Genome-Wide Identification of the CAT Genes and Molecular Characterization of Their Transcriptional Responses to Various Nutrient Stresses in Allotetraploid Rapeseed
Abstract
:1. Background
2. Results
2.1. Identification of BnaCAT Family Members and Construction of Phylogenetic Tree in B. napus
2.2. Molecular Characterization of BnaCATs
2.3. Identification of Evolutionary Selection Pressure on BnaCATs
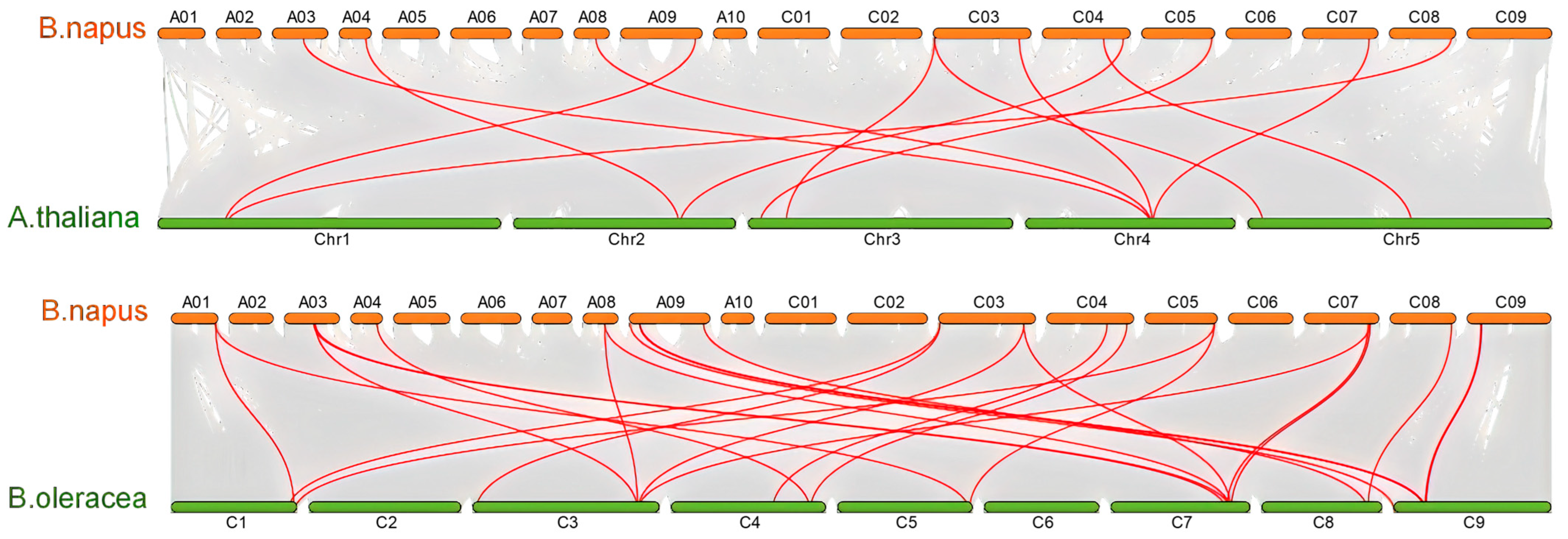
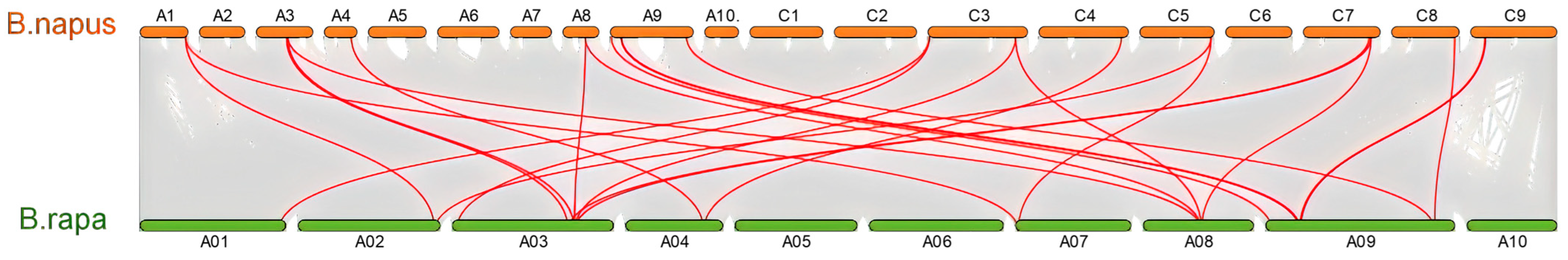
2.4. Chromosomal Distribution and Syntenic Analysis of BnaCATs
2.5. Conserved Motifs, Gene Structure Analysis of BnaCATs
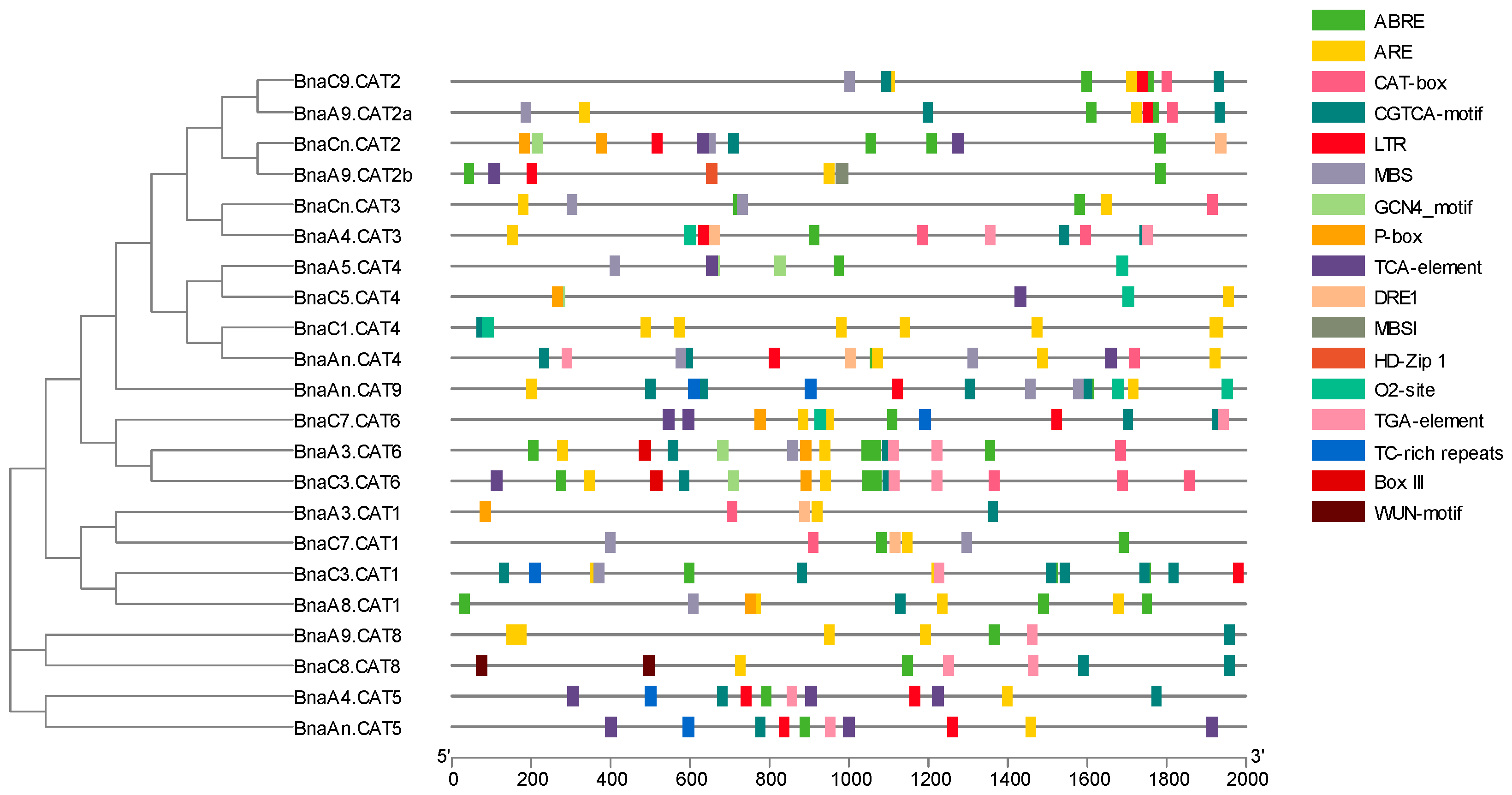
2.6. Cis-Element Analysis of the Promoter Regions of the BnaCATs
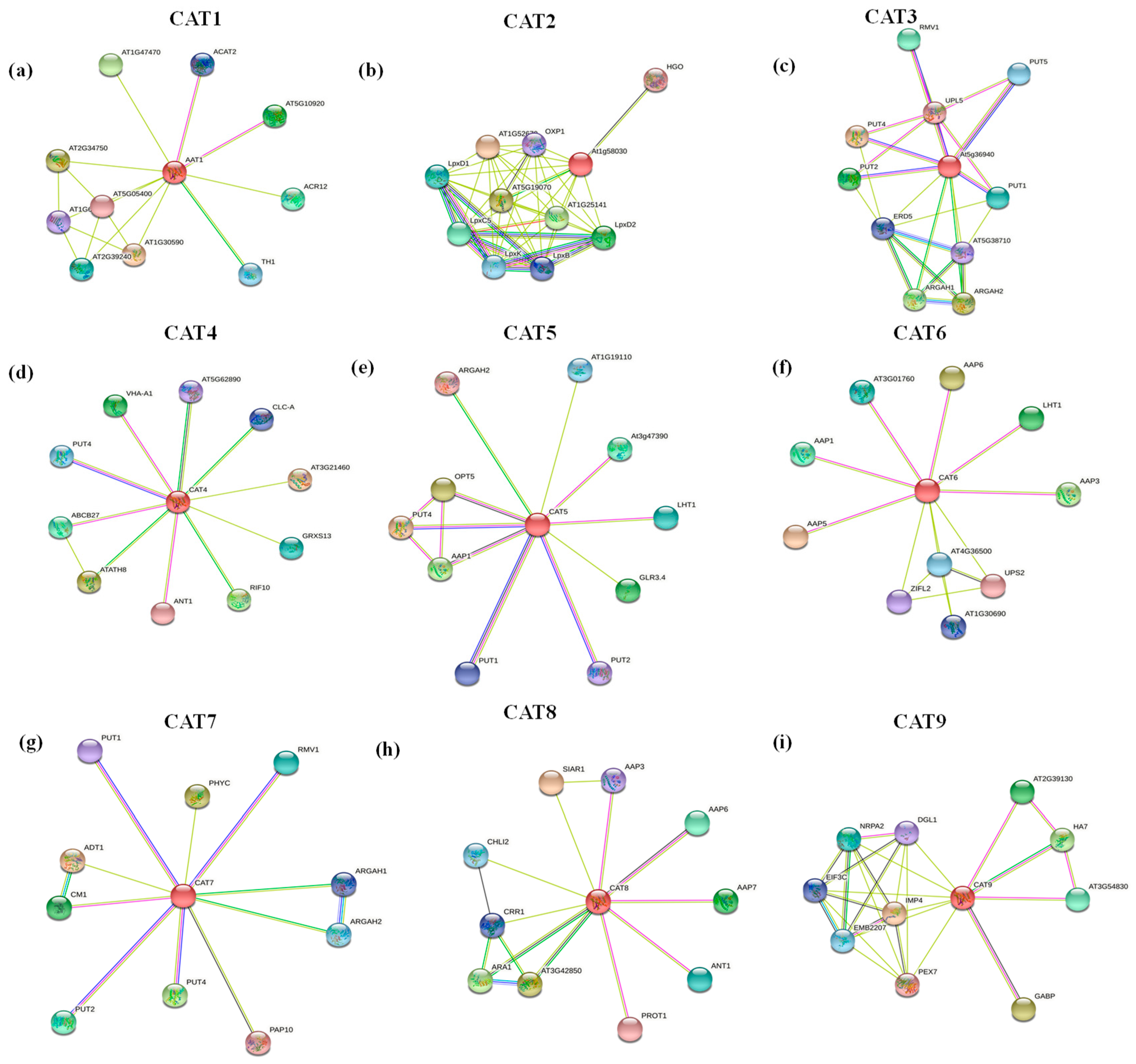
2.7. Protein–Protein Interaction Analysis of BnaCATs
2.8. Expression Profiles of BnaCATs in Response to Diverse Nutrient Stresses
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Identification of Members of the CAT Gene Family
5.2. Gene Nomenclature of CATs in B. napus
5.3. Multiple Sequence Alignment and Phylogeny Analysis of BnaCATs
5.4. Molecular Characterization of BnaCATs
5.5. Analysis of Evolutionary Selection Pressure and Functional Divergence of BnaCATs
5.6. Chromosomal Distribution and Gene Duplication
