Kinetic Features of Degradation of R-Loops by RNase H1 from Escherichia coli
Abstract
:1. Introduction
2. Results
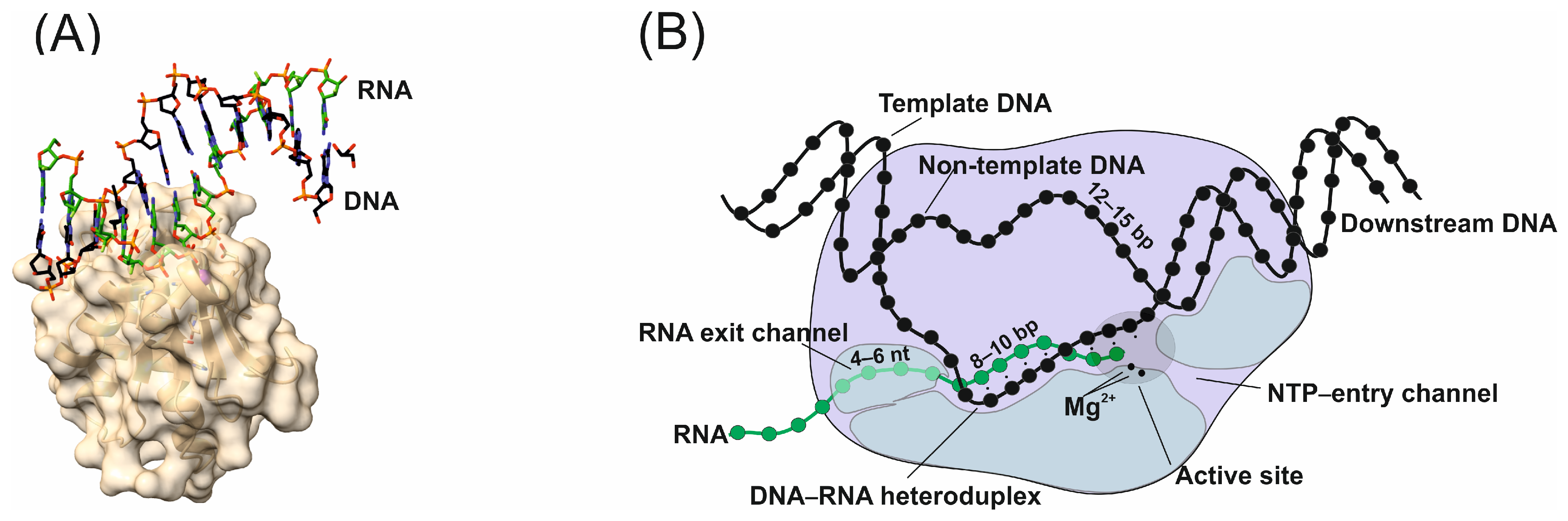
2.1. Design of R-Loops
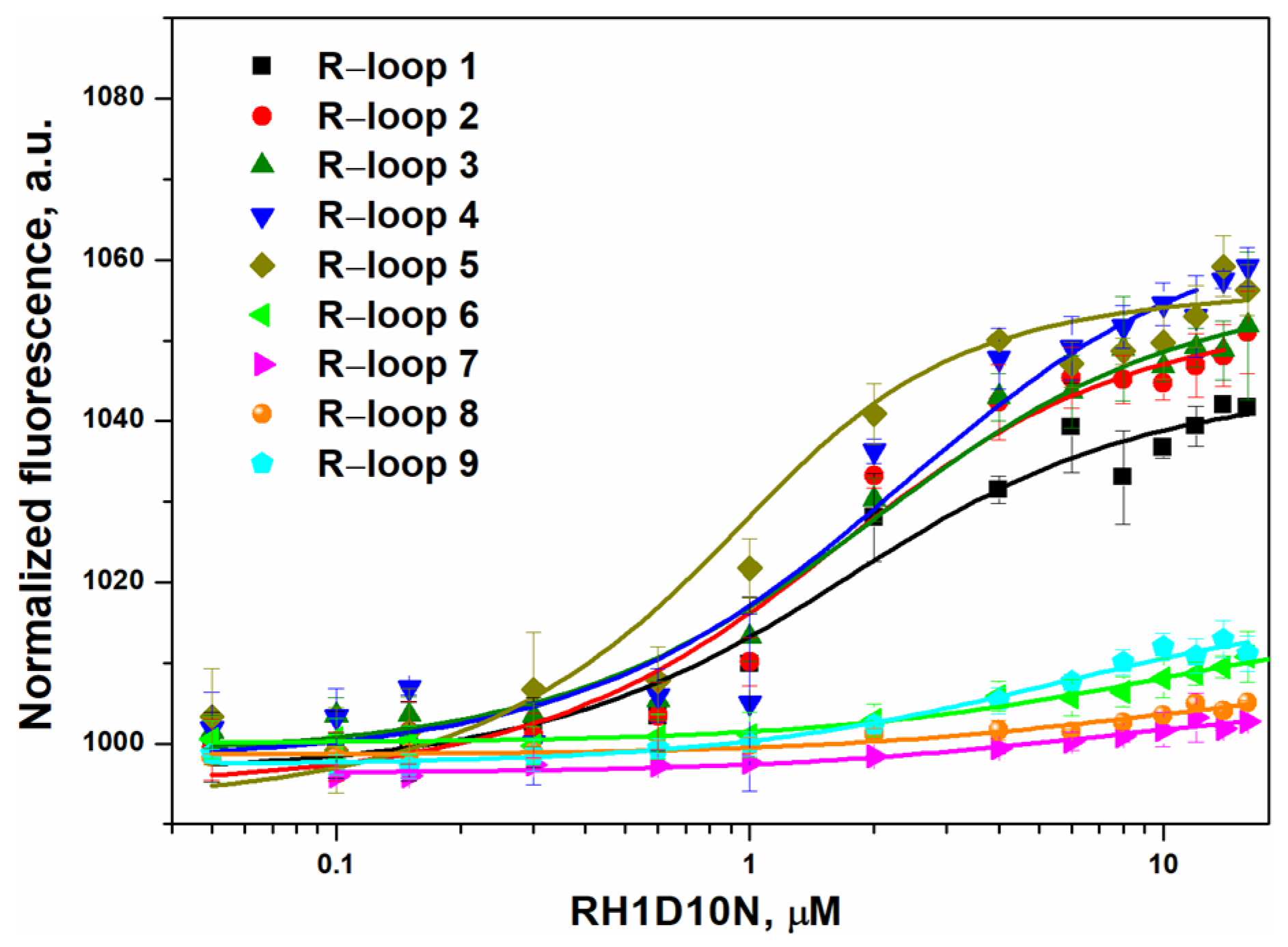
2.2. R-Loop-Binding Assays
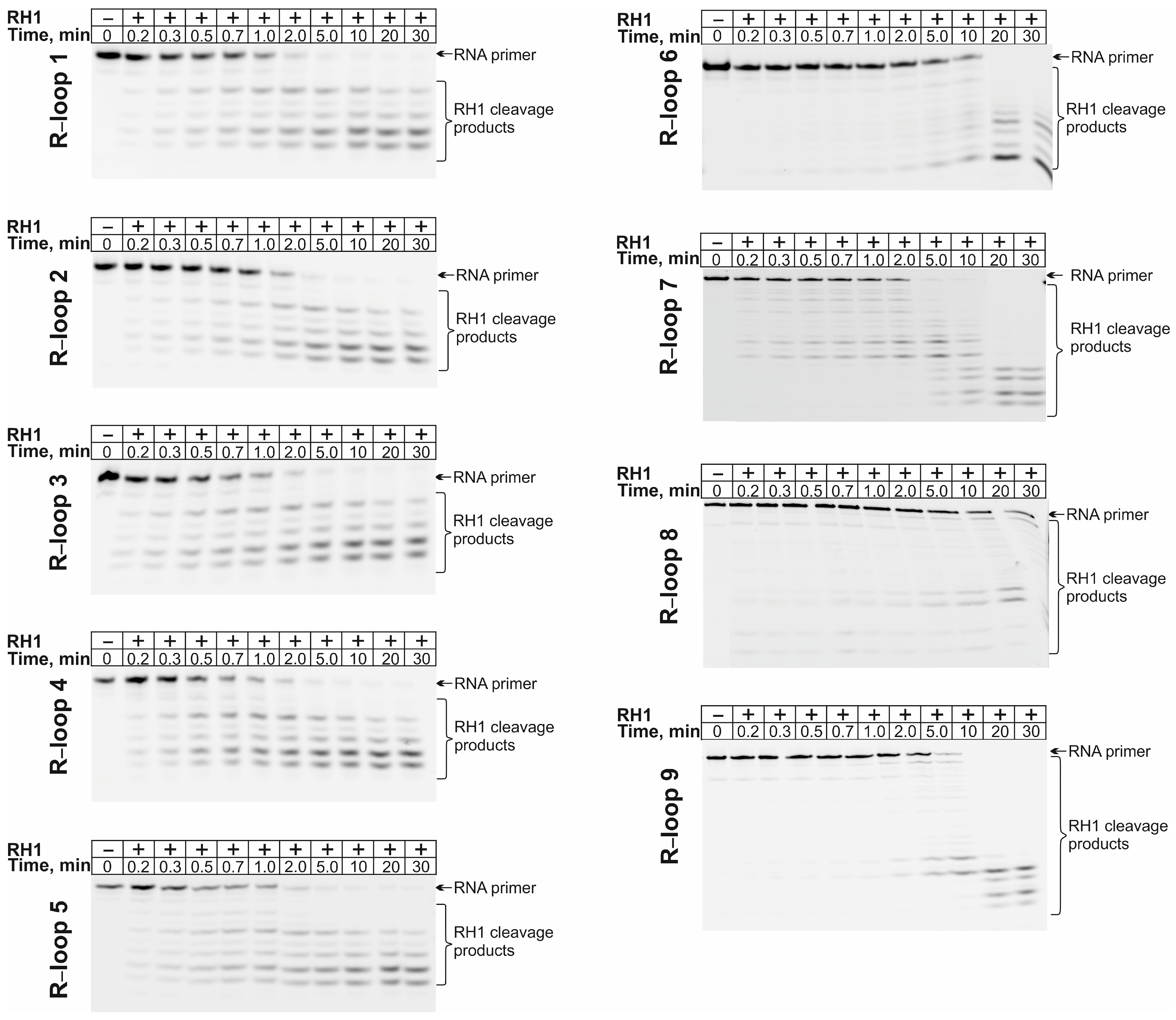
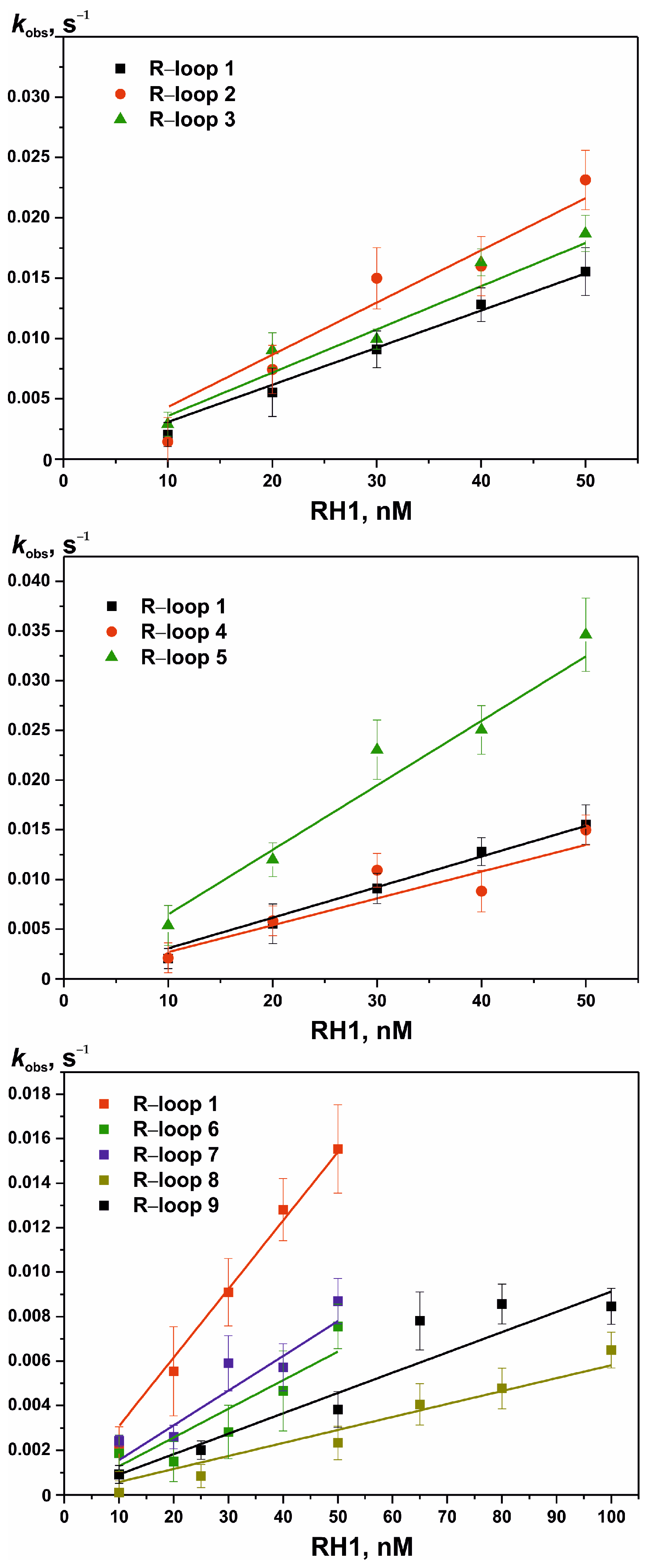
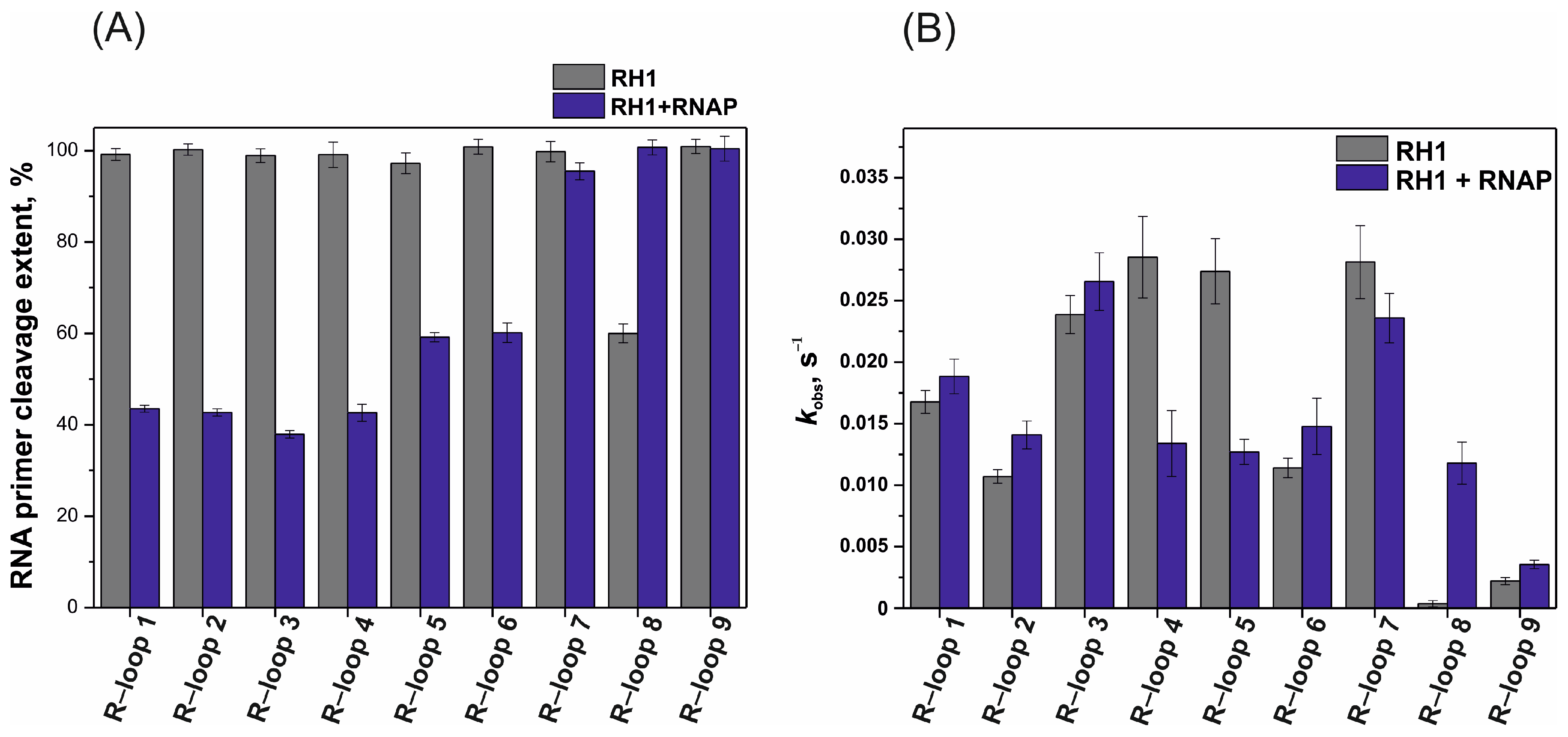
2.3. RNase H Activity Assay
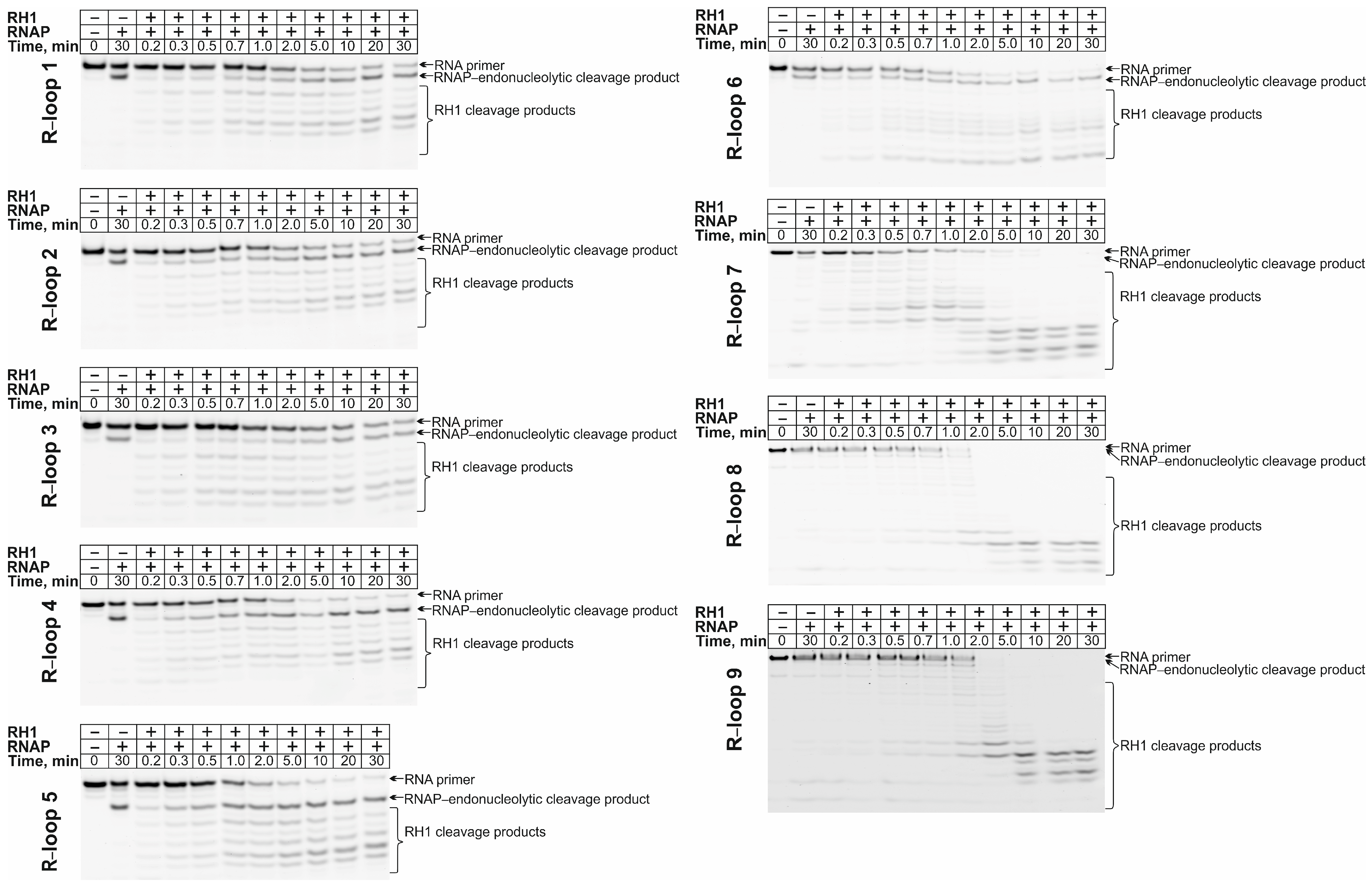
2.4. RNase H Activity Assayactivity Assay in the Presence of the RNAP
3. Discussion
4. Materials and Methods
4.1. Enzymes and Oligonucleotides
4.2. Preparation of R-Loops
4.3. R-Loop-Binding Assays
4.4. RNase H Activity Assay
4.5. RNase H Activity Assay in the Presence of RNAP
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuzminov, A. Bacterial nucleoid is a riddle wrapped in a mystery inside an enigma. J. Bacteriol. 2024, 206, e0021123. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, T.; Ishihama, A. Biosynthesis of RNA polymerase in Escherichia coli. J. Mol. Biol. 1977, 115, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.J.; Martin, C.M.; Sun, Z.; Cagliero, C.; Zhou, Y.N. Nucleolus-like compartmentalization of the transcription machinery in fast-growing bacterial cells. Crit. Rev. Biochem. Mol. Biol. 2016, 52, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.E.; Jin, D.J. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 2003, 50, 1493–1505. [Google Scholar] [CrossRef] [PubMed]
- Libby, E.A.; Roggiani, M.; Goulian, M. Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Stracy, M.; Lesterlin, C.; de Leon, F.G.; Uphoff, S.; Zawadzki, P.; Kapanidis, A.N. Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc. Natl. Acad. Sci. USA 2015, 112, E4390–E4399. [Google Scholar] [CrossRef]
- Stracy, M.; Kapanidis, A.N. Single-molecule and super-resolution imaging of transcription in living bacteria. Methods 2017, 120, 103–114. [Google Scholar] [CrossRef]
