y.; Stewart, C.N. Less Is More: Strategies to Remove Marker Genes from Transgenic Plants. BMC Biotechnol. 2013, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically Modified Crops: Current Status and Future Prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef] [PubMed]
- Waldron, C.; Murphy, E.B.; Roberts, J.L.; Gustafson, G.D.; Armour, S.L.; Malcolm, S.K. Resistance to Hygromycin B—A New Marker for Plant Transformation Studies. Plant Mol. Biol. 1985, 5, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Twyman, R.M.; Stöger, E.; Kohli, A.; Capell, T.; Christou, P. Selectable and Screenable Markers for Rice Transformation. In Testing for Genetic Manipulation in Plants; Springer: Berlin/Heidelberg, Germany, 2002; pp. 1–17. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 4 March 2024).
- Modgil, M.; Sharma, R. Effect of Antibiotics on Regeneration and Elimination of Bacteria during Gene Transfer in Apple. Acta Hortic. 2009, 839, 353–359. [Google Scholar] [CrossRef]
- Tripathi, J.N.; Oduor, R.O.; Tripathi, L. A High-Throughput Regeneration and Transformation Platform for Production of Genetically Modified Banana. Front. Plant Sci. 2015, 6, 1025. [Google Scholar] [CrossRef]
- Nirala, N.K.; Das, D.K.; Srivastava, P.S.; Sopory, S.K.; Upadhyaya, K.C. Expression of a Rice Chitinase Gene Enhances Antifungal Potential in Transgenic Grapevine (Vitis vinifera L.). Vitis—J. Grapevine Res. 2010, 49, 181–187. [Google Scholar]
- Lebedev, V.G.; Taran, S.A.; Shmatchenko, V.V.; Dolgov, S.V.; Lunin, V.G.; Skryabin, K.G. Molecular Breeding of Pear. Acta Hortic. 2002, 587, 217–223. [Google Scholar] [CrossRef]
- Wang, M.L.; Uruu, G.; Xiong, L.; He, X.; Nagai, C.; Cheah, K.T.; Hu, J.S.; Nan, G.L.; Sipes, B.S.; Atkinson, H.J.; et al. Production of Transgenic Pineapple (Ananas Cosmos (L.) Merr.) Plants via Adventitious Bud Regeneration. Vitr. Cell. Dev. Biol.—Plant 2009, 45, 112–121. [Google Scholar] [CrossRef]
- Liu, Y.S.; Wang, H.Y.; Zhao, Y.J.; Jin, Y.B.; Li, C.; Ma, F.-W. Establishment of an Efficient Regeneration and Genetic Transformation System for Malus Prunifolia Borkh. ‘Fupingqiuzi’. J. Integr. Agric. 2022, 21, 2615–2627. [Google Scholar] [CrossRef]
- Wada, M.; Nishitani, C.; Komori, S. Stable and Efficient Transformation of Apple. Plant Biotechnol. 2020, 37, 163–170. [Google Scholar] [CrossRef]
- Timerbaev, V.; Mitiouchkina, T.; Pushin, A.; Dolgov, S. Production of Marker-Free Apple Plants Expressing the Supersweet Protein Gene Driven by Plant Promoter. Front. Plant Sci. 2019, 10, 388. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Feng, F.; Liang, D.; Cheng, L.; Ma, F.; Shi, S. Overexpression of a Malus Vacuolar Na+/H+ Antiporter Gene (MdNHX1) in Apple Rootstock M.26 and Its Influence on Salt Tolerance. Plant Cell. Tissue Organ Cult. 2010, 102, 337–345. [Google Scholar] [CrossRef]
- Martinelli, F.; Perrone, A.; Dandekar, A.M. Development of a Protocol for Genetic Transformation of Malus spp. Caryologia 2021, 74, 9–19. [Google Scholar] [CrossRef]
- Chen, J.; Tomes, S.; Gleave, A.P.; Hall, W.; Luo, Z.; Xu, J.; Yao, J. Significant Improvement of Apple (Malus Domestica Borkh.) Transgenic Plant Production by Pre-Transformation with a Baby Boom Transcription Factor. Hortic. Res. 2022, 9, uhab014. [Google Scholar] [CrossRef] [PubMed]
- Charrier, A.; Vergne, E.; Dousset, N.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient Targeted Mutagenesis in Apple and First Time Edition of Pear Using the CRISPR-Cas9 System. Front. Plant Sci. 2019, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Flachowsky, H.; Peil, A.; Sopanen, T.; Elo, A.; Hanke, V. Overexpression of BpMADS4 from Silver Birch (Betula Pendula Roth.) Induces Early-Flowering in Apple (Malus x Domestica Borkh.). Plant Breed. 2007, 126, 137–145. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Y.; Wu, Y.; Cheng, H.; Li, Y.; Zhao, Y.; Li, Y. Transgenic Plants from Fragmented Shoot Tips of Apple (Malus Baccata (L.) Borkhausen) via Agrobacterium-Mediated Transformation. Sci. Hortic. 2011, 128, 450–456. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, D.; Wang, C.; Wang, X.; Li, J.; Wang, R. Construction of High Efficiency Regeneration and Transformation Systems of Pyrus ussuriensis Maxim. Plant Cell. Tissue Organ Cult. 2017, 131, 139–150. [Google Scholar] [CrossRef]
- Matsuda, N.; Gao, M.; Isuzugawa, K.; Takashina, T.; Nishimura, K. Development of an Agrobacterium-Mediated Transformation Method for Pear (Pyrus communis L.) with Leaf-Section and Axillary Shoot-Meristem Explants. Plant Cell Rep. 2005, 24, 45–51. [Google Scholar] [CrossRef]
- Yancheva, S.D.; Shlizerman, L.A.; Golubowicz, S.; Yabloviz, Z.; Perl, A.; Hanania, U.; Flaishman, M.A. The Use of Green Fluorescent Protein (GFP) Improves Agrobacterium-Mediated Transformation of “Spadona” Pear (Pyrus communis L.). Plant Cell Rep. 2006, 25, 183–189. [Google Scholar] [CrossRef]
- Nakajima, I.; Sato, Y.; Saito, T.; Moriguchi, T.; Yamamoto, T. Agrobacterium-Mediated Genetic Transformation Using Cotyledons in Japanese Pear (Pyrus Pyrifolia). Breed. Sci. 2013, 63, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhang, S.; Liu, Y.; Chen, Y.; Zhai, R.; Yang, C.; Wang, Z.; Ma, F.; Xu, L. Efficient Agrobacterium-Mediated Genetic Transformation Using Cotyledons, Hypocotyls and Roots of ‘Duli’ (Pyrus Betulifolia Bunge). Sci. Hortic. 2022, 296, 110906. [Google Scholar] [CrossRef]
- Peng, A.; Zou, X.; Xu, L.; He, Y.; Lei, T.; Yao, L.; Li, Q.; Chen, S. Improved Protocol for the Transformation of Adult Citrus sinensis Osbeck ‘Tarocco’ Blood Orange Tissues. Vitr. Cell. Dev. Biol.—Plant 2019, 55, 659–667. [Google Scholar] [CrossRef]
- Almeida, W.A.B.; Mourão Filho, F.A.A.; Pino, L.E.; Boscariol, R.L.; Rodriguez, A.P.M.; Mendes, B.M.J. Genetic Transformation and Plant Recovery from Mature Tissues of Citrus sinensis L. Osbeck. Plant Sci. 2003, 164, 203–211. [Google Scholar] [CrossRef]
- Canton, M.; Wu, H.; Dutt, M.; Zale, J. A New Liquid Selection System for Mature Citrus Transformation. Sci. Hortic. 2022, 293, 110672. [Google Scholar] [CrossRef]
- Khan, E.U.; Fu, X.Z.; Liu, J.H. Agrobacterium-Mediated Genetic Transformation and Regeneration of Transgenic Plants Using Leaf Segments as Explants in Valencia Sweet Orange. Plant Cell. Tissue Organ Cult. 2012, 109, 383–390. [Google Scholar] [CrossRef]
- Dutt, M.; Grosser, J.W. Evaluation of Parameters Affecting Agrobacterium-Mediated Transformation of Citrus. Plant Cell. Tissue Organ Cult. 2009, 98, 331–340. [Google Scholar] [CrossRef]
- Fávero, P.; de Alves Mourão Filho, F.A.; Stipp, L.C.L.; Mendes, B.M.J. Genetic Transformation of Three Sweet Orange Cultivars from Explants of Adult Plants. Acta Physiol. Plant. 2012, 34, 471–477. [Google Scholar] [CrossRef]
- Al Bachchu, M.A.; Jin, S.B.; Park, J.W.; Sun, H.J.; Yun, S.H.; Lee, H.Y.; Lee, D.S.; Hong, Q.C.; Kim, Y.W.; Riu, K.Z.; et al. Agrobacterium-Mediated Transformation Using Embryogenic Calli in Satsuma Mandarin (Citrus unshiu Marc.) cv. Miyagawa Wase. Hortic. Environ. Biotechnol. 2011, 52, 170–175. [Google Scholar] [CrossRef]
- Mezzetti, B.; Pandolfini, T.; Navacchi, O.; Landi, L. Genetic Transformation of Vitis Vinifera via Organogenesis. BMC Biotechnol. 2002, 2, 18. [Google Scholar] [CrossRef]
- Xie, X.; Agüero, C.B.; Wang, Y.; Walker, M.A. Genetic Transformation of Grape Varieties and Rootstocks via Organogenesis. Plant Cell. Tissue Organ Cult. 2016, 126, 541–552. [Google Scholar] [CrossRef]
- Li, Z.T.; Dhekney, S.; Dutt, M.; Van Aman, M.; Tattersall, J.; Kelley, K.T.; Gray, D.J. Optimizing Agrobacterium-Mediated Transformation of Grapevine. Vitr. Cell. Dev. Biol.—Plant 2006, 42, 220–227. [Google Scholar] [CrossRef]
- Dhekney, S.A.; Li, Z.T.; Zimmerman, T.W.; Gray, D.J. Factors Influencing Genetic Transformation and Plant Regeneration of Vitis. Am. J. Enol. Vitic. 2009, 60, 285–292. [Google Scholar] [CrossRef]
- Bornhoff, B.A.; Harst, M.; Zyprian, E.; Töpfer, R. Transgenic Plants of Vitis vinifera cv. Seyval Blanc. Plant Cell Rep. 2005, 24, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, P.; Hanania, U.; Sahar, N.; Mawassi, M.; Gafny, R.; Sela, I.; Tanne, E.; Perl, A. Improvement of Agrobacterium-Mediated Transformation Efficiency and Transgenic Plant Regeneration of Vitis vinifera L. by Optimizing Selection Regimes and Utilizing Cryopreserved Cell Suspensions. Plant Sci. 2005, 168, 565–571. [Google Scholar] [CrossRef]
