Genome-Wide Analysis of CSL Family Genes Involved in Petiole Elongation, Floral Petalization, and Response to Salinity Stress in Nelumbo nucifera
Abstract
1. Introduction
2. Results
2.1. Identification of CSL Family Members in N. nucifera
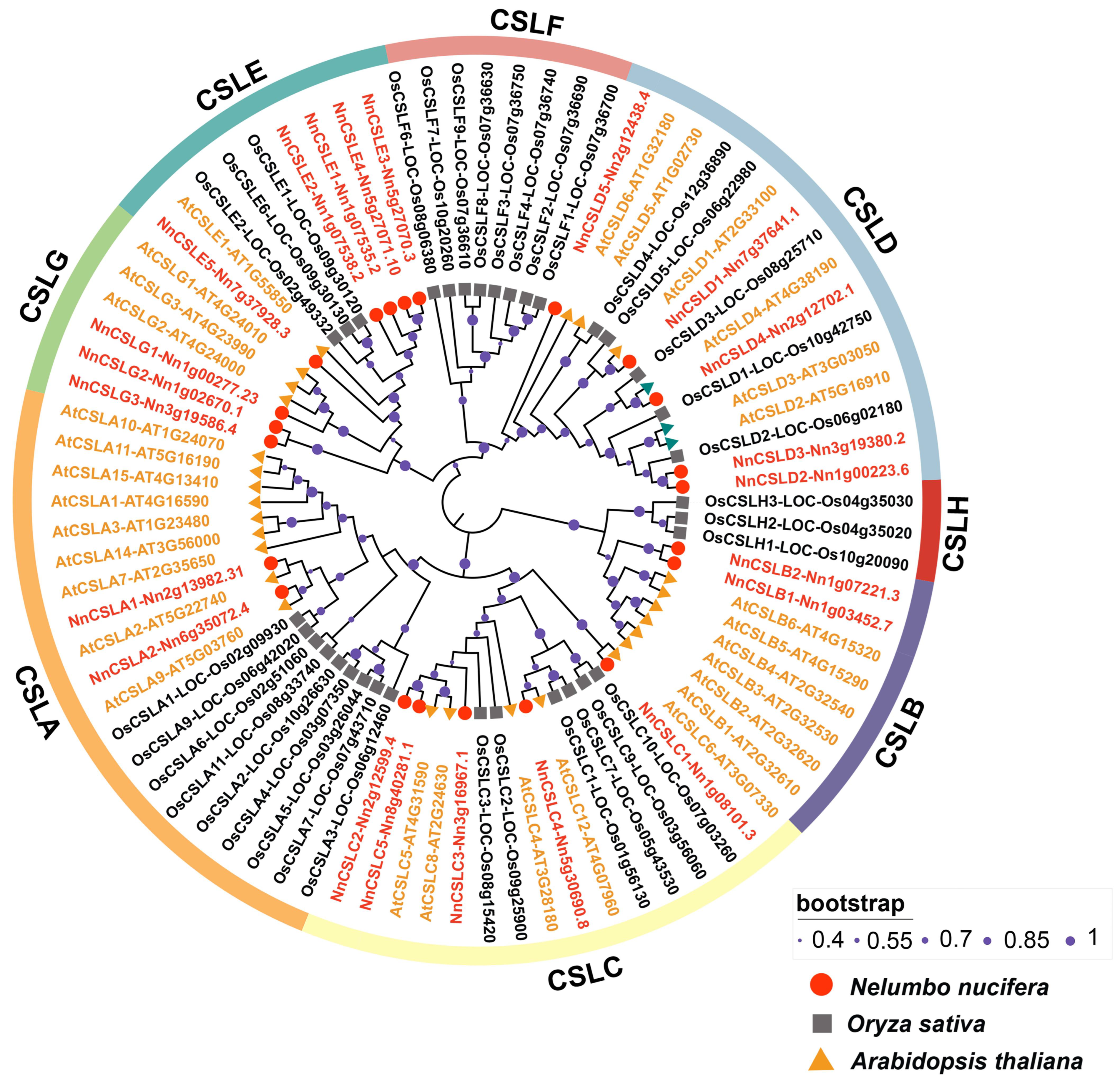
2.2. Phylogenetic Analysis and Classification of the NnCSL Family
2.3. Structure Analysis of CSL Family Members in N. nucifera
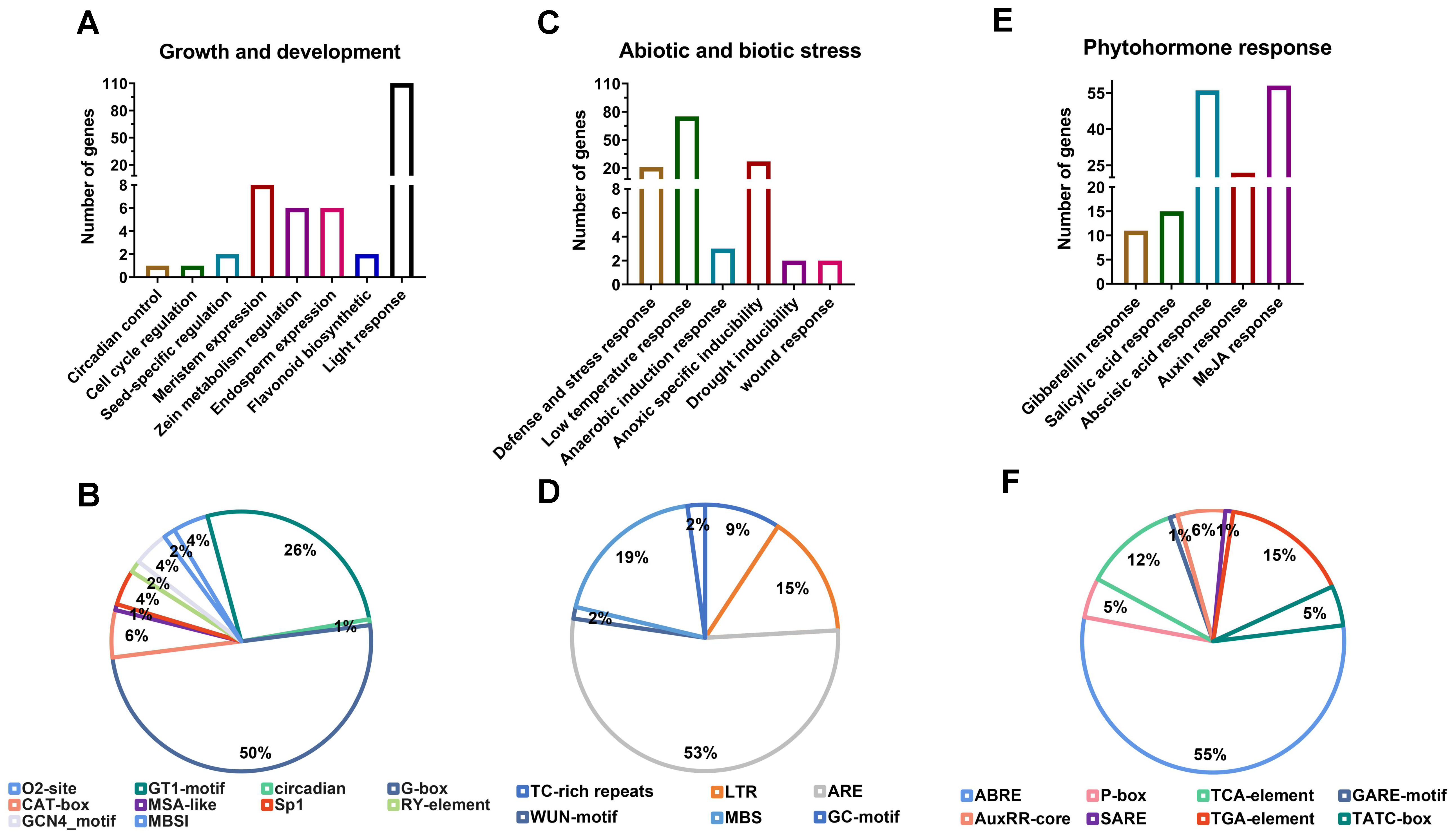
2.4. Cis-Acting Elements and Transcription Factor Binding Sites Analysis in the Promoter Region of NnCSL Family Genes
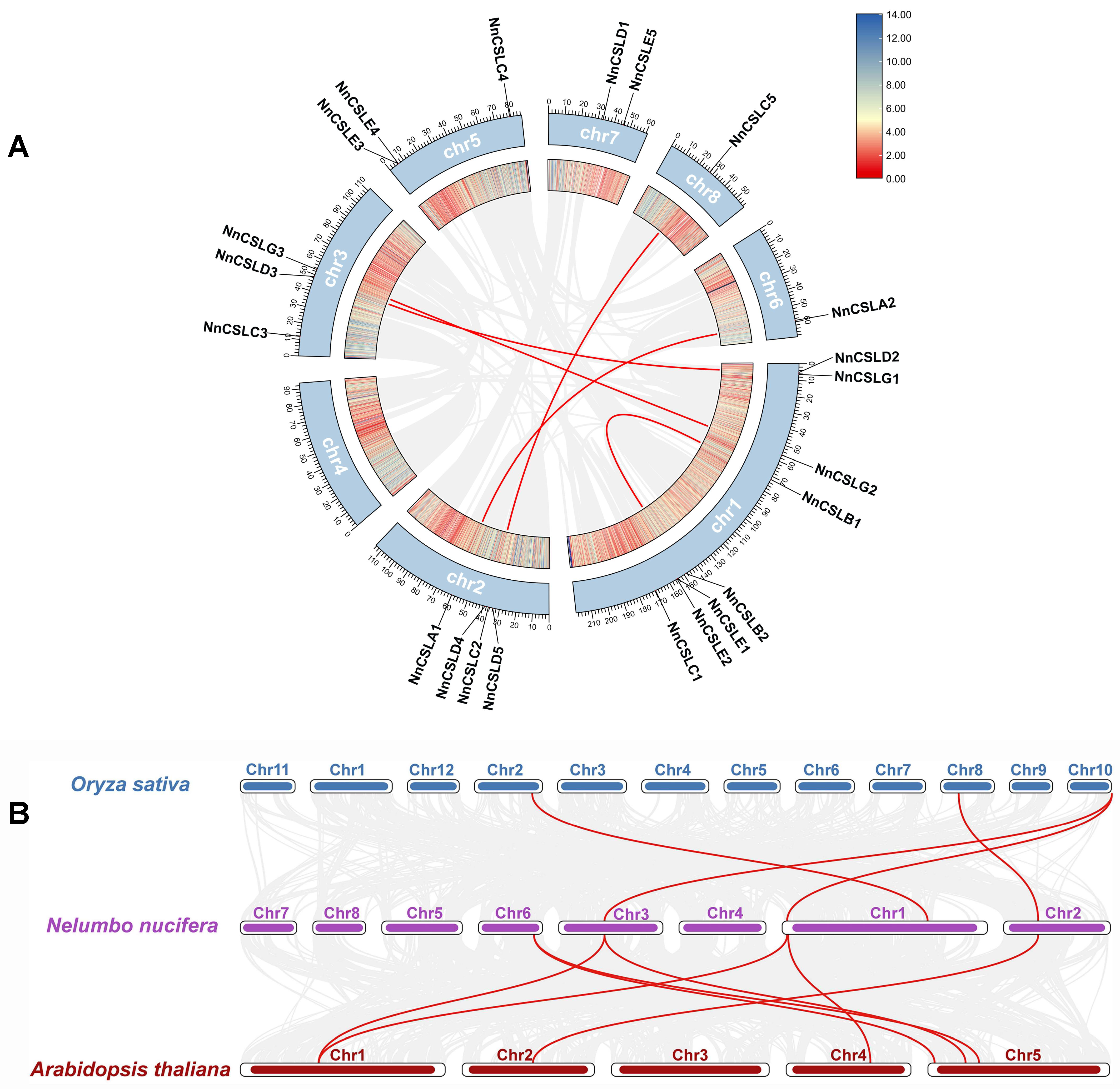
2.5. Gene Replication Events and Collinearity Analysis of CSL Family Genes
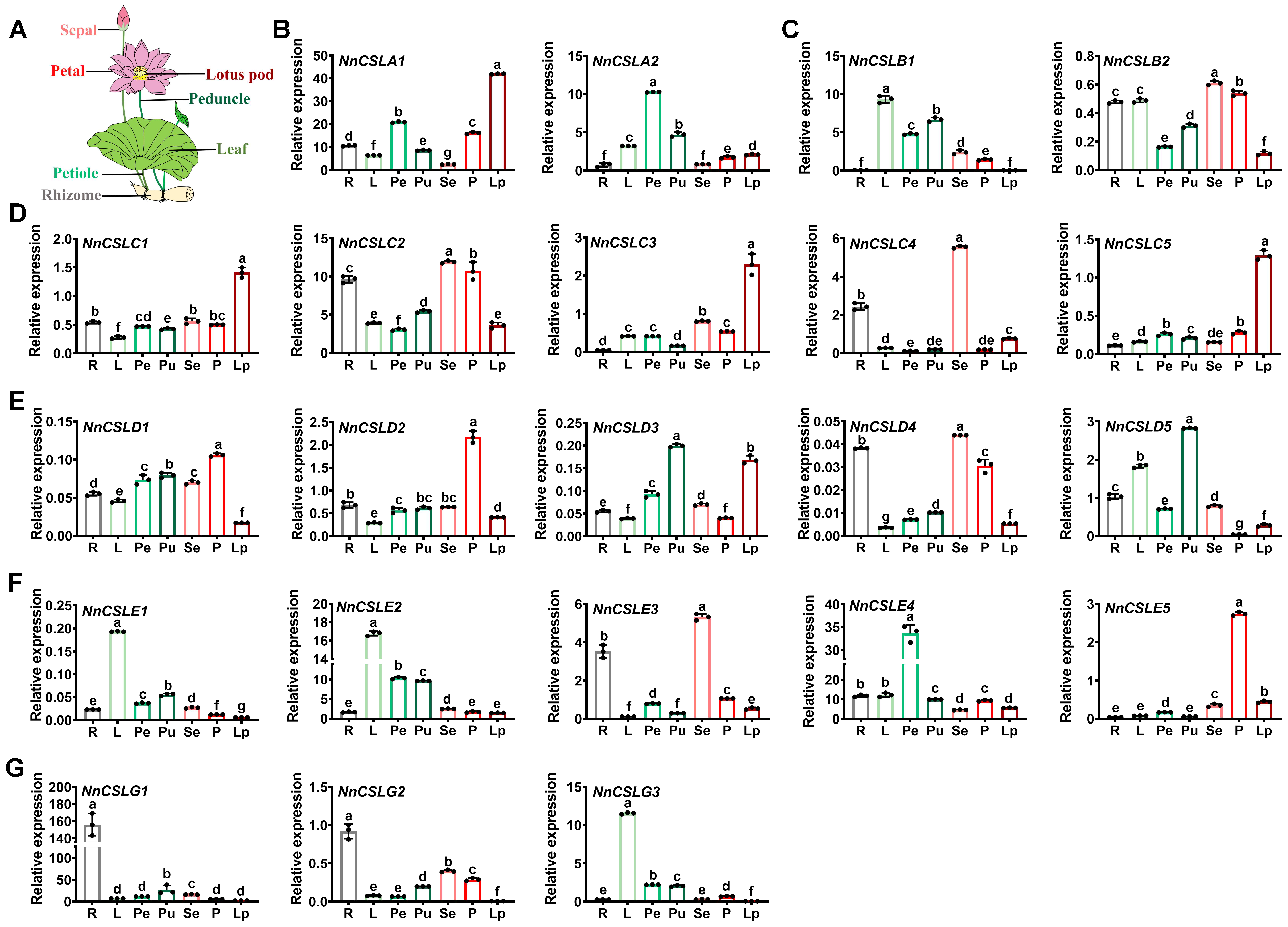
2.6. Tissue Expression Patterns of NnCSL Genes in N. nucifera
2.7. Identification of NnCSL Genes as Candidates for Petiole Elongation in N. nucifera
2.8. Identification of NnCSL Genes as Candidates for Floral Petalization in N. nucifera
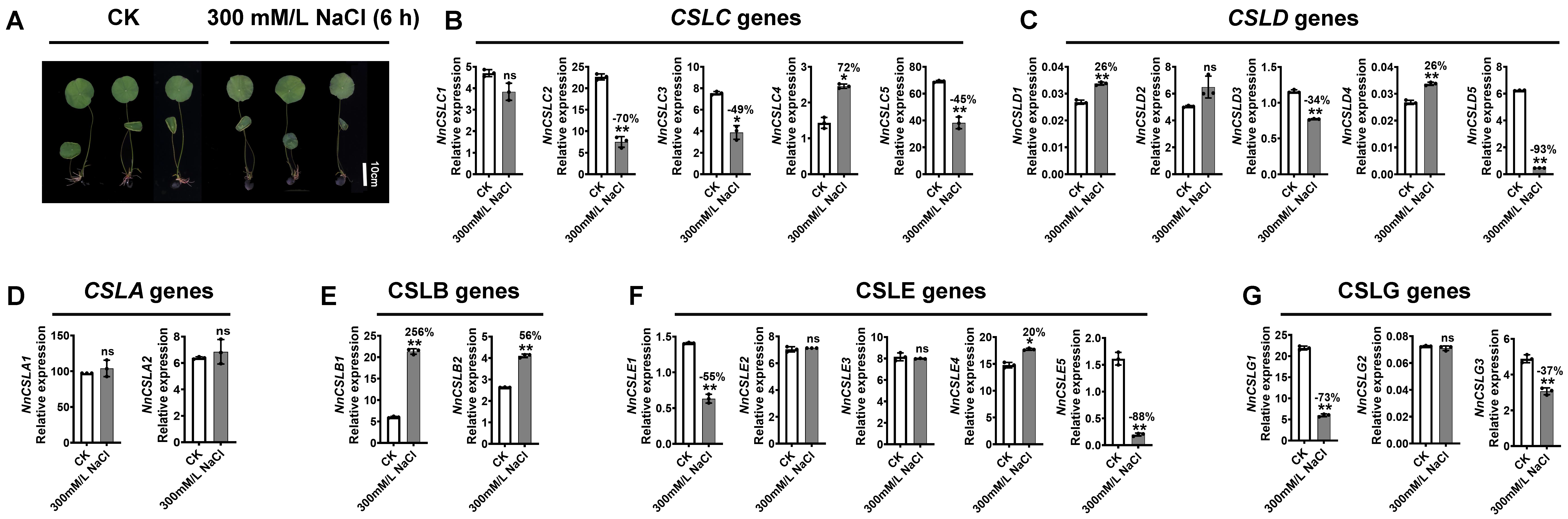
2.9. NnCSL Genes Responding to Salinity Stress Antagonize Growth and Development in N. nucifera
3. Discussion
3.1. Characteristics of the Lotus CSL Gene Family Indicative of Stronger Dicotyledonous Attributes
3.2. Roles of NnCSLs in Plant Growth and Development
3.3. Roles of NnCSLs in Response to Salinity Stress Antagonize Growth and Development
3.4. Potential Complex Regulatory Networks of NnCSLs Involved in Plant Growth and Abiotic Stress Responses
4. Materials and Methods
4.1. Plant Materials and Sample Collection
4.2. Salinity Stress Treatment
4.3. Identification and Property Analysis of NnCSLs Family Genes
4.4. Phylogenetic Analysis
4.5. Chromosome Localization, Gene Structure, Motif Distribution, and Conserved Domains of NnCSL Family Genes
4.6. Secondary and 3D Structure Analysis of NnCSL Family Genes
4.7. Cis-Acting Elements Prediction and Transcription Factor Binding Sites Analysis of NnCSLs Promoter
4.8. Gene Duplication and Synteny Analyses
4.9. RNA-Seq and Analysis
4.10. Nucleic Acid Isolation and qRT-PCR Analysis
4.11. Heatmap of Enriched Correlations Between Candidate NnCSL Genes and TFs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doblin, M.S.; Melis, L.D.; Newbigin, E.; Bacic, A.; Read, S.M. Pollen tubes of nicotiana alata express two genes from different B-Glucan synthase families. Plant Physiol. 2001, 125, 2040–2052. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Wilson, S.M.; Hrmova, M.; Harvey, A.J.; Shirley, N.J.; Medhurst, A.; Stone, B.A.; Newbigin, E.J.; Bacic, A.; Fincher, G.B. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-Beta-D-Glucans. Science 2006, 311, 1940–1942. [Google Scholar] [CrossRef]
- Hu, H.Z.; Zhang, R.; Feng, S.Q.; Wang, Y.M.; Wang, Y.T.; Fan, C.F.; Li, Y.; Liu, Z.Y.; Schneider, R.; Xia, T.; et al. Three AtCesA6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnol. J. 2018, 16, 976–988. [Google Scholar] [CrossRef]
- Purushotham, P.; Ho, R.; Zimmer, J. Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 2020, 369, 1089–1094. [Google Scholar] [CrossRef]
- McFarlane, H.E.; Döring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Somerville, C. Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 2006, 22, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Fan, C.F.; Hu, H.Z.; Li, Y.; Sun, D.; Wang, Y.M.; Peng, L.C. Genetic modification of plant cell walls to enhance biomass yield and biofuel production in bioenergy crops. Biotechnol. Adv. 2016, 34, 997–1017. [Google Scholar] [CrossRef]