- McHenry, C.S. DNA Replicases from a Bacterial Perspective. Annu. Rev. Biochem. 2011, 80, 403–436. [Google Scholar] [CrossRef]
- Xu, Z.-Q.; E Dixon, N. Bacterial replisomes. Curr. Opin. Struct. Biol. 2018, 53, 159–168. [Google Scholar] [CrossRef]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef]
- Merrikh, H.; Zhang, Y.; Grossman, A.D.; Wang, J.D. Replication–transcription conflicts in bacteria. Nat. Rev. Microbiol. 2012, 10, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Trautinger, B.W.; Jaktaji, R.P.; Rusakova, E.; Lloyd, R.G. RNA Polymerase Modulators and DNA Repair Activities Resolve Conflicts between DNA Replication and Transcription. Mol. Cell 2005, 19, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Tehranchi, A.K.; Blankschien, M.D.; Zhang, Y.; Halliday, J.A.; Srivatsan, A.; Peng, J.; Herman, C.; Wang, J.D. The Transcription Factor DksA Prevents Conflicts between DNA Replication and Transcription Machinery. Cell 2010, 141, 595–605. [Google Scholar] [CrossRef]
- Brewer, B.J. When polymerases collide: Replication and the transcriptional organization of the E. coli chromosome. Cell 1988, 53, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Srivatsan, A.; Tehranchi, A.; MacAlpine, D.M.; Wang, J.D. Co-Orientation of Replication and Transcription Preserves Genome Integrity. PLoS Genet. 2010, 6, e1000810. [Google Scholar] [CrossRef]
- Merrikh, H.; Machón, C.; Grainger, W.H.; Grossman, A.D.; Soultanas, P. Co-directional replication–transcription conflicts lead to replication restart. Nature 2011, 470, 554–557. [Google Scholar] [CrossRef]
- Dutta, D.; Shatalin, K.; Epshtein, V.; Gottesman, M.E.; Nudler, E. Linking RNA Polymerase Backtracking to Genome Instability in E. coli. Cell 2011, 146, 533–543. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. Transcription–replication conflicts: How they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 2016, 17, 553–563. [Google Scholar] [CrossRef]