- Ahmed, M.; Khan, N.; Hafiz, I.A.; Abbasi, N.A.; Anjum, M.A.; Hussain, S. Effect of Various Factors on the Efficiency of Agrobacterium-Mediated Transformation of Grape (Vitis vinifera L.). Vegetos 2015, 28, 171–178. [Google Scholar] [CrossRef]
- Iocco, P.; Franks, T.; Thomas, M.R. Genetic Transformation of Major Wine Grape Cultivars of Vitis vinifera L. Transgenic Res. 2001, 10, 105–112. [Google Scholar] [CrossRef]
- Torregrosa, L.; Iocco, P.; Thomas, M.R. Influence of Agrobacterium Strain, Culture Medium, and Cultivar on the Transformation Efficiency of Vitis vinifera L. Am. J. Enol. Vitic. 2002, 53, 183–190. [Google Scholar] [CrossRef]
- Guan, X.; Zhao, H.; Xu, Y.; Wang, Y. Studies on Gene Transfer of Shoot Apical Meristems by Agrobacterium- Mediated Genetic Transformation in a Progeny of Chinese Wild Vitis pseudo reticulata. Vitis—J. Grapevine Res. 2013, 52, 185–192. [Google Scholar]
- Sabbadini, S.; Pandolfini, T.; Girolomini, L.; Molesini, B.; Navacchi, O. Peach (Prunus persica L.) BT—Agrobacterium Protocols: Volume 2; Wang, K., Ed.; Springer: New York, NY, USA, 2015; pp. 205–215. ISBN 978-1-4939-1658-0. [Google Scholar]
- Pérez-Clemente, R.M.; Pérez-Sanjuán, A.; García-Férriz, L.; Beltrán, J.P.; Cañas, L.A. Transgenic Peach Plants (Prunus persica L.) Produced by Genetic Transformation of Embryo Sections Using the Green Fluorescent Protein (GFP) as an In Vivo Marker. Mol. Breed. 2004, 14, 419–427. [Google Scholar] [CrossRef]
- Zong, X.; Xu, L.; Tan, Y.; Wei, H. Development of Genetically Modified Sweet Cherry Rootstock ‘Gisela 6’ with Overexpression of PcMPK3-HA Gene by Agrobacterium-Mediated Genetic Transformation. Plant Cell. Tissue Organ Cult. 2022, 151, 375–384. [Google Scholar] [CrossRef]
- Dolgov, S.V. Genetic Transformation of Sour Cherry (Cerasus vulgaris Mill.). In Transgenic Trees; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 29–38. ISBN 978-3-642-59609-4. [Google Scholar]
- Song, G.Q.; Sink, K.C.; Walworth, A.E.; Cook, M.A.; Allison, R.F.; Lang, G.A. Engineering Cherry Rootstocks with Resistance to Prunus Necrotic Ring Spot Virus through RNAi-Mediated Silencing. Plant Biotechnol. J. 2013, 11, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Song, G.Q.; Sink, K.C. Transformation of Montmorency Sour Cherry (Prunus cerasus L.) and Gisela 6 (P. cerasus × P. canescens) Cherry Rootstock Mediated by Agrobacterium tumefaciens. Plant Cell Rep. 2006, 25, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Sgamma, T.; Thomas, B.; Muleo, R. Ethylene Inhibitor Silver Nitrate Enhances Regeneration and Genetic Transformation of Prunus avium (L.) cv. Stella. Plant Cell. Tissue Organ Cult. 2015, 120, 79–88. [Google Scholar] [CrossRef]
- Tian, L.; Canli, F.A.; Wang, X.; Sibbald, S. Genetic Transformation of Prunus domestica L. Using the Hpt Gene Coding for Hygromycin Resistance as the Selectable Marker. Sci. Hortic. 2009, 119, 339–343. [Google Scholar] [CrossRef]
- Sidorova, T.; Mikhailov, R.; Pushin, A.; Miroshnichenko, D.; Dolgov, S. Agrobacterium-Mediated Transformation of Russian Commercial Plum cv. “Startovaya” (Prunus domestica L.) with Virus-Derived Hairpin Rna Construct Confers Durable Resistance to PPV Infection in Mature Plants. Front. Plant Sci. 2019, 10, 286. [Google Scholar] [CrossRef]
- Urtubia, C.; Devia, J.; Castro, Á.; Zamora, P.; Aguirre, C.; Tapia, E.; Barba, P.; Dell’Orto, P.; Moynihan, M.R.; Petri, C.; et al. Agrobacterium-Mediated Genetic Transformation of Prunus salicina. Plant Cell Rep. 2008, 27, 1333–1340. [Google Scholar] [CrossRef]
- Petri, C.; López-Noguera, S.; Alburquerque, N.; Egea, J.; Burgos, L. An Antibiotic-Based Selection Strategy to Regenerate Transformed Plants from Apricot Leaves with High Efficiency. Plant Sci. 2008, 175, 777–783. [Google Scholar] [CrossRef]
- Mourenets, L.Y.; Pushin, A.S.; Dolgov, S.V. Agrobacterium-Mediated Genetic Transformation of 146-2 Russian Apricot and Plum Rootstock with the Gfp Gene. Acta Hortic. 2020, 1282, 95–100. [Google Scholar] [CrossRef]
- Petri, C.; Wang, H.; Burgos, L.; Sánchez-Navarro, J.; Alburquerque, N. Production of Transgenic Apricot Plants from Hypocotyl Segments of Mature Seeds. Sci. Hortic. 2015, 197, 144–149. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Kaiser, B.N.; Franks, T.; Collins, G.; Sedgley, M. Improved Methods in Agrobacterium-Mediated Transformation of Almond Using Positive (Mannose/Pmi) or Negative (Kanamycin Resistance) Selection-Based Protocols. Plant Cell Rep. 2006, 25, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.; Miguel, C.; Oliveira, M.M. Improved Conditions for Agrobacterium-Mediated Transformation of Almond. Acta Hortic. 2007, 738, 575–582. [Google Scholar] [CrossRef]
- Costa, M.S.; Miguel, C.; Oliveira, M.M. An Improved Selection Strategy and the Use of Acetosyringone in Shoot Induction Medium Increase Almond Transformation Efficiency by 100-Fold. Plant Cell. Tissue Organ Cult. 2006, 85, 205–209. [Google Scholar] [CrossRef]
- Grucha, A.; Zurawicz, E. Conditions of Transformation and Regeneration of ‘Induka’ and ‘Elista’ Strawberry Plants. Plant Cell Tissue Organ Cult. 2004, 79, 153–160. [Google Scholar] [CrossRef]
- Cappelletti, R.; Sabbadini, S.; Mezzetti, B. Strawberry (Fragaria × Ananassa). Methods Mol. Biol. 2015, 1224, 217–227. [Google Scholar] [CrossRef]
- Barceló, M.; El-Mansouri, I.; Mercado, J.A.; Quesada, M.A.; Pliego Alfaro, F. Regeneration and Transformation via Agrobacterium Tumefaciens of the Strawberry Cultivar Chandler. Plant Cell. Tissue Organ Cult. 1998, 54, 29–36. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, Q.; Meng, N.; Song, H.; Li, C.; Hu, G.; Wu, J.; Lin, S.; Zhang, Z. Over-Expression of EjLFY-1 Leads to an Early Flowering Habit in Strawberry (Fragaria × Ananassa) and Its Asexual Progeny. Front. Plant Sci. 2017, 8, 496. [Google Scholar] [CrossRef]
- Pantazis, C.J.; Fisk, S.; Mills, K.; Flinn, B.S.; Shulaev, V.; Veilleux, R.E.; Dan, Y. Development of an Efficient Transformation Method by Agrobacterium tumefaciens and High Throughput Spray Assay to Identify Transgenic Plants for Woodland Strawberry (Fragaria vesca) Using NPTII Selection. Plant Cell Rep. 2013, 32, 329–337. [Google Scholar] [CrossRef]
- Haddadi, F.; Aziz, M.A.; Abdullah, S.N.A.; Tan, S.G.; Kamaladini, H. An Efficient Agrobacterium-Mediated Transformation of Strawberry cv. Camarosa by a Dual Plasmid System. Molecules 2015, 20, 3647–3666. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Q.; Davis, R.E. Transgene Expression in Strawberries Driven by a Heterologous Phloem-Specific Promoter. Plant Cell Rep. 2004, 23, 224–230. [Google Scholar] [CrossRef]
- Monticelli, S.; Gentile, A.; Damiano, C. Regeneration and Agrobacterium-Mediated Transformation in Stipules of Strawberry. Acta Hortic. 2002, 567, 105–107. [Google Scholar] [CrossRef]
- Fitch, M.M.M.; Manshardt, R.M.; Gonsalves, D.; Slightom, J.L. Transgenic Papaya Plants from Agrobacterium-Mediated Transformation of Somatic Embryos. Plant Cell Rep. 1993, 12, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Souza Júnior, M.T.; Nickel, O.; Gonsalves, D. Development of Virus Resistant Transgenic Papayas Expressing the Coat Protein Gene from a Brazilian Isolate of Papaya Ringspot Virus. Fitopatol. Bras. 2005, 30, 357–365. [Google Scholar] [CrossRef]
- Dhekney, S.A.; Litz, R.E.; Moraga Amador, D.A.; Yadav, A.K. Potential for Introducing Cold Tolerance into Papaya by Transformation with C-Repeat Binding Factor (CBF) Genes. Vitr. Cell. Dev. Biol.—Plant 2007, 43, 195–202. [Google Scholar] [CrossRef]
- Litz, R.E. Biotechnology and Mango Improvement. Acta Hortic. 2004, 645, 85–92. [Google Scholar] [CrossRef]
- Mathews, H.; Litz, R.E.; Wilde, H.D.; Merkle, S.A.; Wetzstein, H.Y. Stable Integration and Expression of β-Glucuronidase and NPT II Genes in Mango Somatic Embryos. Vitr.—Plant 1992, 28, 172–178. [Google Scholar] [CrossRef]
- Becker, D.K.; Dugdale, B.; Smith, M.K.; Harding, R.M.; Dale, J.L. Genetic Transformation of Cavendish Banana (Musa spp. AAA Group) cv “Grand Nain” via Microprojectile Bombardment. Plant Cell Rep. 2000, 19, 229–234. [Google Scholar] [CrossRef]
- Shivani; Tiwari, S. Enhanced Agrobacterium-Mediated Transformation Efficiency of Banana Cultivar Grand Naine by Reducing Oxidative Stress. Sci. Hortic. 2019, 246, 675–685. [Google Scholar] [CrossRef]
- Sagi, L.; Panis, B.; Remy, S.; Schoofs, H.; De Smet, K.; Swennen, R.; Cammue, B.P.A. Genetic transformation of banana and plantain (Musa spp.) via particle bombardment. Bio/technology 1995, 13, 481–485. [Google Scholar] [CrossRef]