- Zhang, R.; Hu, Z.; Wang, Y.T.; Hu, H.Z.; Li, F.C.; Li, M.; Ragauskas, A.; Xia, T.; Han, H.Y.; Tang, J.F.; et al. Single-molecular insights into the breakpoint of cellulose nanofibers assembly during saccharification. Nat. Commun. 2023, 14, 1100. [Google Scholar] [CrossRef]
- Zhao, H.Q.; Wang, S.; Yang, R.J.; Yang, D.M.; Zhao, Y.J.; Kuang, J.H.; Chen, L.Q.; Zhang, R.; Hu, H.Z. Side chain of confined xylan affects cellulose integrity leading to bending stem with reduced mechanical strength in ornamental plants. Carbohydr. Polym. 2024, 329, 121787. [Google Scholar] [CrossRef]
- Hu, H.Z.; Zhang, R.; Zhao, Y.; Yang, J.; Zhao, H.Q.; Zhao, L.; Wang, L.; Cheng, Z.P.; Zhao, W.Y.; Wang, B.; et al. Cell wall remodeling confers plant architecture with distinct wall structure in Nelumbo nucifera. Plant J. 2024, 20, 1392–1409. [Google Scholar] [CrossRef]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a systems approach to understanding plant cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Richmond, T.A.; Somerville, C.R. The cellulose synthase superfamily. Plant Physiol. 2000, 124, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Li, L.G.; Sun, Y.H.; Chiang, V.L. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in populus trichocarpa. Plant Physiol. 2006, 142, 1233–1245. [Google Scholar] [CrossRef]
- Doblin, M.S.; Pettolino, F.; Bacic, A. Plant cell walls: The skeleton of the plant world. Funct. Plant Biol. 2010, 37, 357–381. [Google Scholar] [CrossRef]
- Wang, L.Q.; Guo, K.; Li, Y.; Tu, Y.Y.; Hu, H.Z.; Wang, B.R.; Cui, X.C.; Peng, L.C. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010, 10, 282. [Google Scholar] [CrossRef]
- Little, A.; Schwerdt, J.G.; Shirley, N.J.; Khor, S.F.; Neumann, K.; O’Donovan, L.A.; Lahnstein, J.; Collins, H.M.; Henderson, M.; Fincher, G.B.; et al. Revised phylogeny of the Cellulose Synthase gene superfamily: Insights into cell wall evolution. Plant Physiol. 2018, 177, 1124–1141. [Google Scholar] [CrossRef]
- Cao, S.J.; Cheng, H.; Zhang, J.S.; Aslam, M.; Yan, M.; Hu, A.; Lin, L.L.; Ojolo, S.P.; Zhao, H.M.; Priyadarshani, S.V.G.N.; et al. Genome-wide identification, expression pattern analysis and evolution of the Ces/Csl gene superfamily in Pineapple (Ananas comosus). Plants 2019, 8, 275. [Google Scholar] [CrossRef]
- Li, G.H.; Liu, X.; Liang, Y.X.; Zhang, Y.; Cheng, X.; Cai, Y.P. Genome-wide characterization of the cellulose synthase gene superfamily in Pyrus bretschneideri and reveal its potential role in stone cell formation. Funct. Integr. Genom. 2020, 20, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Daras, G.; Templalexis, D.; Avgeri, F.; Tsitsekian, D.; Karamanou, K.; Rigas, S. Updating insights into the catalytic domain properties of plant Cellulose Synthase (CesA) and Cellulose Synthase-like (Csl) proteins. Molecules 2021, 26, 4335. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Liu, J.; Takáč, T.; Chen, H.; Li, X.; Meng, J.; Tan, Y.; Ning, T.; He, Z.; Yi, G.; et al. Genome-wide identification of banana Csl gene family and their different responses to low temperature between chilling-sensitive and tolerant cultivars. Plants 2021, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Z.; Zhao, K.K.; Chen, Y.; Wei, Q.Z.; Chen, X.Y.; Wan, H.J.; Sun, C.B. Species-specific gene expansion of the cellulose synthase gene superfamily in the orchidaceae family and functional divergence of mannan synthesis-related genes in Dendrobium officinale. Front. Plant Sci. 2022, 13, 777332. [Google Scholar] [CrossRef]