- Foster, P.L.; Niccum, B.A.; Lee, H. DNA Replication-Transcription Conflicts Do Not Significantly Contribute to Spontaneous Mutations Due to Replication Errors in Escherichia coli. mBio 2021, 12, e0250321. [Google Scholar] [CrossRef]
- Lovett, S.T. DNA polymerase III protein, HolC, helps resolve replication/transcription conflicts. Microb. Cell 2021, 8, 143–145. [Google Scholar] [CrossRef]
- Lang, K.S.; Merrikh, H. Topological stress is responsible for the detrimental outcomes of head-on replication-transcription conflicts. Cell Rep. 2021, 34, 108797. [Google Scholar] [CrossRef] [PubMed]
- Browning, K.R.; Merrikh, H. Replication–Transcription Conflicts: A Perpetual War on the Chromosome. Annu. Rev. Biochem. 2024, 93, 21–46. [Google Scholar] [CrossRef] [PubMed]
- Vlachos-Breton, É.; Drolet, M. R-Loop Detection in Bacteria. Methods Mol. Biol. 2022, 2528, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Drolet, M.; Bi, X.; Liu, L. Hypernegative supercoiling of the DNA template during transcription elongation in vitro. J. Biol. Chem. 1994, 269, 2068–2074. [Google Scholar] [CrossRef] [PubMed]
- Phoenix, P.; Raymond, M.-A.; Massé, É.; Drolet, M. Roles of DNA Topoisomerases in the Regulation of R-loop Formation in Vitro. J. Biol. Chem. 1997, 272, 1473–1479. [Google Scholar] [CrossRef]
- Massé, E.; Phoenix, P.; Drolet, M. DNA Topoisomerases Regulate R-loop Formation during Transcription of the rrnB Operon in Escherichia coli. J. Biol. Chem. 1997, 272, 12816–12823. [Google Scholar] [CrossRef]
- Massé, E.; Drolet, M. Escherichia coli DNA Topoisomerase I Inhibits R-loop Formation by Relaxing Transcription-induced Negative Supercoiling. J. Biol. Chem. 1999, 274, 16659–16664. [Google Scholar] [CrossRef]
- Drolet, M.; Phoenix, P.; Menzel, R.; Massé, E.; Liu, L.F.; Crouch, R.J. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 1995, 92, 3526–3530. [Google Scholar] [CrossRef]