- Rustagi, A.; Jain, S.; Kumar, D.; Shekhar, S.; Jain, M.; Bhat, V.; Sarin, N.B. High Efficiency Transformation of Banana [Musa acuminata L. cv. Matti (AA)] for Enhanced Tolerance to Salt and Drought Stress Through Overexpression of a Peanut Salinity-Induced Pathogenesis-Related Class 10 Protein. Mol. Biotechnol. 2015, 57, 27–35. [Google Scholar] [CrossRef]
- Tripathi, L.; Tripathi, J.N.; Hughes, J.D.A. Agrobacterium-Mediated Transformation of Plantain (Musa spp.) Cultivar Agbagba. Afr. J. Biotechnol. 2005, 4, 1378–1383. [Google Scholar]
- Ma, J.; He, Y.-H.; Wu, C.-H.; Liu, H.-P.; Hu, Z.-Y.; Sun, G.-M. Effective Agrobacterium mediated Transformation of Pineapple with CYP1A1 by Kanamycin Selection Technique. Afr. J. Biotechnol. 2012, 11, 2555–2562. [Google Scholar] [CrossRef]
- Gangopadhyay, G.; Roy, S.K.; Gangopadhyay, S.B.; Mukherjee, K.K. Agrobacterium-Mediated Genetic Transformation of Pineapple Var. Queen Using a Novel Encapsulation-Based Antibiotic Selection Technique. Plant Cell. Tissue Organ Cult. 2009, 97, 295–302. [Google Scholar] [CrossRef]
- Vardi, A.; Bleichman, S.; Aviv, D. Genetic Transformation of Citrus Protoplasts and Regeneration of Transgenic Plants. Plant Sci. 1990, 69, 199–206. [Google Scholar] [CrossRef]
- Hayford, M.B.; Medford, J.I.; Hoffman, N.L.; Rogers, S.G.; Klee, H.J. Development of a Plant Transformation Selection System Based on Expression of Genes Encoding Gentamicin Acetyltransferases. Plant Physiol. 1988, 86, 1216–1222. [Google Scholar] [CrossRef]
- De Block, M.; Herrera-Estrella, L.; Van Montagu, M.; Schell, J.; Zambryski, P. Expression of Foreign Genes in Regenerated Plants and in Their Progeny. EMBO J. 1984, 3, 1681–1689. [Google Scholar] [CrossRef]
- Gambino, G.; Gribaudo, I. Genetic Transformation of Fruit Trees: Current Status and Remaining Challenges. Transgenic Res. 2012, 21, 1163–1181. [Google Scholar] [CrossRef]
- Wohlleben, W.; Arnold, W.; Broer, I.; Hillemann, D.; Strauch, E.; Pühler, A. Nucleotide Sequence of the Phosphinothricin N-Acetyltransferase Gene from Streptomyces Viridochromogenes Tü494 and Its Expression in Nicotiana tabacum. Gene 1988, 70, 25–37. [Google Scholar] [CrossRef]
- Thompson, C.J.; Rao Moval, N.; Tizard, R.; Crameri, R.; Davies, J.E.; Lauwereys, M.; Botterman, J. Characterization of the Herbicide-resistance Gene Bar from Streptomyces Hygroscopicus. EMBO J. 1987, 6, 2519–2523. [Google Scholar] [CrossRef]
- Lea, P.J.; Miflin, B.J. Glutamate Synthase and the Synthesis of Glutamate in Plants. Plant Physiol. Biochem. 2003, 41, 555–564. [Google Scholar] [CrossRef]
- Szankowski, I.; Briviba, K.; Fleschhut, J.; Schönherr, J.; Jacobsen, H.J.; Kiesecker, H. Transformation of Apple (Malus domestica Borkh.) with the Stilbene Synthase Gene from Grapevine (Vitis vinifera L.) and a PGIP Gene from Kiwi (Actinidia deliciosa). Plant Cell Rep. 2003, 22, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Arcos, Y.; Godoy, F.; Flores-Ortiz, C.; Arenas-M, A.; Stange, C. Boosting Carotenoid Content in Malus Domestica Var. Fuji by Expressing AtDXR through an Agrobacterium-Mediated Transformation Method. Biotechnol. Bioeng. 2020, 117, 2209–2222. [Google Scholar] [CrossRef] [PubMed]
- Jardak, R.; Boubakri, H.; Zemni, H.; Gandoura, S.; Mejri, S.; Mliki, A.; Ghorbel, A. Establishment of an In Vitro Regeneration System and Genetic Transformation of the Tunisian “Maltese Half-Blood” (Citrus sinensis): An Agro-Economically Important Variety. 3 Biotech 2020, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Sripaoraya, S.; Marchant, R.; Power, J.B.; Davey, M.R. Herbicide-Tolerant Transgenic Pineapple (Ananas Comosus) Produced by Microprojectile Bombardment. Ann. Bot. 2001, 88, 597–603. [Google Scholar] [CrossRef]
- Espinosa, P.; Lorenzo, J.C.; Iglesias, A.; Yabor, L.; Menéndez, E.; Borroto, J.; Hernández, L.; Arencibia, A.D. Production of Pineapple Transgenic Plants Assisted by Temporary Immersion Bioreactors. Plant Cell Rep. 2002, 21, 136–140. [Google Scholar] [CrossRef]
- Steinrücken, H.C.; Amrhein, N. The Herbicide Glyphosate Is a Potent Inhibitor of 5-Enolpyruvylshikimic Acid-3-Phosphate Synthase. Biochem. Biophys. Res. Commun. 1980, 94, 1207–1212. [Google Scholar] [CrossRef]
- Heap, I.; Duke, S.O. Overview of Glyphosate-Resistant Weeds Worldwide. Pest Manag. Sci. 2018, 74, 1040–1049. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Folta, K.M. Assessment of Promoters and a Selectable Marker for Development of Strawberry Intragenic Vectors. Plant Cell. Tissue Organ Cult. 2017, 128, 259–271. [Google Scholar] [CrossRef]
- Merritt, B.A.; Zhang, X.; Triplett, E.W.; Mou, Z.; Orbović, V. Selection of Transgenic Citrus Plants Based on Glyphosate Tolerance Conferred by a Citrus 5-Enolpyruvylshikimate-3-Phosphate Synthase Variant. Plant Cell Rep. 2021, 40, 1947–1956. [Google Scholar] [CrossRef]
- Yao, J.-L.; Tomes, S.; Gleave, A.P. Transformation of Apple (Malus Domestica) Using Mutants of Apple Acetolactate Synthase as a Selectable Marker and Analysis of the T-DNA Integration Sites. Plant Cell Rep. 2013, 32, 703–714. [Google Scholar] [CrossRef]
- Wuddineh, W.A.; Xu, X.; Zhong, G.-Y. Amino Acid Substitutions in Grapevine (Vitis vinifera) Acetolactate Synthase Conferring Herbicide Resistance. Plant Cell. Tissue Organ Cult. 2023, 154, 75–87. [Google Scholar] [CrossRef]
- Malabarba, J.; Chevreau, E.; Dousset, N.; Veillet, F.; Moizan, J.; Vergne, E. New Strategies to Overcome Present CRISPR/Cas9 Limitations in Apple and Pear: Efficient Dechimerization and Base Editing. Int. J. Mol. Sci. 2021, 22, 319. [Google Scholar] [CrossRef] [PubMed]
- Alquézar, B.; Bennici, S.; Carmona, L.; Gentile, A.; Peña, L. Generation of Transfer-DNA-Free Base-Edited Citrus Plants. Front. Plant Sci. 2022, 13, 835282. [Google Scholar] [CrossRef] [PubMed]
- Wehrmann, A.; Van Vliet, A.; Opsomer, C.; Botterman, J.; Schulz, A. The Similarities of Bar and Pat Gene Products Make Them Equally Applicable for Plant Engineers. Nat. Biotechnol. 1996, 14, 1274–1278. [Google Scholar] [CrossRef]
- Schütte, G.; Eckerstorfer, M.; Rastelli, V.; Reichenbecher, W.; Vassalli, S.R.; Lehto, M.R.; Gabrielle, A.; Saucy, W.; Mertens, M. Herbicide Resistance and Biodiversity: Agronomic and Environmental Aspects of Genetically Modified Herbicide—Resistant Plants. Environ. Sci. Eur. 2017, 29, 1–12. [Google Scholar] [CrossRef]
- Miles, J.S.; Guest, J.R. Nucleotide Sequence and Transcriptional Start Point of the Phosphomannose Isomerase Gene (ManA) of Escherichia coli. Gene 1984, 32, 41–48. [Google Scholar] [CrossRef]
- Reed, J.; Privalle, L.; Powell, M.L.; Meghji, M.; Dawson, J.; Dunder, E.; Suttie, J.; Wenck, A.; Launis, K.; Kramer, C.; et al. Phosphomannose Isomerase: An Efficient Selectable Marker for Plant Transformation. Vitr. Cell. Dev. Biol.—Plant 2001, 37, 127–132. [Google Scholar] [CrossRef]
- Degenhardt, J.; Poppe, A.; Montag, J.; Szankowski, I. The Use of the Phosphomannose-Isomerase/Mannose Selection System to Recover Transgenic Apple Plants. Plant Cell Rep. 2006, 25, 1149–1156. [Google Scholar] [CrossRef]
- Boscariol, R.L.; Almeida, W.A.B.; Derbyshire, M.T.V.C.; Mourão Filho, F.A.A.; Mendes, B.M.J. The Use of the PMI/Mannose Selection System to Recover Transgenic Sweet Orange Plants (Citrus sinensis L. Osbeck). Plant Cell Rep. 2003, 22, 122–128. [Google Scholar] [CrossRef]
- Ballester, A.; Cervera, M.; Peña, L. Evaluation of Selection Strategies Alternative to NptII in Genetic Transformation of Citrus. Plant Cell Rep. 2008, 27, 1005–1015. [Google Scholar] [CrossRef]
- Wu, H.; Acanda, Y.; Canton, M.; Zale, J. Efficient Biolistic Transformation of Immature Citrus Rootstocks Using Phosphomannose-Isomerase Selection. Plants 2019, 8, 390. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Canton, M.; Mahmoud, L.M.; Weber, K.R.; Michalczyk, G.Z.; Dutt, M.; Zale, J.M. Identification and Characterization of Two Putative Citrus Phosphomannose Isomerase (CsPMI) Genes as Selectable Markers for Mature Citrus Transformation. Horticulturae 2022, 8, 204. [Google Scholar] [CrossRef]
- Wang, H.; Petri, C.; Burgos, L.; Alburquerque, N. Phosphomannose-Isomerase as a Selectable Marker for Transgenic Plum (Prunus domestica L.). Plant Cell. Tissue Organ Cult. 2013, 113, 189–197. [Google Scholar] [CrossRef]
- Sidorova, T.; Mikhailov, R.; Pushin, A.; Miroshnichenko, D.; Dolgov, S. A Non-Antibiotic Selection Strategy Uses the Phosphomannose-Isomerase (PMI) Gene and Green Fluorescent Protein (GFP) Gene for Agrobacterium-Mediated Transformation of Prunus domestica L. Leaf Explants. Plant Cell. Tissue Organ Cult. 2017, 128, 197–209. [Google Scholar] [CrossRef]