- Huang, H.X.; Zhao, S.; Chen, J.L.; Li, T.X.; Guo, G.G.; Xu, M.; Liao, S.F.; Wang, R.X.; Lan, J.Y.; Su, Y.X.; et al. Genome-wide identification and functional analysis of cellulose synthase gene superfamily in Fragaria vesca. Front. Plant Sci. 2022, 13, 1044029. [Google Scholar] [CrossRef]
- Niu, N.; Zhang, Y.; Li, S.J.; Meng, X.R.; Liu, M.J.; Wang, H.B.; Zhao, J. Genome-wide characterization of the cellulose synthase gene family in Ziziphus jujuba reveals its function in cellulose biosynthesis during fruit development. Int. J. Biol. Macromol. 2023, 239, 124360. [Google Scholar] [CrossRef]
- Zhang, S.S.; Hu, H.B.; Cui, S.M.; Yan, L.; Wu, B.; Wei, S.J. Genome-wide identification and functional analysis of the cellulose synthase-like gene superfamily in common oat (Avena sativa L.). Phytochemistry 2024, 218, 113940. [Google Scholar] [CrossRef]
- Bernal, A.J.; Jensen, J.K.; Harholt, J.; Sørensen, S.; Moller, I.; Blaukopf, C.; Johansen, B.; de Lotto, R.; Pauly, M.; Scheller, H.V.; et al. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 2007, 52, 791–802. [Google Scholar] [CrossRef]
- Park, S.; Szumlanski, A.L.; Gu, F.; Guo, F.; Nielsen, E. A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 2011, 13, 973–980. [Google Scholar] [CrossRef]
- Verhertbruggen, Y.; Yin, L.; Oikawa, A.; Scheller, H.V. Mannan synthase activity in the CSLD family. Plant Signal. Behav. 2011, 6, 1620–1623. [Google Scholar] [CrossRef]
- Hu, H.Z.; Zhang, R.; Dong, S.C.; Li, Y.; Fan, C.F.; Wang, Y.T.; Xia, T.; Chen, P.; Wang, L.Q.; Feng, S.S.; et al. AtCSLD3 and GhCSLD3 mediate root growth and cell elongation downstream of the ethylene response pathway in Arabidopsis. J. Exp. Bot. 2018, 69, 1065–1080. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.Z.; Zhang, R.; Tang, Y.W.; Peng, C.L.; Wu, L.M.; Feng, S.Q.; Chen, P.; Wang, Y.T.; Du, X.Z.; Peng, L.C. Cotton CSLD3 restores cell elongation and cell wall integrity mainly by enhancing primary cellulose production in the Arabidopsis cesa6 mutant. Plant Mol. Biol. 2019, 101, 389–401. [Google Scholar] [CrossRef]
- Yang, J.J.; Bak, G.; Burgin, T.; Barnes, W.J.; Mayes, H.B.; Peña, M.J.; Urbanowicz, B.R.; Nielsen, E. Biochemical and genetic analysis identify CSLD3 as a beta-1,4-glucan synthase that functions during plant cell wall synthesis. Plant Cell 2020, 32, 1749–1767. [Google Scholar] [CrossRef] [PubMed]
- Bernal, A.J.; Yoo, C.M.; Mutwil, M.; Jensen, J.K.; Hou, G.; Blaukopf, C.; Sørensen, I.; Blancaflor, E.B.; Scheller, H.V.; Willats, W.G.T. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 2008, 148, 1238–1253. [Google Scholar] [CrossRef]
- Li, X.; Tang, C.; Li, X.Q.; Zhu, X.X.; Cai, Y.L.; Wang, P.; Zhang, S.L.; Wu, J.Y. Cellulose accumulation mediated by PbrCSLD5, a cellulose synthase-like protein, results in cessation of pollen tube growth in Pyrus bretschneideri. Physiol. Plant. 2022, 174, 13700. [Google Scholar] [CrossRef] [PubMed]
- Favery, B.; Ryan, E.; Foreman, J.; Linstead, P.; Boudonck, K.; Steer, M.; Shaw, P.; Dolan, L. LKOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001, 15, 79–89. [Google Scholar] [CrossRef]