- Hraiky, C.; Raymond, M.-A.; Drolet, M. RNase H Overproduction Corrects a Defect at the Level of Transcription Elongation during rRNA Synthesis in the Absence of DNA Topoisomerase I in Escherichia coli. J. Biol. Chem. 2000, 275, 11257–11263. [Google Scholar] [CrossRef]
- Baaklini, I.; Hraiky, C.; Rallu, F.; Tse-Dinh, Y.; Drolet, M. RNase HI overproduction is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol. Microbiol. 2004, 54, 198–211. [Google Scholar] [CrossRef]
- Baaklini, I.; Usongo, V.; Nolent, F.; Sanscartier, P.; Hraiky, C.; Drlica, K.; Drolet, M. Hypernegative Supercoiling Inhibits Growth by Causing RNA Degradation. J. Bacteriol. 2008, 190, 7346–7356. [Google Scholar] [CrossRef] [PubMed]
- Martel, M.; Balleydier, A.; Sauriol, S.A.; Drolet, M. Constitutive stable DNA replication in Escherichia coli cells lacking type 1A topoisomerase activity. DNA Repair 2015, 35, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Brochu, J.; Vlachos-Breton, É.; Sutherland, S.; Martel, M.; Drolet, M. Topoisomerases I and III inhibit R-loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli. PLoS Genet. 2018, 14, e1007668. [Google Scholar] [CrossRef] [PubMed]
- Hausen, P.; Stein, H.; Ribonuclease, H. An Enzyme Degrading the RNA Moiety of DNA-RNA Hybrids. Eur. J. Biochem. 1970, 14, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Hyjek, M.; Figiel, M.; Nowotny, M. RNases H: Structure and mechanism. DNA Repair 2019, 84, 102672. [Google Scholar] [CrossRef]
- Itoh, T.; Tomizawa, J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. USA 1980, 77, 2450–2454. [Google Scholar] [CrossRef]
- Masukata, H.; Tomizawa, J.-I. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell 1990, 62, 331–338. [Google Scholar] [CrossRef]
- Yu, K.; Chedin, F.; Hsieh, C.-L.; Wilson, T.E.; Lieber, M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003, 4, 442–451. [Google Scholar] [CrossRef]