5.7. Protein Motif, Gene Structure, and Cis-Element Analyses
5.8. Transcriptional Analysis of BnaCATs Under Diverse Nutrient Stresses
5.9. Plant Materials and Treatments
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raza, A.; Su, W.; Gao, A.; Mehmood, S.S.; Hussain, M.A.; Nie, W.; Lv, Y.; Zou, X.; Zhang, X. Catalase (CAT) Gene Family in Rapeseed (Brassica napus L.): Genome-Wide Analysis, Identification, and Expression Pattern in Response to Multiple Hormones and Abiotic Stress Conditions. Int. J. Mol. Sci. 2021, 22, 4281. [Google Scholar] [CrossRef] [PubMed]
- Sarcheshmeh, M.K.; Abedi, A.; Aalami, A. Genome-wide survey of catalase genes in Brassica rapa, Brassica oleracea, and Brassica napus: Identification, characterization, molecular evolution, and expression profiling of BnCATs in response to salt and cadmium stress. Protoplasma 2023, 260, 899–917. [Google Scholar] [CrossRef]
- Yang, G.; Wei, Q.; Huang, H.; Xia, J. Amino Acid Transporters in Plant Cells: A Brief Review. Plants 2020, 9, 967. [Google Scholar] [CrossRef]
- Duan, Y.; Zhu, X.; Shen, J.; Xing, H.; Zou, Z.; Ma, Y.; Wang, Y.; Fang, W. Genome-wide identification, characterization and expression analysis of the amino acid permease gene family in tea plants (Camellia sinensis). Genomics 2020, 112, 2866–2874. [Google Scholar] [CrossRef]