- Wang, X.; Cnops, G.; Vanderhaeghen, R.; De Block, S.; Van Montagu, M.; Van Lijsebettens, M. AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol. 2001, 126, 575–586. [Google Scholar] [CrossRef]
- Kim, C.M.; Park, S.H.; Je, B.I.; Park, S.H.; Park, S.J.; Piao, H.L.; Eun, M.Y.; Dolan, L.; Han, C.D. OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol. 2007, 143, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Penning, B.W.; Hunter, C.T.; Tayengwa, R.; Eveland, A.L.; Dugard, C.K.; Olek, A.T.; Vermerris, W.; Koch, K.E.; McCarty, D.R.; Davis, M.F.; et al. Genetic resources for maize cell wall biology. Plant Physiol. 2009, 151, 1703–1728. [Google Scholar] [CrossRef]
- Karas, B.J.; Ross, L.; Novero, M.; Amyot, L.; Shrestha, A.; Inada, S.; Nakano, M.; Sakai, T.; Bonetta, D.; Sato, S.; et al. Intragenic complementation at the Lotus japonicus CELLULOSE SYNTHASE-LIKE D1 locus rescues root hair defects. Plant Physiol. 2021, 186, 2037–2050. [Google Scholar] [CrossRef]
- Li, M.; Xiong, G.Y.; Li, R.; Cui, J.J.; Tang, D.; Zhang, B.C.; Pauly, M.; Cheng, Z.K.; Zhou, Y.H. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesisand plant growth. Plant J. 2009, 60, 1055–1069. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.J.; Liu, Y.Q.; Zhang, F.X.; Song, Y.L.; Wang, Z.Y.; Peng, Y.K.; Sun, Z.X. OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotechnol. J. 2011, 9, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Eiguchi, M.; Hibara, K.I.; Ito, J.I.; Nagato, Y. Rice SLENDER LEAF 1 gene encodes Cellulose Synthase-like D4 and is specifically expressed in M-Phase cells to regulate cell proliferation. J. Exp. Bot. 2013, 64, 2049–2061. [Google Scholar] [CrossRef]
- Li, F.C.; Xie, G.S.; Huang, J.F.; Zhang, R.; Yu, L.; Zhang, M.M.; Wang, Y.T.; LI, A.; Li, X.K.; Xia, T.; et al. OsCESA9 conserved-site mutation leads to largely enhanced plant lodging resistance and biomass enzymatic saccharification by reducing cellulose DP and crystallinity in rice. Plant Biotechnol. J. 2017, 15, 1093–1104. [Google Scholar] [CrossRef]
- Qiao, L.; Wu, Q.L.; Yuan, L.Z.; Huang, X.D.; Yang, Y.T.; Li, Q.Y.; Shahzad, N.; Li, H.F.; Li, W.Q. SMALL PLANT AND ORGAN 1 (SPO1) encoding a cellulose synthase-like protein D4 (OsCSLD4) is an important regulator for plant architecture and organ size in rice. Int. J. Mol. Sci. 2023, 24, 16974. [Google Scholar] [CrossRef]
- Gu, F.W.; Bringmann, M.; Combs, J.R.; Yang, J.Y.; Bergmann, D.C.; Nielsen, E. Arabidopsis CSLD5 functions in cell plate formation in a cell cycle-dependent manner. Plant Cell 2016, 28, 1722–1737. [Google Scholar] [CrossRef]
- Zhu, J.H.; Lee, B.H.; Dellinger, M.; Cui, X.P.; Zhang, C.Q.; Wu, S.; Nothnagel, E.A.; Zhu, J.K. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010, 63, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Li, Z.X.; Wang, Y.Y.; Xiao, M.G.; Liu, H.; Quan, R.D.; Zhang, H.W.; Hang, R.F.; Zhu, L.; Zhang, Z. Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 2022, 20, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Liepman, A.H.; Cavalier, D.M. The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: Recent advances and future perspectives. Front. Plant Sci. 2012, 3, 109. [Google Scholar] [CrossRef]
- Liepman, A.H.; Wilkerson, C.G.; Keegstra, K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA 2005, 102, 2221–2226. [Google Scholar] [CrossRef]