- Holmes, J.B.; Akman, G.; Wood, S.R.; Sakhuja, K.; Cerritelli, S.M.; Moss, C.; Bowmaker, M.R.; Jacobs, H.T.; Crouch, R.J.; Holt, I.J. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc. Natl. Acad. Sci. USA 2015, 112, 9334–9339. [Google Scholar] [CrossRef]
- Park, J.; Baruch-Torres, N.; Yin, Y.W. Structural and Molecular Basis for Mitochondrial DNA Replication and Transcription in Health and Antiviral Drug Toxicity. Molecules 2023, 28, 1796. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Proudfoot, N.J.; Gromak, N. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination. Mol. Cell 2011, 42, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Ohle, C.; Tesorero, R.; Schermann, G.; Dobrev, N.; Sinning, I.; Fischer, T. Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 2016, 167, 1001–1013.e7. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.; Puget, N.; Lin, Y.-L.; Clouaire, T.; Aguirrebengoa, M.; Rocher, V.; Pasero, P.; Canitrot, Y.; Legube, G. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; García-Muse, T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Manley, J.L. Inactivation of the SR Protein Splicing Factor ASF/SF2 Results in Genomic Instability. Cell 2005, 122, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774–786.e19. [Google Scholar] [CrossRef]
- Hamperl, S.; Cimprich, K.A. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair 2014, 19, 84–94. [Google Scholar] [CrossRef]
- Hamperl, S.; Cimprich, K.A. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell 2016, 167, 1455–1467. [Google Scholar] [CrossRef]
- Barroso, S.; Herrera-Moyano, E.; Muñoz, S.; García-Rubio, M.; Gómez-González, B.; Aguilera, A. The DNA damage response acts as a safeguard against harmful DNA–RNA hybrids of different origins. EMBO Rep. 2019, 20, e47250. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Patidar, P.L.; Motea, E.A.; Dang, T.T.; Manley, J.L.; Boothman, D.A. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016, 12, e1006107. [Google Scholar] [CrossRef]