- Miki, B.; McHugh, S. Selectable Marker Genes in Transgenic Plants: Applications, Alternatives and Biosafety. J. Biotechnol. 2004, 107, 193–232. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Agbayani, R.; McCafferty, H.; Albert, H.H.; Moore, P.H. Effective Selection of Transgenic Papaya Plants with the PMI/Man Selection System. Plant Cell Rep. 2005, 24, 426–432. [Google Scholar] [CrossRef]
- Haldrup, A.; Petersen, S.G.; Okkels, F.T. Positive Selection: A Plant Selection Principle Based on Xylose Isomerase, an Enzyme Used in the Food Industry. Plant Cell Rep. 1998, 18, 76–81. [Google Scholar] [CrossRef]
- Haldrup, A.; Petersen, S.G.; Okkels, F.T. The Xylose Isomerase Gene from Thermoanaerobacterium Thermosulfurogenes Allows Effective Selection of Transgenic Plant Cells Using D-Xylose as the Selection Agent. Plant Mol. Biol. 1998, 37, 287–296. [Google Scholar] [CrossRef]
- Zhao, H.; Fan, W.; Zhu, Y.; Zeng, H.; Huang, T.; Peng, M. Study on Sensitivity of Banana (Musa spp.) to D-Mannose and D-Xylose. J. Fruit Sci. 2010, 27, 233–237. [Google Scholar]
- Gordon-Kamm, B.; Sardesai, N.; Arling, M.; Lowe, K.; Hoerster, G.; Betts, S.; Jones, T. Using Morphogenic Genes to Improve Recovery and Regeneration of Transgenic Plants. Plants 2019, 8, 38. [Google Scholar] [CrossRef]
- Endo, S.; Kasahara, T.; Sugita, K.; Matsunaga, E.; Ebinuma, H. The Isopentenyl Transferase Gene Is Effective as a Selectable Marker Gene for Plant Transformation in Tobacco (Nicotiana tabacum cv. Petite Havana SRI). Plant Cell Rep. 2001, 20, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.; Cervera, M.; Peña, L. Efficient Production of Transgenic Citrus Plants Using Isopentenyl Transferase Positive Selection and Removal of the Marker Gene by Site-Specific Recombination. Plant Cell Rep. 2007, 26, 39–45. [Google Scholar] [CrossRef] [PubMed]
- López-Noguera, S.; Petri, C.; Olmos, E.; Burgos, L. Regeneration-Promoting Genes Improve Transformation Efficiency in Apricot. Acta Hortic. 2006, 725, 95–99. [Google Scholar] [CrossRef]
- Peña, L.; Cervera, M.; Fagoaga, C.; Pérez, R.; Romero, J.; Juárez, J.; Pina, J.A.; Navarro, L. Agrobacterum-Mediated Transformation of Citrus. Transgenic Crop. World 2004, 145–156. [Google Scholar] [CrossRef]
- Petri, C.; Wang, H.; Alburquerque, N.; Faize, M.; Burgos, L. Agrobacterium-Mediated Transformation of Apricot (Prunus armeniaca L.) Leaf Explants. Plant Cell Rep. 2008, 27, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muino, J.M.; Angenent, G.C.; Boutilier, K. The BABY BOOM Transcription Factor Activates the LEC1-ABI3-FUS3-LEC2 Network to Induce Somatic Embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef]
- Xiao, X.; Zhang, C.; Liu, Y.; Wang, X.; You, C. Functional Identification of Apple Baby Boom in Genetic Transformation and Somatic Embryogenesis. Vitr. Cell. Dev. Biol.—Plant 2023, 59, 1–13. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS Fusions: Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher Plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Schaart, J.G.; Salentijn, E.M.J.; Krens, F.A. Tissue-Specific Expression of the β-Glucuronidase Reporter Gene in Transgenic Strawberry (Fragaria × Ananassa) Plants. Plant Cell Rep. 2002, 21, 313–319. [Google Scholar] [CrossRef]
- Feng, J.; Dai, C.; Luo, H.; Han, Y.; Liu, Z.; Kang, C. Reporter Gene Expression Reveals Precise Auxin Synthesis Sites during Fruit and Root Development in Wild Strawberry. J. Exp. Bot. 2019, 70, 563–574. [Google Scholar] [CrossRef]
- Gago, J.; Grima-Pettenati, J.; Gallego, P.P. Vascular-Specific Expression of GUS and GFP Reporter Genes in Transgenic Grapevine (Vitis vinifera L. cv. Albariño) Conferred by the EgCCR Promoter of Eucalyptus Gunnii. Plant Physiol. Biochem. 2011, 49, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Kandel, R.; Bergey, D.R.; Dutt, M.; Sitther, V.; Li, Z.T.; Gray, D.J.; Dhekney, S.A. Evaluation of a Grapevine-Derived Reporter Gene System for Precision Breeding of Vitis. Plant Cell. Tissue Organ Cult. 2016, 124, 599–609. [Google Scholar] [CrossRef]
- Benyon, L.S.; Stover, E.; Bowman, K.D.; Niedz, R.; Shatters, R.G.; Zale, J.; Belknap, W. GUS Expression Driven by Constitutive and Phloem-Specific Promoters in Citrus Hybrid US-802. Vitr. Cell. Dev. Biol.—Plant 2013, 49, 255–265. [Google Scholar] [CrossRef]
- Miyata, L.Y.; Harakava, R.; Stipp, L.C.L.; Mendes, B.M.J.; Appezzato-da-Glória, B.; de Assis Alves Mourão Filho, F. GUS Expression in Sweet Oranges (Citrus sinensis L. Osbeck) Driven by Three Different Phloem-Specific Promoters. Plant Cell Rep. 2012, 31, 2005–2013. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Yu, T.A.; Cheng, Y.H.; Yeh, S.D. Transgenic Papaya Plants from Agrobacterium-Mediated Transformation of Petioles of In Vitro Propagated Multishoots. Plant Cell Rep. 1996, 15, 459–464. [Google Scholar] [CrossRef]
- Cao, X.; Liu, Q.; Rowland, L.J.; Hammerschlag, F.A. GUS Expression in Blueberry (Vaccinium spp.): Factors Influencing Agrobacterium-Mediated Gene Transfer Efficiency. Plant Cell Rep. 1998, 18, 266–270. [Google Scholar] [CrossRef]
- Padilla, I.M.G.; Golis, A.; Gentile, A.; Damiano, C.; Scorza, R. Evaluation of Transformation in Peach Prunus Persica Explants Using Green Fluorescent Protein (GFP) and Beta-Glucuronidase (GUS) Reporter Genes. Plant Cell Tissue Organ Cult. 2006, 84, 309–314. [Google Scholar] [CrossRef]
- Ow, D.W.; Wood, K.V.; DeLuca, M.; De Wet, J.R.; Helinski, D.R.; Howell, S.H. Transient and Stable Expression of the Firefly Luciferase Gene in Plant Cells and Transgenic Plants. Science 1986, 234, 856–859. [Google Scholar] [CrossRef]
- Chang, J.R.; Geider, K. The Use of Luciferase as a Reporter for Response of Plant Cells to the Fireblight Pathogen Erwinia amylovora. Plant Cell Rep. 1995, 14, 497–500. [Google Scholar] [CrossRef]
- Spolaore, S.; Trainotti, L.; Casadoro, G. A Simple Protocol for Transient Gene Expression in Ripe Fleshy Fruit Mediated by Agrobacterium. J. Exp. Bot. 2001, 52, 845–850. [Google Scholar] [CrossRef]
- Villao, L.; Sánchez, E.; Romero, C.; Galarza, L.; Flores, J.; Santos-Ordóñez, E. Activity Characterization of the Plantain Promoter from the Heavy Metal-Associated Isoprenylated Plant Gene (MabHIPP) Using the Luciferase Reporter Gene. Plant Gene 2019, 19, 100187. [Google Scholar] [CrossRef]
- Ahlandsberg, S.; Sathish, P.; Sun, C.; Jansson, C. Green Fluorescent Protein as a Reporter System in the Transformation of Barley Cultivars. Physiol. Plant. 1999, 107, 194–200. [Google Scholar] [CrossRef]
- Donaldson, L. Autofluorescence in Plants. Molecules 2020, 25, 2393. [Google Scholar] [CrossRef] [PubMed]
- Sen, T.; Mamontova, A.V.; Titelmayer, A.V.; Shakhov, A.M.; Astafiev, A.A.; Acharya, A.; Lukyanov, K.A.; Krylov, A.I.; Bogdanov, A.M. Influence of the First Chromophore-Forming Residue on Photobleaching and Oxidative Photoconversion of EGFP and EYFP. Int. J. Mol. Sci. 2019, 20, 5229. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Lu, H.; Tang, D.; Hassan, M.M.; Li, Y.; Chen, J.-G.; Tuskan, G.A.; Yang, X. Expanding the Application of a UV-Visible Reporter for Transient Gene Expression and Stable Transformation in Plants. Hortic. Res. 2021, 8, 234. [Google Scholar] [CrossRef]
- Howard, A.R.; Heppler, M.L.; Ju, H.-J.; Krishnamurthy, K.; Payton, M.E.; Verchot-Lubicz, J. Potato Virus X TGBp1 Induces Plasmodesmata Gating and Moves between Cells in Several Host Species Whereas CP Moves Only in N. Benthamiana Leaves. Virology 2004, 328, 185–197. [Google Scholar] [CrossRef]
- Capriotti, L.; Ricci, A.; Molesini, B.; Mezzetti, B.; Pandolfini, T.; Piunti, I.; Sabbadini, S. Efficient Protocol of de Novo Shoot Organogenesis from Somatic Embryos for Grapevine Genetic Transformation. Front. Plant Sci. 2023, 14, 1172758. [Google Scholar] [CrossRef]
- Sabbadini, S.; Ricci, A.; Limera, C.; Baldoni, D.; Capriotti, L.; Mezzetti, B. Factors Affecting the Regeneration, via Organogenesis, and the Selection of Transgenic Calli in the Peach Rootstock Hansen 536 (Prunus persica × Prunus amygdalus) to Express an RNAi Construct against PPV Virus. Plants 2019, 8, 178. [Google Scholar] [CrossRef]
- Ricci, A.; Sabbadini, S.; Prieto, H.; Padilla, I.M.G.; Dardick, C.; Li, Z.; Scorza, R.; Limera, C.; Mezzetti, B.; Perez-Jimenez, M.; et al. Genetic Transformation in Peach (Prunus persica L.): Challenges and Ways Forward. Plants 2020, 9, 971. [Google Scholar] [CrossRef]