- Dhugga, K.S.; Barreiro, R.; Whitten, B.; Stecca, K.; Hazebroek, J.; Randhawa, G.S.; Dolan, M.; Kinney, A.J.; Tomes, D.; Nichols, S.; et al. Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science 2004, 303, 363–366. [Google Scholar] [CrossRef]
- Goubet, F.; Barton, C.J.; Mortimer, J.C.; Yu, X.L.; Zhang, Z.N.; Miles, G.P.; Richens, J.; Liepman, A.H.; Seffen, K.; Dupree, P. Cell wall glucomannan in Arabidopsis is synthesised by CSLA Glycosyltransferases, and influences the progression of embryogenesis. Plan. J. 2009, 60, 527–538. [Google Scholar] [CrossRef]
- Kim, S.J.; Chandrasekar, B.; Rea, A.C.; Danhof, L.; Durfee, S.Z.; Thrower, N.; Shepard, Z.S.; Pauly, M.; Brandizzi, F.; Keegstra, k. The synthesis of xyloglucan, an abundant plant cell wall polysaccharide, requires CSLC function. Proc. Natl. Acad. Sci. USA 2020, 117, 20316–20324. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gao, Y.H.; Zhang, B.C.; Yang, H.L.; Tian, Y.B.; Huang, Y.H.; Yin, C.C.; Tao, J.J.; Wei, W.; Zhang, W.K.; et al. CELLULOSE SYNTHASE-LIKE C proteins modulate cell wall establishment during ethylene-mediated root growth inhibition in rice. Plant Cell 2024, 36, 3751–3769. [Google Scholar] [CrossRef] [PubMed]
- Daher, F.B.; Serra, L.; Carter, R.; Jönsson, H.; Robinson, S.; Meyerowitz, E.M.; Gray, W.M. Xyloglucan deficiency leads to a reduction in turgor pressure and changes in cell wall properties, affecting early seedling establishment. Curr. Biol. 2024, 34, 2094–2106.e6. [Google Scholar] [CrossRef]
- Vega-Sánchez, M.E.; Verhertbruggen, Y.; Christensen, U.; Chen, X.W.; Sharma, V.; Varanasi, P.; Jobling, S.A.; Talbot, M.; White, R.G.; Joo, M.; et al. Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 2012, 159, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Zemelis-Durfee, S.; Mckinley, B.; Sokoloski, R.; Aufdemberge, W.; Mullet, J.; Brandizzi, F. Cell- and development-specific degradation controls the levels of mixed-linkage glucan in sorghum leaves. Plant J. 2023, 116, 360–374. [Google Scholar] [CrossRef]
- Purushotham, P.; Ho, R.; Yu, L.; Fincher, G.B.; Bulone, V.; Zimmer, J. Mechanism of mixed-linkage glucan biosynthesis by barley cellulose synthase-like CslF6 (1,3;1,4)-β-glucan synthase. Sci. Adv. 2022, 8, eadd1596. [Google Scholar] [CrossRef]
- Little, A.; Lahnstein, J.; Jeffery, D.W.; Khor, S.F.; Schwerdt, J.G.; Shirley, N.J.; Hooi, M.; Xing, X.; Burton, R.A.; Bulone, V. A novel (1,4)-β-Linked glucoxylan is synthesized by members of the cellulose synthase-like F gene family in land plants. ACS Cent. Sci. 2019, 5, 73–84. [Google Scholar] [CrossRef]
- Lou, H.Y.; Tucker, M.R.; Shirley, N.J.; Lahnstein, J.; Yang, X.J.; Ma, C.; Schwerdt, J.; Fusi, R.; Burton, R.A.; Band, L.R.; et al. The cellulose synthase-like F3 (CslF3) gene mediates cell wall polysaccharide synthesis and affects root growth and differentiation in barley. Plant J. 2022, 110, 1681–1699. [Google Scholar] [CrossRef]
- Wang, Q.Z.; Zhang, X.Y. Lotus Flower Cultivars in China; China Forestry Publishing House: Beijing, China, 2005. [Google Scholar]
- Lin, Z.Y.; Zhang, C.; Cao, D.D.; Damaris, R.N.; Yang, P.F. The latest studies on lotus (Nelumbo nucifera)-an emerging horticultural model plant. Int. J. Mol. Sci. 2019, 20, 3680. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, D.; Xu, J.; Jiang, J.; Jiang, H.W. The protective effect of cold acclimation on the low temperature stress of the lotus (Nelumbo nucifera). Hortic. Sci. 2022, 49, 29–37. [Google Scholar] [CrossRef]