- Sollier, J.; Cimprich, K.A. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015, 25, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Stork, C.T.; Garcia-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009, 276, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Huertas, P.; Aguilera, A. Cotranscriptionally Formed DNA:RNA Hybrids Mediate Transcription Elongation Impairment and Transcription-Associated Recombination. Mol. Cell 2003, 12, 711–721. [Google Scholar] [CrossRef]
- Paulsen, R.D.; Soni, D.V.; Wollman, R.; Hahn, A.T.; Yee, M.-C.; Guan, A.; Hesley, J.A.; Miller, S.C.; Cromwell, E.F.; Solow-Cordero, D.E.; et al. A Genome-wide siRNA Screen Reveals Diverse Cellular Processes and Pathways that Mediate Genome Stability. Mol. Cell 2009, 35, 228–239. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Sakhuja, K.; Crouch, R.J. RNase H1, the Gold Standard for R-Loop Detection. Methods Mol Biol. 2022, 2528, 91–114. [Google Scholar] [CrossRef]
- Wagner, C.B.; Luke, B. DNA–RNA Hybrids at Telomeres in Budding Yeast. Methods Mol Biol. 2022, 2528, 145–157. [Google Scholar] [CrossRef]
- Zhang, X.; Hao, Y.; Fu, X.-D. Mapping R-Loops Using Catalytically Inactive RNaseH1 (R-ChIP); Springer: Berlin/Heidelberg, Germany, 2022; pp. 359–372. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335–361. [Google Scholar] [CrossRef]
- Cueny, R.R.; McMillan, S.D.; Keck, J.L. G-quadruplexes in bacteria: Insights into the regulatory roles and interacting proteins of non-canonical nucleic acid structures. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 539–561. [Google Scholar] [CrossRef]
- Yadav, P.; Kim, N.; Kumari, M.; Verma, S.; Sharma, T.K.; Yadav, V.; Kumar, A. G-Quadruplex Structures in Bacteria: Biological Relevance and Potential as an Antimicrobial Target. J. Bacteriol. 2021, 203. [Google Scholar] [CrossRef] [PubMed]