- Elomaa, P.; Uimari, A.; Mehto, M.; Albert, V.A.; Laitinen, R.A.E.; Teeri, T.H. Activation of Anthocyanin Biosynthesis in Gerbera Hybrida (Asteraceae) Suggests Conserved Protein-Protein and Protein-Promoter Interactions between the Anciently Diverged Monocots and Eudicots. Plant Physiol. 2003, 133, 1831–1842. [Google Scholar] [CrossRef]
- Polturak, G.; Breitel, D.; Grossman, N.; Sarrion-Perdigones, A.; Weithorn, E.; Pliner, M.; Orzaez, D.; Granell, A.; Rogachev, I.; Aharoni, A. Elucidation of the First Committed Step in Betalain Biosynthesis Enables the Heterologous Engineering of Betalain Pigments in Plants. New Phytol. 2016, 210, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Krens, F.A.; Schaart, J.G.; van der Burgh, A.M.; TinnenbroekCapel, I.E.M.; Groenwold, R.; Kodde, L.P.; Broggini, G.A.L.; Gessler, C.; Schouten, H.J. Cisgenic Apple Trees; Development, Characterization, and Performance. Front. Plant Sci. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed]
- Kortstee, A.J.; Khan, S.A.; Helderman, C.; Trindade, L.M.; Wu, Y.; Visser, R.G.F.; Brendolise, C.; Allan, A.; Schouten, H.J.; Jacobsen, E. Anthocyanin Production as a Potential Visual Selection Marker during Plant Transformation. Transgenic Res. 2011, 20, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Salazar-González, J.A.; Castro-Medina, M.; Bernardino-Rivera, L.E.; Martínez-Terrazas, E.; Casson, S.A.; Urrea-López, R. In-Planta Transient Transformation of Avocado (Persea Americana) by Vacuum Agroinfiltration of Aerial Plant Parts. Plant Cell. Tissue Organ Cult. 2023, 152, 635–646. [Google Scholar] [CrossRef]
- Zhang, Y.; Patankar, H.; Aljedaani, F.; Blilou, I. A Framework for Date Palm (Phoenix dactylifera L.) Tissue Regeneration and Stable Transformation. Physiol. Plant. 2024, 176, e14189. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, R.; Liu, X.; Zhao, X.; Hyden, B.; Han, Y.; Zhang, X.; Wang, J.; Chen, H. Establishment of Genetic Transformation System of Peach Callus. Sci. Hortic. 2024, 323, 112501. [Google Scholar] [CrossRef]
- Rosellini, D. Selectable Markers and Reporter Genes: A Well Furnished Toolbox for Plant Science and Genetic Engineering. CRC. Crit. Rev. Plant Sci. 2012, 31, 401–453. [Google Scholar] [CrossRef]
- Sabbadini, S.; Capocasa, F.; Battino, M.; Mazzoni, L.; Mezzetti, B. Improved Nutritional Quality in Fruit Tree Species through Traditional and Biotechnological Approaches. Trends Food Sci. Technol. 2021, 117, 125–138. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Kanchiswamy, C.N. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Ballester, A.; Cervera, M.; Peña, L. Selectable Marker-Free Transgenic Orange Plants Recovered under Non-Selective Conditions and through PCR Analysis of All Regenerants. Plant Cell. Tissue Organ Cult. 2010, 102, 329–336. [Google Scholar] [CrossRef]
- Petri, C.; Hily, J.-M.; Vann, C.; Dardick, C.; Scorza, R. A High-Throughput Transformation System Allows the Regeneration of Marker-Free Plum Plants (Prunus domestica). Ann. Appl. Biol. 2011, 159, 302–315. [Google Scholar] [CrossRef]
- Mmbando, G.S. Recent Advances in Antibiotic-Free Markers; Novel Technologies to Enhance Safe Human Food Production in the World. Mol. Biotechnol. 2022, 65, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Dutt, M.; Li, Z.T.; Dhekney, S.A.; Gray, D.J. A Co-Transformation System to Produce Transgenic Grapevines Free of Marker Genes. Plant Sci. 2008, 175, 423–430. [Google Scholar] [CrossRef]
- Stougaard, J. Substrate-Dependent Negative Selection in Plants Using a Bacterial Cytosine Deaminase Gene. Plant J. 1993, 3, 755–761. [Google Scholar] [CrossRef]
- Dale, E.C.; Ow, D.W. Intra- and Intramolecular Site-Specific Recombination in Plant Cells Mediated by Bacteriophage P1 Recombinase. Gene 1990, 91, 79–85. [Google Scholar] [CrossRef]
- Petri, C.; López-Noguera, S.; Wang, H.; García-Almodóvar, C.; Alburquerque, N.; Burgos, L. A Chemical-Inducible Cre-LoxP System Allows for Elimination of Selection Marker Genes in Transgenic Apricot. Plant Cell. Tissue Organ Cult. 2012, 110, 337–346. [Google Scholar] [CrossRef]
- Chong-Pérez, B.; Kosky, R.G.; Reyes, M.; Rojas, L.; Ocaña, B.; Tejeda, M.; Pérez, B.; Angenon, G. Heat Shock Induced Excision of Selectable Marker Genes in Transgenic Banana by the Cre-Lox Site-Specific Recombination System. J. Biotechnol. 2012, 159, 265–273. [Google Scholar] [CrossRef]
- Chong-Pérez, B.; Reyes, M.; Rojas, L.; Ocaña, B.; Ramos, A.; Kosky, R.G.; Angenon, G. Excision of a Selectable Marker Gene in Transgenic Banana Using a Cre/Lox System Controlled by an Embryo Specific Promoter. Plant Mol. Biol. 2013, 83, 143–152. [Google Scholar] [CrossRef]
- Peng, A.; Xu, L.; He, Y.; Lei, T.; Yao, L.; Chen, S.; Zou, X. Efficient Production of Marker-Free Transgenic ‘Tarocco’ Blood Orange (Citrus sinensis Osbeck) with Enhanced Resistance to Citrus Canker Using a Cre/LoxP Site-Recombination System. Plant Cell. Tissue Organ Cult. 2015, 123, 1–13. [Google Scholar] [CrossRef]
- Peng, A.; Zhang, J.; Zou, X.; He, Y.; Xu, L.; Lei, T.; Yao, L.; Li, Q.; Chen, S. Pyramiding the Antimicrobial PR1aCB and AATCB Genes in ‘Tarocco’ Blood Orange (Citrus sinensis Osbeck) to Enhance Citrus Canker Resistance. Transgenic Res. 2021, 30, 635–647. [Google Scholar] [CrossRef]
- He, Y.; Xu, L.; Peng, A.; Lei, T.; Li, Q.; Yao, L.; Jiang, G.; Chen, S.; Li, Z.; Zou, X. Production of Marker-Free Transgenic Plants from Mature Tissues of Navel Orange Using a Cre/LoxP Site-Recombination System. Hortic. Plant J. 2023, 9, 473–480. [Google Scholar] [CrossRef]
- Sadowski, P.D. The Flp Recombinase of Th 2-Μm Plasmid of Saccharomyces Cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 1995, 51, 53–91. [Google Scholar] [CrossRef] [PubMed]
- Herzog, K.; Flachowsky, H.; Deising, H.B.; Hanke, M.V. Heat-Shock-Mediated Elimination of the NptII Marker Gene in Transgenic Apple (Malus × domestica Borkh.). Gene 2012, 498, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Würdig, J.; Flachowsky, H.; Hanke, M.V. Studies on Heat Shock Induction and Transgene Expression in Order to Optimize the Flp/FRT Recombinase System in Apple (Malus × domestica Borkh.). Plant Cell. Tissue Organ Cult. 2013, 115, 457–467. [Google Scholar] [CrossRef]
- Würdig, J.; Flachowsky, H.; Saß, A.; Peil, A.; Hanke, M.V. Improving Resistance of Different Apple Cultivars Using the Rvi6 Scab Resistance Gene in a Cisgenic Approach Based on the Flp/FRT Recombinase System. Mol. Breed. 2015, 35, 1–18. [Google Scholar] [CrossRef]
- Kost, T.D.; Gessler, C.; Jänsch, M.; Flachowsky, H.; Patocchi, A.; Broggini, G.A.L. Development of the First Cisgenic Apple with Increased Resistance to Fire Blight. PLoS ONE 2015, 10, e0143980. [Google Scholar] [CrossRef]
- Dalla Costa, L.; Piazza, S.; Campa, M.; Flachowsky, H.; Hanke, M.V.; Malnoy, M. Efficient Heat-Shock Removal of the Selectable Marker Gene in Genetically Modified Grapevine. Plant Cell. Tissue Organ Cult. 2016, 124, 471–481. [Google Scholar] [CrossRef]
- Araki, H.; Jearnpipatkul, A.; Tatsumi, H.; Sakurai, T.; Ushio, K.; Muta, T.; Oshima, Y. Molecular and Functional Organization of Yeast Plasmid PSR1. J. Mol. Biol. 1985, 182, 191–203. [Google Scholar] [CrossRef]
- Schaart, J.G.; Krens, F.A.; Pelgrom, K.T.B.; Mendes, O.; Rouwendal, G.J.A. Effective Production of Marker-Free Transgenic Strawberry Plants Using Inducible Site-Specific Recombination and a Bifunctional Selectable Marker Gene. Plant Biotechnol. J. 2004, 2, 233–240. [Google Scholar] [CrossRef]
- Vanblaere, T.; Szankowski, I.; Schaart, J.; Schouten, H.; Flachowsky, H.; Broggini, G.A.L.; Gessler, C. The Development of a Cisgenic Apple Plant. J. Biotechnol. 2011, 154, 304–311. [Google Scholar] [CrossRef]
- Vanblaere, T.; Flachowsky, H.; Gessler, C.; Broggini, G.A.L. Molecular Characterization of Cisgenic Lines of Apple “Gala” Carrying the Rvi6 Scab Resistance Gene. Plant Biotechnol. J. 2014, 12, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.G.; Schaart, J.G.; Groenwold, R.; Jacobsen, E.; Schouten, H.J.; Krens, F.A. Functional Analysis and Expression Profiling of HcrVf1 and HcrVf2 for Development of Scab Resistant Cisgenic and Intragenic Apples. Plant Mol. Biol. 2011, 75, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Righetti, L.; Djennane, S.; Berthelot, P.; Cournol, R.; Wilmot, N.; Loridon, K.; Vergne, E.; Chevreau, E. Elimination of the NptII Marker Gene in Transgenic Apple and Pear with a Chemically Inducible R/Rs Recombinase. Plant Cell. Tissue Organ Cult. 2014, 117, 335–348. [Google Scholar] [CrossRef]