- Vartapetian, B.B.; Andreeva, I.N.; Generozova, I.P.; Polyakova, L.I.; Maslova, I.P.; Dolgikh, Y.I.; Stepanova, A.Y. Functional electron microscopy in studies of plant response and adaptation to anaerobic stress. Ann. Bot. 2003, 91, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zhang, Q.Y.; Li, M.H.; Zhai, J.W.; Wu, S.S.; Ahmad, S.; Lan, S.; Peng, D.H.; Liu, Z.J. Genome-wide identification and expression pattern analysis of TIFY family genes reveal Their potential roles in Phalaenopsis aphrodite flower opening. Int. J. Mol. Sci. 2024, 25, 5422. [Google Scholar] [CrossRef]
- Su, Y.T.; Fang, J.Y.; Zeeshan Ul Haq, M.; Yang, W.L.; Yu, J.; Yang, D.M.; Liu, Y.; Wu, Y.G. Genome-wide identification and expression analysis of the casparian strip membrane domain protein-like gene family in Peanut (Arachis hypogea L.) revealed its crucial role in growth and multiple stress tolerance. Plants 2024, 13, 2077. [Google Scholar] [CrossRef]
- Wu, H.P.; Zhang, R.L.; Diao, X.M. Genome-wide characterization and haplotypic variation analysis of the IDD gene family in Foxtail Millet (Setaria italica). Int. J. Mol. Sci. 2024, 25, 8804. [Google Scholar] [CrossRef]
- Chen, J.T.; Aroca, R.; Romano, D. Molecular aspects of plant salinity stress and tolerance. Int. J. Mol. Sci. 2021, 22, 4918. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Gabaldón, T.; Koonin, E.V. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013, 14, 360–366. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Chen, C.; Xiong, G.; Tan, X.Y.; Yang, K.Z.; Wang, Z.C.; Zhou, Y.; Ye, D.; Chen, L.Q. Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J. Exp. Bot. 2011, 62, 5161–5177. [Google Scholar] [CrossRef]
- Du, P.; Wang, Q.; Yuan, D.Y.; Chen, S.S.; Su, Y.N.; Li, L.; Chen, S.; He, X.J. WRKY transcription factors and OBERON histone-binding proteins form complexes to balance plant growth and stress tolerance. EMBO J. 2023, 42, e113639. [Google Scholar] [CrossRef] [PubMed]
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, e2000034. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Q.; Song, X.Q.; Zhou, H.J.; Wei, K.L.; Jiang, C.; Wang, J.N.; Cao, Y.; Tang, F.; Zhao, S.T.; Lu, M.Z. KNAT2/6b, a class I KNOX gene, impedes xylem differentiation by regulating NAC domain transcription factors in poplar. New Phytol. 2020, 225, 1531–1544. [Google Scholar] [CrossRef]
- Han, K.J.; Zhao, Y.; Sun, Y.H.; Li, Y. NACs, generalist in plant life. Plant Biotechnol. J. 2023, 21, 2433–2457. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Kobayashi, Y.; Koyama, H.; Sahoo, L. Cowpea NAC1/NAC2 transcription factors improve growth and tolerance to drought and heat in transgenic cowpea through combined activation of photosynthetic and antioxidant mechanisms. J. Integr. Plant Biol. 2023, 65, 25–44. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L.J.; Lei, J.; Chai, S.S.; Jin, X.J.; Zou, Y.Y.; Sun, X.Q.; Mei, Y.Q.; Cheng, X.L.; Yang, X.S.; et al. IbMYB308, a sweet potato R2R3-MYB gene, improves salt stress tolerance in transgenic tobacco. Genes 2022, 13, 1476. [Google Scholar] [CrossRef]
- Wang, T.T.; Jin, Y.; Deng, L.X.; Li, F.; Wang, Z.Y.; Zhu, Y.Y.; Wu, Y.F.; Qu, H.Y.; Zhang, S.N.; Liu, Y.; et al. The transcription factor MYB110 regulates plant height, lodging resistance, and grain yield in rice. Plant Cell 2024, 36, 298–323. [Google Scholar] [CrossRef]