- Kanaya, S.; Kohara, A.; Miura, Y.; Sekiguchi, A.; Iwai, S.; Inoue, H.; Ohtsuka, E.; Ikehara, M. Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J. Biol. Chem. 1990, 265, 4615–4621. [Google Scholar] [CrossRef] [PubMed]
- Katayanagi, K.; Miyagawa, M.; Matsushima, M.; Ishikawa, M.; Kanaya, S.; Ikehara, M.; Matsuzaki, T.; Morikawa, K. Three-dimensional structure of ribonuclease H from E. coli. Nature 1990, 347, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Haruki, M.; Noguchi, E.; Kanaya, S.; Crouch, R.J. Kinetic and Stoichiometric Analysis for the Binding of Escherichia coli Ribonuclease HI to RNA-DNA Hybrids Using Surface Plasmon Resonance. J. Biol. Chem. 1997, 272, 22015–22022. [Google Scholar] [CrossRef] [PubMed]
- Kanaya, E.; Kanaya, S. Kinetic Analysis of Escherichia Coli Ribonuclease HI Using Oligomeric DNA/RNA Substrates Suggests an Alternative Mechanism for the Interaction between the Enzyme and the Substrate. Eur. J. Biochem. 1995, 231, 557–562. [Google Scholar] [CrossRef]
- Iwai, S.; Wakasa, M.; Ohtsuka, E.; Kanaya, S.; Kidera, A.; Nakamura, H. Interaction of the Basic Protrusion ofEscherichia coliRibonuclease HI with its Substrate. J. Mol. Biol. 1996, 263, 699–706. [Google Scholar] [CrossRef]
- Nowotny, M.; Gaidamakov, S.A.; Crouch, R.J.; Yang, W. Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis. Cell 2005, 121, 1005–1016. [Google Scholar] [CrossRef]
- Pallan, P.S.; Prakash, T.P.; de Leon, A.R.; Egli, M. Limits of RNA 2′-OH Mimicry by Fluorine: Crystal Structure of Bacillus halodurans RNase H Bound to a 2′-FRNA:DNA Hybrid. Biochemistry 2016, 55, 5321–5325. [Google Scholar] [CrossRef]
- Esyunina, D.; Turtola, M.; Pupov, D.; Bass, I.; Klimašauskas, S.; Belogurov, G.; Kulbachinskiy, A. Lineage-specific variations in the trigger loop modulate RNA proofreading by bacterial RNA polymerases. Nucleic Acids Res. 2016, 44, 1298–1308. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Novopashina, D.S.; Fedorova, O.S.; Kuznetsov, N.A. Effect of the Substrate Structure and Metal Ions on the Hydrolysis of Undamaged RNA by Human AP Endonuclease APE1. Acta Naturae 2020, 12, 74–85. [Google Scholar] [CrossRef]