- Kleidon, J.; Brinin, A.; Paul, J.Y.; Harding, R.; Dale, J.; Dugdale, B. Production of Selectable Marker Gene-Free Cavendish Banana (Musa spp.) Using a Steroid-Inducible Recombinase Platform. Transgenic Res. 2020, 29, 81–93. [Google Scholar] [CrossRef] [PubMed]
- López-Noguera, S.; Petri, C.; Burgos, L. Combining a Regeneration-Promoting Ipt Gene and Site-Specific Recombination Allows a More Efficient Apricot Transformation and the Elimination of Marker Genes. Plant Cell Rep. 2009, 28, 1781–1790. [Google Scholar] [CrossRef]
- Zou, X.; Peng, A.; Xu, L.; Liu, X.; Lei, T.; Yao, L.; He, Y.; Chen, S. Efficient Auto-Excision of a Selectable Marker Gene from Transgenic Citrus by Combining the Cre/LoxP System and Ipt Selection. Plant Cell Rep. 2013, 32, 1601–1613. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Campa, M.; Miranda, S.; Licciardello, C.; Lashbrooke, J.G.; Dalla Costa, L.; Guan, Q.; Spök, A.; Malnoy, M. Application of New Breeding Techniques in Fruit Trees. Plant Physiol. 2024, 194, 1304–1322. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Dalla Costa, L.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to Produce T-DNA Free CRISPRed Fruit Trees via Agrobacterium Tumefaciens Stable Gene Transfer. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Pompili, V.; Dalla Costa, L.; Piazza, S.; Pindo, M.; Malnoy, M. Reduced Fire Blight Susceptibility in Apple Cultivars Using a High-Efficiency CRISPR/Cas9-FLP/FRT-Based Gene Editing System. Plant Biotechnol. J. 2020, 18, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Mezzetti, B.; Navacchi, O.; Sabbadini, S. In Vitro Shoot Regeneration from Leaves of Pyrus communis L. Rootstock and Cultivars. Plant Biotechnol. Rep. 2023, 17, 341–352. [Google Scholar] [CrossRef]
- Ricci, A.; Sabbadini, S.; Navacchi, O.; Burgos, L.; Mezzetti, B. Approaches Applied to Induce Regeneration, via Somatic Embryogenesis, in the Peach Rootstock ‘GF677’ (P. persica × P. amygdalus) and in Different Peach Cultivars Using Leaves and Anthers as Starting Explants. Acta Hortic. 2023, 1359, 241–247. [Google Scholar] [CrossRef]
- Gambino, G.; Nuzzo, F.; Moine, A.; Chitarra, W.; Pagliarani, C.; Petrelli, A.; Boccacci, P.; Delliri, A.; Velasco, R.; Nerva, L.; et al. Genome Editing of a Recalcitrant Wine Grape Genotype by Lipofectamine-Mediated Delivery of CRISPR/Cas9 Ribonucleoproteins to Protoplasts. Plant J. 2024, 119, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Bertini, E.; D’Incà, E.; Fasoli, M.; Zenoni, S. DNA-Free Genome Editing in Grapevine Using CRISPR/Cas9 Ribonucleoprotein Complexes Followed by Protoplast Regeneration. Hortic. Res. 2023, 10, uhac240. [Google Scholar] [CrossRef]
- Pavese, V.; Moglia, A.; Abbà, S.; Milani, A.; Torello Marinoni, D.; Corredoira, E.; Martìnez, M.; Botta, R. First Report on Genome Editing via Ribonucleoprotein (RNP) in Castanea sativa Mill. Int. J. Mol. Sci. 2022, 23, 5762. [Google Scholar] [CrossRef]
- Nerva, L.; Costa, L.D.; Ciacciulli, A.; Sabbadini, S.; Pavese, V.; Dondini, L.; Vendramin, E.; Caboni, E.; Perrone, I.; Moglia, A.; et al. The Role of Italy in the Use of Advanced Plant Genomic Techniques on Fruit Trees: State of the Art and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 977. [Google Scholar] [CrossRef]
- Liu, G.; Li, H.; Fu, D. Applications of Virus-Induced Gene Silencing for Identification of Gene Function in Fruit. Food Qual. Saf. 2021, 5, fyab018. [Google Scholar] [CrossRef]
- Shan-E-Ali Zaidi, S.; Mansoor, S. Viral Vectors for Plant Genome Engineering. Front. Plant Sci. 2017, 8, 539. [Google Scholar] [CrossRef]
- Kim, D.Y.; Lee, J.M.; Moon, S.B.; Chin, H.J.; Park, S.; Lim, Y.; Kim, D.; Koo, T.; Ko, J.H.; Kim, Y.S. Efficient CRISPR Editing with a Hypercompact Cas12f1 and Engineered Guide RNAs Delivered by Adeno-Associated Virus. Nat. Biotechnol. 2022, 40, 94–102. [Google Scholar] [CrossRef]
- Karvelis, T.; Druteika, G.; Bigelyte, G.; Budre, K.; Zedaveinyte, R.; Silanskas, A.; Kazlauskas, D.; Venclovas, C.; Siksnys, V. Transposon-Associated TnpB Is a Programmable RNA-Guided DNA Endonuclease. Nature 2021, 599, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Altae-Tran, H.; Kannan, S.; Demircioglu, F.E.; Oshiro, R.; Nety, S.P.; McKay, L.J.; Dlakić, M.; Inskeep, W.P.; Makarova, K.S.; Macrae, R.K.; et al. The Widespread IS200/IS605 Transposon Family Encodes Diverse Programmable RNA-Guided Endonucleases. Science 2021, 374, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.R.; Cui, Z.H.; Li, J.W.; Hao, X.Y.; Zhao, L.; Wang, Q.C. In Vitro Thermotherapy-Based Methods for Plant Virus Eradication. Plant Methods 2018, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Hensel, G.; Malnoy, M.; Mach, F.E.; Stoger, E.; Sprink, T.; Metje-Sprink, J.; Menz, J.; Modrzejewski, D. DNA-Free Genome Editing: Past, Present and Future. Front. Plant Sci. 2019, 9, 1957. [Google Scholar] [CrossRef]
- Philips, J.G.; Martin-Avila, E.; Robold, A.V. Horizontal Gene Transfer from Genetically Modified Plants—Regulatory Considerations. Front. Bioeng. Biotechnol. 2022, 10, 971402. [Google Scholar] [CrossRef]
- Elsas, J.D.; Bailey, M.J. The Ecology of Transfer of Mobile Genetic Elements. FEMS Microbiol. Ecol. 2006, 42, 187–197. [Google Scholar] [CrossRef]
- Pontiroli, A.; Simonet, P.; Frostegard, A.; Vogel, T.M.; Monier, J.-M. Fate of Transgenic Plant DNA in the Environment. Environ. Biosafety Res. 2007, 6, 15–35. [Google Scholar] [CrossRef]
- Fuchs, R.L.; Ream, J.E.; Hammond, B.G.; Naylor, M.W.; Leimgruber, R.M.; Berberich, S.A. Safety Assessment of the Neomycin Phosphotransferase II (NPTII) Protein. Nat. Biotechnol. 1993, 11, 1543–1547. [Google Scholar] [CrossRef]
- Hily, J.M.; Demanèche, S.; Poulicard, N.; Tannières, M.; Djennane, S.; Beuve, M.; Vigne, E.; Demangeat, G.; Komar, V.; Gertz, C.; et al. Metagenomic-Based Impact Study of Transgenic Grapevine Rootstock on Its Associated Virome and Soil Bacteriome. Plant Biotechnol. J. 2018, 16, 208–220. [Google Scholar] [CrossRef]
- EC Commission. Implementing Regulation (EU) No 503/2013 of 3 April 2013 on Applications for Authorisation of Genetically Modified Food and Feed in Accordance with Regulation (EC) No 1829/2003 of the European Parliament and of the Council and Amending Commi. Off. J. Eur. Union 2013, L157, 1–48. [Google Scholar]
- FDA. Guidance for Industry: Use of Antibiotic Resistance Marker Genes in Transgenic Plants; U.S. Food and Drug Administration: Silver Spring, MD, USA, 1998.
- Messeguer, J. Gene Flow Assessment in Transgenic Plants. Plant Cell. Tissue Organ Cult. 2003, 73, 201–212. [Google Scholar] [CrossRef]
- Clark, M.; Maselko, M. Transgene Biocontainment Strategies for Molecular Farming. Front. Plant Sci. 2020, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Shukla, A.; Thakur, S.; Rathore, M.; Singh, N.P. Estimation of Neomycin Phosphotransferase-II (NPT-II) Protein in Vegetative and Reproductive Tissues of Transgenic Chickpea (Cicer arietinum L.) and Biosafety Perspectives. J. Plant Biochem. Biotechnol. 2020, 29, 568–570. [Google Scholar] [CrossRef]
- Siegrist, M.; Hartmann, C. Consumer Acceptance of Novel Food Technologies. Nat. Food 2020, 1, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A.K. Global Regulation of Genetically Modified Crops amid the Gene Edited Crop Boom–a Review. Front. Plant Sci. 2021, 12, 630396. [Google Scholar] [CrossRef]
- Vega Rodríguez, A.; Rodríguez-Oramas, C.; Sanjuán Velázquez, E.; Hardisson de la Torre, A.; Rubio Armendáriz, C.; Carrascosa Iruzubieta, C. Myths and Realities about Genetically Modified Food: A Risk-Benefit Analysis. Appl. Sci. 2022, 12, 2861. [Google Scholar] [CrossRef]
- Breyer, D.; Kopertekh, L.; Reheul, D. Alternatives to Antibiotic Resistance Marker Genes for In Vitro Selection of Genetically Modified Plants—Scientific Developments, Current Use, Operational Access and Biosafety Considerations. CRC Crit. Rev. Plant Sci. 2014, 33, 286–330. [Google Scholar] [CrossRef]
- Bohle, F.; Schneider, R.; Mundorf, J.; Zühl, L.; Simon, S.; Engelhard, M. Where Does the EU-Path on New Genomic Techniques Lead Us? Front. Genome Ed. 2024, 6, 1377117. [Google Scholar] [CrossRef]
- European Parliament Council of the European Union Annexes of the Proposal on Plants Obtained by Certain New Genomic Techniques and Their Food and Feed, and Amending Regulation (EU) 2017/625: COM(2023) 411 Annexes 1 to 3. 2023. Available online: https://food.ec.europa.eu/system/files/2023-07/gmo_biotech_ngt_ia_report.pdf (accessed on 20 September 2024).