- Tsuda, K.; Maeno, A.; Otake, A.; Kato, K.; Tanaka, W.; Hibara, K.I.; Nonomura, K.I. YABBY and diverged KNOX1 genes shape nodes and internodes in the stem. Science 2024, 384, 1241–1247. [Google Scholar] [CrossRef]
- Nan, G.L.; Teng, C.; Fernandes, J.; O’Connor, L.; Meyers, B.C.; Walbot, V. A cascade of bHLH-regulated pathways programs maize anther development. Plant Cell 2022, 34, 1207–1225. [Google Scholar] [CrossRef]
- Gao, H.; Song, W.; Severing, E.; Vayssières, A.; Huettel, B.; Franzen, R.; Richter, R.; Chai, J.J.; Coupland, G. PIF4 enhances DNA binding of CDF2 to co-regulate target gene expression and promote Arabidopsis hypocotyl cell elongation. Nat. Plants 2022, 8, 1082–1093. [Google Scholar] [CrossRef]
- Wang, H.P.; Chen, W.Q.; Xu, Z.Y.; Chen, M.F.; Yu, D.Q. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023, 28, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Mahato, H.; Choudhury, U.; Thakur, R.S.; Debnath, P.; Ansari, N.G.; Sane, V.A.; Sane, A.P. The tomato EAR-motif repressor, SlERF36, accelerates growth transitions and reduces plant life cycle by regulating GA levels and responses. Plant Biotechnol. J. 2024, 22, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Colquhoun, T.A.; Schwieterman, M.L.; Wedde, A.E.; Schimmel, B.C.; Marciniak, D.M.; Verdonk, J.C.; Kim, J.Y.; Oh, Y.; Gális, I.; Baldwin, I.T.; et al. EOBII controls flower opening by functioning as a general transcriptomic switch. Plant Physiol. 2011, 156, 974–984. [Google Scholar] [CrossRef]
- Schubert, R.; Dobritzsch, S.; Gruber, C.; Hause, G.; Athmer, B.; Schreiber, T.; Marillonnet, S.; Okabe, Y.; Ezura, H.; Acosta, I.F.; et al. Tomato MYB21 acts in ovules to mediate jasmonate-regulated fertility. Plant Cell 2019, 31, 1043–1062. [Google Scholar] [CrossRef]
- Song, S.S.; Qi, T.C.; Huang, H.; Ren, Q.C.; Wu, D.W.; Chang, C.Q.; Peng, W.; Liu, Y.L.; Peng, J.R.; Xie, D.X. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Battat, M.; Eitan, A.; Rogachev, I.; Hanhineva, K.; Fernie, A.; Tohge, T.; Beekwilder, J.; Aharoni, A.A. MYB triad controls primary and phenylpropanoid metabolites for pollen coat patterning. Plant Physiol. 2019, 180, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Chopy, M.; Binaghi, M.; Cannarozzi, G.; Halitschke, R.; Boachon, B.; Heutink, R.; Bomzan, D.P.; Jäggi, L.; van Geest, G.; Verdonk, J.C.; et al. A single MYB transcription factor with multiple functions during flower development. New Phytol. 2023, 239, 2007–2025. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Li, J.W.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]




| Gene Name | Gene ID | LOC | ORF (bp) | AA(aa) | Mw (KDa) | PI | Instability Index | Aliphatic Index | GRAVY | TMHs | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NnCSLA1 | Nn2g13982.31 | 104612509 | 1638 | 546 | 61.906 | 8.84 | 43.55 | 101.39 | 0.208 | 5 | Plasma membrane |
| NnCSLA2 | Nn6g35072.4 | 104587567 | 1602 | 533 | 61.23 | 9.11 | 36.13 | 99.77 | 0.155 | 5 | Plasma membrane |
| NnCSLB1 | Nn1g03452.7 | 104588296 | 2304 | 767 | 85.923 | 6.47 | 45.6 | 96.62 | 0.067 | 8 | Plasma membrane |
| NnCSLB2 | Nn1g07221.3 | 104594052 | 2100 | 699 | 78.988 | 5.86 | 45.24 | 97.87 | 0.028 | 2 | Plasma membrane |
| NnCSLC1 | Nn1g08101.3 | 104589898 | 2079 | 692 | 79.686 | 8.63 | 43.58 | 101.01 | 0.022 | 5 | Plasma membrane |