| Shorthand | Nucleotide Sequence |
|---|---|
| R-loop 1 |  |
| R-loop 2 |  |
| R-loop 3 |  |
| R-loop 4 |  |
| R-loop 5 |  |
| R-loop 6 |  |
| R-loop 7 |  |
| R-loop 8 |  |
| R-loop 9 | 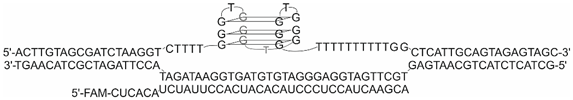 |
| Substrate | Kd, µM | k1, M−1⋅s−1, ×10−5 |
|---|---|---|
| R-loop 1 | 1.2 ± 0.4 | 28 ± 3 |
| R-loop 2 | 1.1 ± 0.4 | 38 ± 8 |
| R-loop 3 | 1.5 ± 0.4 | 36 ± 2 |
| R-loop 4 | 1.8 ± 0.4 | 27 ± 2 |
| R-loop 5 | 0.35 ± 0.15 | 65 ± 5 |
| R-loop 6 | 9.7 ± 2.7 | 14 ± 3 |
| R-loop 7 | 6.7 ± 3.1 | 16 ± 2 |
| R-loop 8 | 9.3 ± 4.8 | 5.8 ± 0.5 |
| R-loop 9 | 5.1 ± 1.4 | 9.1 ± 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuznetsova, A.A.; Kosarev, I.A.; Timofeyeva, N.A.; Novopashina, D.S.; Kuznetsov, N.A. Kinetic Features of Degradation of R-Loops by RNase H1 from Escherichia coli. Int. J. Mol. Sci. 2024, 25, 12263. https://doi.org/10.3390/ijms252212263
Kuznetsova AA, Kosarev IA, Timofeyeva NA, Novopashina DS, Kuznetsov NA. Kinetic Features of Degradation of R-Loops by RNase H1 from Escherichia coli. International Journal of Molecular Sciences. 2024; 25(22):12263. https://doi.org/10.3390/ijms252212263
Chicago/Turabian StyleKuznetsova, Aleksandra A., Iurii A. Kosarev, Nadezhda A. Timofeyeva, Darya S. Novopashina, and Nikita A. Kuznetsov. 2024. "Kinetic Features of Degradation of R-Loops by RNase H1 from Escherichia coli" International Journal of Molecular Sciences 25, no. 22: 12263. https://doi.org/10.3390/ijms252212263
APA StyleKuznetsova, A. A., Kosarev, I. A., Timofeyeva, N. A., Novopashina, D. S., & Kuznetsov, N. A. (2024). Kinetic Features of Degradation of R-Loops by RNase H1 from Escherichia coli. International Journal of Molecular Sciences, 25(22), 12263. https://doi.org/10.3390/ijms252212263