- Schouten, H.J.; Krens, F.A.; Jacobsen, E. Cisgenic Plants Are Similar to Traditionally Bred Plants: International Regulations for Genetically Modified Organisms Should Be Altered to Exempt Cisgenesis. EMBO Rep. 2006, 7, 750–753. [Google Scholar] [CrossRef]
- Lusser, M.; Davies, H.V. Comparative Regulatory Approaches for Groups of New Plant Breeding Techniques. N. Biotechnol. 2013, 30, 437–446. [Google Scholar] [CrossRef]
- Rommens, C.M. Intragenic Crop Improvement: Combining the Benefits of Traditional Breeding and Genetic Engineering. J. Agric. Food Chem. 2007, 55, 4281–4288. [Google Scholar] [CrossRef] [PubMed]
- Súnico, V.; Higuera, J.J.; Molina-Hidalgo, F.J.; Blanco-Portales, R.; Moyano, E.; Rodríguez-Franco, A.; Muñoz-Blanco, J.; Caballero, J.L. The Intragenesis and Synthetic Biology Approach towards Accelerating Genetic Gains on Strawberry: Development of New Tools to Improve Fruit Quality and Resistance to Pathogens. Plants 2022, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Qu, J.; Wang, C.; Liu, M.; Zhang, C.; Zhang, X.; Guo, C.; Wu, C.; Yang, G.; Huang, J.; et al. An Efficient Genetic Transformation System Mediated by Rhizobium Rhizogenes in Fruit Trees Based on the Transgenic Hairy Root to Shoot Conversion. Plant Biotechnol. J. 2024, 22, 2093–2103. [Google Scholar] [CrossRef] [PubMed]
- Vanderschuren, H.; Chatukuta, P.; Weigel, D.; Mehta, D. A New Chance for Genome Editing in Europe. Nat. Biotechnol. 2023, 41, 1378–1380. [Google Scholar] [CrossRef] [PubMed]
- El-Mounadi, K.; Luisa Morales-Floriano, M.; Garcia-Ruiz, H. Principles, Applications, and Biosafety of Plant Genome Editing Using CRISPR-Cas9. Front. Plant Sci. 2020, 11, 56. [Google Scholar] [CrossRef]
- Enfissi, E.; Drapal, M.; Perez-Fons, L.; Nogueira, M.; Berry, H.; Almeida, J.; Fraser, P. New Plant Breeding Techniques and Their Regulatory Implications: An Opportunity to Advance Metabolomics Approaches. J. Plant Physiol. 2021, 258, 153378. [Google Scholar] [CrossRef]
- Vasudevan, S.N.; Pooja, S.K.; Raju, T.J.; Damini, C.S. Cisgenics and Intragenics: Boon or Bane for Crop Improvement. Front. Plant Sci. 2023, 14, 1275145. [Google Scholar] [CrossRef]
- Maori, E.; Galanty, Y.; Pignocchi, C.; Chaparro Garcia, A.; Meir, O. Modifying the Specificity of Plant Non-Coding RNA Molecules for Silencing Gene Expression. EP3684930, 4 May 2022. [Google Scholar]
- Capriotti, L.; Molesini, B.; Pandolfini, T.; Jin, H.; Baraldi, E.; Cecchin, M.; Mezzetti, B.; Sabbadini, S. Correction to: RNA Interference-Based Strategies to Control Botrytis Cinerea Infection in Cultivated Strawberry. Plant Cell Rep. 2024, 43, 225. [Google Scholar] [CrossRef]
- Huang, X.; Jia, H.; Xu, J.; Wang, Y.; Wen, J.; Wang, N. Transgene-Free Genome Editing of Vegetatively Propagated and Perennial Plant Species in the T0 Generation via a Co-Editing Strategy. Nat. Plants 2023, 9, 1591–1597. [Google Scholar] [CrossRef]
- Yang, L.; Machin, F.; Wang, S.; Saplaoura, E.; Kragler, F. Heritable Transgene-Free Genome Editing in Plants by Grafting of Wild-Type Shoots to Transgenic Donor Rootstocks. Nat. Biotechnol. 2023, 41, 958–967. [Google Scholar] [CrossRef]
- Alburquerque, N.; Pérez-Caselles, C.; Faize, L.; Ilardi, V.; Burgos, L. Trans-Grafting Plum Pox Virus Resistance from Transgenic Plum Rootstocks to Apricot Scions. Front. Plant Sci. 2023, 14, 1216217. [Google Scholar] [CrossRef] [PubMed]
| Genotype and Species | Explant | Kanamycin Concentration (mg/L) (nptII Gene) | Hygromycin B Concentration (mg/L) (hpt Gene) | Reference |
|---|---|---|---|---|
| APPLE | ||||
| Fupingqiuzi (Malus prunifolia) | Leaf | 15 | [21] | |
| Rootstock JM1 (Malus prunifolia) | Leaf | 25 | [22] | |
| Hybrid MELBA (Malus × domestica) | Leaf | 35 | [23] | |
| Rootstock M.26 (Malus × domestica) | Leaf | 50 | [24] | |
| Greensleaves (Malus × domestica) | Leaf | 100 | [25] | |
| Royal Gala (Malus × domestica) | Leaf | 100 | [26] | |
| Gala (Malus × domestica) | Leaf | 100 | [27] | |
| Rootstock MM106 (Malus × domestica) | Leaf | 5 | [16] | |
| Pinova (Malus × domestica) | Axillary shoot | 100 | [28] | |
| Borkhausen (Malus baccata) | Shoot tip | 20 | [29] | |
| PEAR | ||||
| Shanli (Pyrus ussuriensis) | Leaf | 15 | [30] | |
| Burakovka (Pyrus communis) | Leaf | 25 | 5 | [19] |
| Silver bell, La France (Pyrus communis) | Leaf | 30 | [31] | |
| Spadona (Pyrus communis) | Leaf | 50 | [32] | |
| Conference (Pyrus communis) | Leaf | 100 | [27] | |
| Burakovka (Pyrus communis) | Petiole | 25 | 5 | [19] |
| Silver bell, La France (Pyrus communis) | Axillary shoot | 5 | [31] | |
| Japanese pear (Pyrus pyrifolia) | Cotyledon | 5 | [33] | |
| Duli (Pyrus betulifolia) | Cotyledon, hypocotyl, root | 20 | [34] | |
| CITRUS | ||||
| Tarocco (Citrus sinensis) | Internodal segment | 50 | [35] | |
| Hamlin (Citrus sinensis) | Internodal segment | 100 | [36] | |
| US-942 rootstock (C. reticulata × P. trifoliata) Kuharske rootstock (C. sinensis × P. trifoliata) | Internodal segment | 150 200 | [37] | |
| Valencia (Citrus sinensis) | Leaf | 50 | [38] | |
| Carrizo (C. sinensis × P. trifoliata), Duncan (Citrus paradisi), Hamlin (Citrus sinensis), Mexican Lime (Citrus aurantifolia) | Epycotyl | 70 | [39] | |
| Hamlin, Pêra, Valencia (Citrus sinensis) | Lateral branch | 100 | [40] | |
| Miyagawa wase (Citrus unshiu) | Embryogenic callus | 15–25 | [41] | |
| GRAPEVINE | ||||
| Silcora, Thompson Seedless (Vitis vinifera) | Meristematic bulk | 25–75 | [42] | |
| Thompson seedless (Vitis vinifera) | Meristematic bulk | 70 | [7] | |
| Chardonnay, Thompson Seedless, Redglobe, Cabernet Sauvignon (Vitis vinifera), St. George (Vitis rupestris), 101-14 Millardet et de Grasset (V. riparia × V. rupestris) | Meristematic bulk | 100 | [43] | |
| Thompson Seedless (Vitis vinifera) | Somatic embryo | 75 | [44] | |
| Ramsey (Vitis champinii), Gloire (Vitis riparia), St. George (Vitis rupestris), Cabernet franc, Cabernet Sauvignon, Chardonnay, Merlot, Orange Muscat, Pinot noir, Sauvignon blanc, Shiraz, Zinfandel, Superior Seedless, Thompson Seedless (Vitis vinifera), Seyval blanc, 110 Richter Harmony, Conquistador, Freedom (Vitis hybrids) | Somatic embryo | 100 | [45] | |
| Seyval blanc (Vitis vinifera) | Leaf | 100 | [46] | |
| Red Globe (Vitis vinifera) | Embryogenic callus | 80 | [47] | |
| King’s Ruby (Vitis vinifera) | Embryogenic callus | 100 | 10 | [48] |
| Cabernet Sauvignon, Shiraz, Chardonnay, Riesling, Sauvignon Blanc, Chenin Blanc, Muscat Gordo Blanco (Vitis vinifera) | Embryogenic callus | 100 | [49] | |
| Pusa Seedless (Vitis vinifera) | Embryogenic callus | 25 | [18] | |
| Portan, Danuta, Syrah (Vitis vinifera) | Embryogenic callus | 80–100 | 2.5–5.0 | [50] |
| Portan, Danuta, Syrah (Vitis vinifera) | Axillary shoot | 4.0 | 0.8 | [50] |
| Vitis 6-12-2 (V. pseudoreticulata × V. vinifera) | Shoot tip, internode | 3–12 | [51] | |
| PEACH | ||||
| Rootstock GF677 (P. persica × P. amygdalus) | Meristematic bulk | 25–70 | [52] | |
| Miraflores (Prunus persica) | Immature embryo | 40 | [53] | |
| CHERRY | ||||
| Rootstock Gisela 6 (P. cerasus × P. canescens) | Leaf | 20 | [54] | |
| Black Eagle (C. fruticosa × C. avium) | Leaf | 25 | 5–10 | [55] |
| Rootstock Gisela 6, Rootstock Gisela 7 (P. cerasus × P. canescens) | Leaf | 50 | [56] | |
| Montmorency (Prunus cerasus) Rootstock Gisela 6 (P. cerasus × P. canescens) | Leaf | 50 | [57] | |
| Stella (Prunus avium) | Leaf | 10–50 | [58] | |
| PLUM | ||||