| NnCSLC2 | Nn2g12599.4 | 104612365 | 2085 | 694 | 79.761 | 8.71 | 38.34 | 102.85 | 0.096 | 6 | Plasma membrane |
| NnCSLC3 | Nn3g16967.1 | 104595557 | 1989 | 662 | 75.846 | 9.13 | 37.11 | 106.19 | 0.194 | 5 | Plasma membrane |
| NnCSLC4 | Nn5g30690.8 | 104609129 | 2001 | 666 | 76.162 | 8.29 | 39.4 | 95.44 | 0.065 | 6 | Plasma membrane |
| NnCSLC5 | Nn8g40281.1 | 104605331 | 2100 | 699 | 80.629 | 8.81 | 36.45 | 100.3 | 0.072 | 6 | Plasma membrane |
| NnCSLD1 | Nn7g37641.1 | 104598378 | 3249 | 1082 | 120.315 | 6.87 | 43.63 | 80.6 | −0.236 | 6 | Plasma membrane |
| NnCSLD2 | Nn1g00223.6 | 104604803 | 3444 | 1147 | 129.067 | 6.84 | 45.11 | 80.94 | −0.211 | 8 | Plasma membrane |
| NnCSLD3 | Nn3g19380.2 | 104611451 | 3456 | 1151 | 129.122 | 6.64 | 42.26 | 82.17 | −0.189 | 6 | Plasma membrane |
| NnCSLD4 | Nn2g12702.1 | 104599135 | 3378 | 1125 | 126.199 | 6.6 | 42.73 | 79.68 | −0.215 | 8 | Plasma membrane |
| NnCSLD5 | Nn2g12438.4 | 104603242 | 2769 | 938 | 104.642 | 8.24 | 37.64 | 83.73 | −0.16 | 8 | Plasma membrane |
| NnCSLE1 | Nn1g07535.2 | 104594270 | 2028 | 675 | 77.407 | 7.47 | 39.83 | 85.93 | −0.02 | 8 | Plasma membrane |
| NnCSLE2 | Nn1g07538.2 | 104594272 | 2307 | 768 | 87.176 | 8.43 | 45.77 | 92.86 | 0.072 | 6 | Plasma membrane |
| NnCSLE3 | Nn5g27070.3 | 104594007 | 1425 | 474 | 53.794 | 8.92 | 43.05 | 96.88 | 0.07 | 4 | Plasma membrane |
| NnCSLE4 | Nn5g27071.10 | 109114471 | 2199 | 732 | 83.92 | 8.59 | 40.44 | 91.24 | −0.044 | 6 | Plasma membrane |
| NnCSLE5 | Nn7g37928.3 | 104609477 | 2364 | 787 | 89.331 | 8.67 | 40.41 | 86.61 | −0.068 | 8 | Plasma membrane |
| NnCSLG1 | Nn1g00277.23 | 104604823 | 2259 | 752 | 85.034 | 8.7 | 45.83 | 87.65 | 0.075 | 7 | Plasma membrane |
| NnCSLG2 | Nn1g02670.1 | 104610102 | 2034 | 677 | 76.588 | 8.73 | 51.21 | 102.54 | 0.189 | 8 | Plasma membrane |
| NnCSLG3 | Nn3g19586.4 | 104600820 | 2130 | 709 | 80.828 | 7.53 | 40.82 | 95.56 | 0.136 | 8 | Plasma membrane |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Wang, J.; Yang, D.; Xia, W.; Wang, L.; Wang, S.; Zhao, H.; Chen, L.; Hu, H. Genome-Wide Analysis of CSL Family Genes Involved in Petiole Elongation, Floral Petalization, and Response to Salinity Stress in Nelumbo nucifera. Int. J. Mol. Sci. 2024, 25, 12531. https://doi.org/10.3390/ijms252312531
Yang J, Wang J, Yang D, Xia W, Wang L, Wang S, Zhao H, Chen L, Hu H. Genome-Wide Analysis of CSL Family Genes Involved in Petiole Elongation, Floral Petalization, and Response to Salinity Stress in Nelumbo nucifera. International Journal of Molecular Sciences. 2024; 25(23):12531. https://doi.org/10.3390/ijms252312531
Chicago/Turabian StyleYang, Jie, Juan Wang, Dongmei Yang, Wennian Xia, Li Wang, Sha Wang, Hanqian Zhao, Longqing Chen, and Huizhen Hu. 2024. "Genome-Wide Analysis of CSL Family Genes Involved in Petiole Elongation, Floral Petalization, and Response to Salinity Stress in Nelumbo nucifera" International Journal of Molecular Sciences 25, no. 23: 12531. https://doi.org/10.3390/ijms252312531
APA StyleYang, J., Wang, J., Yang, D., Xia, W., Wang, L., Wang, S., Zhao, H., Chen, L., & Hu, H. (2024). Genome-Wide Analysis of CSL Family Genes Involved in Petiole Elongation, Floral Petalization, and Response to Salinity Stress in Nelumbo nucifera. International Journal of Molecular Sciences, 25(23), 12531. https://doi.org/10.3390/ijms252312531