| Stanley (Prunus domestica) | Embryonic axes | 75 | 5 | [59] |
| Startovaya (Prunus domestica) | Leaf | 5 | [60] | |
| Angeleno, Larry Anne (Prunus salicina) | Hypocotyl | 40–75 | [61] | |
| Bluebyrd (Prunus domestica) | Hypocotyl | 80 | [62] | |
| APRICOT | ||||
| Rootstock 146-2 (P. pumila × P. tomentosa) | Leaf | 10–30 | [63] | |
| Canino, Moniquí (Prunus armeniaca) | Hypocotyl | 10 | [64] | |
| ALMOND | ||||
| Ne Plus Ultra (Prunus amygdalus) | Leaf | 7–9 | [65] | |
| Boa Casta (Prunus amygdalus) | Leaf | 10–15 | [66] | |
| Clone VII (Prunus amygdalus) | Leaf | 15–50 | [67] | |
| STRAWBERRY | ||||
| Elista (Fragaria × ananassa), Induka (Fragaria × ananassa) | Leaf | 25 30 | [68] | |
| Sveva, Calypso (Fragaria × ananassa), Alpina W.O. (Fragaria vesca) | Leaf | 25 | [69] | |
| Chandler (Fragaria × ananassa) | Leaf | 25 | [70] | |
| Tudla (Fragaria × ananassa) | Leaf | 30 | [71] | |
| PI 551572 (Fragaria vesca) | Leaf | 30 | [72] | |
| Camarosa (Fragaria × ananassa) | Leaf | 50 | [73] | |
| Hecker, La Sans Rivale (Fragaria × ananassa), Alpine FRA197, Alpine FRA198 (Fragraria vesca) | Leaf, petiole | 50 | [74] | |
| Teodora, Egla (Fragaria × ananassa) | Stipule | 50 | [75] | |
| PAPAYA | ||||
| Kapoho (Carica papaya) | Somatic embryo | 150 | [76] | |
| Sunrise and Sunset (Carica papaya) | Somatic embryo | 150 | [77] | |
| Sunrise (Carica papaya) | Embryogenic culture | 300 | [78] | |
| MANGO | ||||
| Hindi (Mangifera indica) | Embryogenic culture | 100 | [79] | |
| Hindi (Mangifera indica) | Somatic embryo | 100–400 | [80] | |
| Keitt (Mangifera indica) | Somatic embryo | 200 | [80] | |
| BANANA | ||||
| Sukali Ndiizi, Gros Michel, Cavendish, Williams (Musa spp.) | Embryogenic cell | 100 | [17] | |
| Grand Nain (Musa acuminata) | Embryogenic cell | 100 | [81] | |
| Grand nain (Musa acuminata) | Embryogenic cell | 15 | [82] | |
| Bluggoe (Musa spp.) | Embryogenic cell | 50 | [83] | |
| Matti (Musa acuminata) | Multiple shoot clump | 10 | [84] | |
| Agbagba (Musa spp.) | Apical shoot | 25 | [85] | |
| PINEAPPLE | ||||
| Shenwan (Ananas comosus) | Callus | 30–50 | [86] | |
| Queen (Ananus comosus) | Callus | 60 | [87] | |
| Smooth Cayenne (Ananas cosmos) | Leaf, stem disc | 20 | [20] | |
| Genotype and Species | Explant | D-mannose Concentration (g/L) (pmi Gene) | Saccharose Concentration (g/L) | Reference |
|---|---|---|---|---|
| Holsteiner Cox (Malus × domestica) | Leaf | 1–10 | 5–30 | [112] |
| Kuharske (C. sinensis × P. trifoliata) | Mature stem | 7.5 (first step) 15 (second step) | 22.5 (first step) 15 (secondo step) | [116] |
| Carrizo (C. sinensis × P. trifoliata), Swingle (C. paradisi × P. trifoliata) | Epicotyl | 30 | 0.2 | [115] |
| Valencia, Natal, Hamlin, Pera (C. sinensis) | Epicotyl | 13–20 | 0 | [113] |
| Carrizo (C. sinensis × P. trifoliata), Pineapple (C. sinensis) | Epicotyl | 15 12 | 0 5 | [114] |
| Startovaya (Prunus domestica) | Leaf | 15 | 20 | [118] |
| Claudia Verde (Prunus domestica) | Hypocotyl | 1.5–5 | 0.1 | [117] |
| Ne Plus Ultra (Prunus dulcis) | Leaf | 2.5 | 15 | [65] |
| Kapoho (Carica papaya) | Embryogenic callus | 30 | 0 | [120] |
| Genotype and Species | Explant t | Name and Type of Promoter | Gene of Interest (GOI) | Selectable Marker Gene (SMG) | Recombinase Excision Efficiency (%) | Reference |
|---|---|---|---|---|---|---|
| Cre-lox | ||||||
| Helena (Prunus armeniaca) | Leaf | Transactivating XVE factor (Inducible: β-estradiol) | gfp | nptII | 11.3% | [170] |
| Grand Naine (Musa acuminata) | Embryo | Gmhsp17.6-L (soybean) (Inducible: heat-shock) | nptII | hpt and codA | 59.7% | [171] |
| HSP18.2 (A. thaliana) (Inducible: heat-shock) | 40% | |||||
| Grand Naine (Musa acuminata) | Embryo cell | REG-2 (rice embryo globulin gene) (Inducible: tissue specific) | gus | hpt and codA | 41.7% | [172] |
| Jincheng orange (Citrus sinensis) | Epicotyl | CaMV 35S (Constitutive) | gfp | ipt | 81.8% | [190] |
| NosP (nopaline synthase gene) (Constitutive) | 100% | |||||
| Tarocco (Citrus sinensis) | Epicotyl | NosP (Constitutive) | AATCB (cecropin B gene) | ipt | 74.8% 66.7% | [173,174] |
| Navel (Citrus sinensis) | Stem segment | CaMV 35S (Constitutive) | PR1aCB (anti microbial peptide gene) | ipt | 100% | [175] |
| FLP-FRT | ||||||
| Pinova (Malus × domestica) | Leaf | Gmhsp17.6-L (Inducible: heat-shock) | gus | nptII | 37% | [177] |
| Pinova (Malus × domestica) | Leaf | Gmhsp17.6-L (Inducible: heat-shock) | gus | nptII | 1.6% | [178] |
| Gala Galaxy (Malus × domestica) | Leaf | HSP (A. thaliana) (Inducible: heat-shock) | FB_MR5 (Malus × robusta 5 fire blight resistant gene) | nptII | 100% | [180] |
| Brookfield Baigent, Pinova (Malus × domestica) | Axillary shoot | Gmhsp17.6-L (Inducible: heat-shock) | Rvi6 (M. floribunda apple scab resistant gene) | nptII | 100% | [179] |
| Brachetto (Vitis vinifera) | Embryogenic callus | Gmhsp17.6-L (Inducible: heat-shock) | gus | nptII | 100% | [181] |
| R-RS | ||||||
| Calypso (Fragaria × ananassa) | Leaf | glucocorticoid receptor (Inducible: chemical) | gus | nptII and codA | 62% | [183] |
| Gala (Malus × domestica) | Leaf | glucocorticoid receptor (Inducible: chemical) | Rvi6 | nptII and codA | 30% | [184,185] |
| Galaxy (Malus × domestica) | Leaf | CaMV 35S (Constitutive) | Gus | nptII and codA | 19% | [187] |
| Conference (Pyrus communis) | Leaf | 30% | ||||
| Gala (Malus × domestica) | Leaf | glucocorticoid receptor (Inducible: chemical) | Rvi6 | nptII and codA | 28.5% | [186] |
| Melba (Malus × domestica) | Leaf | glucocorticoid receptor (Inducible: chemical) | thaumatin II | nptII and codA | 5.6% | [23] |
| Williams and Grand Naine (Musa acuminata) | Embryo cell | glucocorticoid receptor (Inducible: chemical) | gfp | nptII and codA | 100% | [188] |
| MAT | ||||||
| Carrizo (C. sinensis × P. trifoliata) | Epicotyl | CaMV 35S (Constitutive) | gus | ipt | 32% | [126] |
| Pineapple (Citrus sinensis) | 64% | |||||
| Helena (Prunus armeniaca) | Leaf | CaMV 35S (Constitutive) | gus | ipt | 41% | [189] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Súnico, V.; Piunti, I.; Bhattacharjee, M.; Mezzetti, B.; Caballero, J.L.; Muñoz-Blanco, J.; Ricci, A.; Sabbadini, S. Overview on Current Selectable Marker Systems and Novel Marker Free Approaches in Fruit Tree Genetic Engineering. Int. J. Mol. Sci. 2024, 25, 11902. https://doi.org/10.3390/ijms252211902
Súnico V, Piunti I, Bhattacharjee M, Mezzetti B, Caballero JL, Muñoz-Blanco J, Ricci A, Sabbadini S. Overview on Current Selectable Marker Systems and Novel Marker Free Approaches in Fruit Tree Genetic Engineering. International Journal of Molecular Sciences. 2024; 25(22):11902. https://doi.org/10.3390/ijms252211902
Chicago/Turabian StyleSúnico, Victoria, Irene Piunti, Mamta Bhattacharjee, Bruno Mezzetti, José L. Caballero, Juan Muñoz-Blanco, Angela Ricci, and Silvia Sabbadini. 2024. "Overview on Current Selectable Marker Systems and Novel Marker Free Approaches in Fruit Tree Genetic Engineering" International Journal of Molecular Sciences 25, no. 22: 11902. https://doi.org/10.3390/ijms252211902
APA StyleSúnico, V., Piunti, I., Bhattacharjee, M., Mezzetti, B., Caballero, J. L., Muñoz-Blanco, J., Ricci, A., & Sabbadini, S. (2024). Overview on Current Selectable Marker Systems and Novel Marker Free Approaches in Fruit Tree Genetic Engineering. International Journal of Molecular Sciences, 25(22), 11902. https://doi.org/10.3390/ijms252211902