- Frommer, W.B.; Hummel, S.; Riesmeier, J.W. Expression cloning in yeast of a cDNA encoding a broad specificity amino acid permease from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1993, 90, 5944–5948. [Google Scholar] [CrossRef] [PubMed]
- Wipf, D.; Ludewig, U.; Tegeder, M.; Rentsch, D.; Koch, W.; Frommer, W.B. Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 2002, 27, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Postel, S.; Kemmerling, B.; Ludewig, U. Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant Cell Environ. 2014, 37, 1404–1414. [Google Scholar] [CrossRef]
- Yang, H.; Krebs, M.; Stierhof, Y.D.; Ludewig, U. Characterization of the putative amino acid transporter genes AtCAT2, 3 &4: The tonoplast localized AtCAT2 regulates soluble leaf amino acids. J. Plant Physiol. 2014, 171, 594–601. [Google Scholar]
- Su, Y.H.; Frommer, W.B.; Ludewig, U. Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol. 2004, 136, 3104–3113. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, L.; Li, Z. Molecular cloning and identification of a putative tomato cationic amino acid transporter-2 gene that is highly expressed in stamens. Plant Cell Tissue Organ Cult. 2013, 112, 55–63. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, L.; Yun, L.; Ji, L.; Li, G.; Ji, M.; Shi, Y.; Zheng, X. Catalase (CAT) Gene Family in Wheat (Triticum aestivum L.): Evolution, Expression Pattern and Function Analysis. Int. J. Mol. Sci. 2022, 23, 542. [Google Scholar] [CrossRef] [PubMed]
- Aleem, M.; Aleem, S.; Sharif, I.; Aleem, M.; Shahzad, R.; Khan, M.I.; Batool, A.; Sarwar, G.; Farooq, J.; Iqbal, A.; et al. Whole-Genome Identification of APX and CAT Gene Families in Cultivated and Wild Soybeans and Their Regulatory Function in Plant Development and Stress Response. Antioxidants 2022, 11, 1626. [Google Scholar] [CrossRef]
- Hu, L.; Yang, Y.; Jiang, L.; Liu, S. The catalase gene family in cucumber: Genome-wide identification and organization. Genet. Mol. Biol. 2016, 39, 408–415. [Google Scholar] [CrossRef]
- Polidoros, A.N.; Mylona, P.V.; Scandalios, J.G. Transgenic tobacco plants expressing the maize Cat2 gene have altered catalase levels that affect plant-pathogen interactions and resistance to oxidative stress. Transgenic Res. 2001, 10, 555–569. [Google Scholar] [CrossRef]
- Hammes, U.Z.; Nielsen, E.; Honaas, L.A.; Taylor, C.G.; Schachtman, D.P. AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis. Plant J. 2006, 48, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Stierhof, Y.D.; Ludewig, U. The putative Cationic Amino Acid Transporter 9 is targeted to vesicles and may be involved in plant amino acid homeostasis. Front. Plant Sci. 2015, 6, 212. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Hurgobin, B.; Golicz, A.A.; Chan, C.K.; Yuan, Y.; Lee, H.; Renton, M.; Meng, J.; Li, R.; Long, Y.; et al. Assembly and comparison of two closely related Brassica napus genomes. Plant Biotechnol. J. 2017, 15, 1602–1610. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Sun, F.; Fan, G.; Hu, Q.; Zhou, Y.; Guan, M.; Tong, C.; Li, J.; Du, D.; Qi, C.; Jiang, L.; et al. The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 2017, 92, 452–468. [Google Scholar] [CrossRef]
- Ding, X.; Li, J.; Pan, Y.; Zhang, Y.; Ni, L.; Wang, Y.; Zhang, X. Genome-Wide Identification and Expression Analysis of the UGlcAE Gene Family in Tomato. Int. J. Mol. Sci. 2018, 19, 1583. [Google Scholar] [CrossRef]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genomics 2019, 20, 227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.H.; Zhou, T.; Liao, Q.; Yao, J.Y.; Liang, G.H.; Song, H.X.; Guan, C.Y.; Hua, Y.P. Integrated physiologic, genomic and transcriptomic strategies involving the adaptation of allotetraploid rapeseed to nitrogen limitation. BMC Plant Biol. 2018, 18, 322. [Google Scholar] [CrossRef]
- Ding, L.; Wang, K.J.; Jiang, G.M.; Biswas, D.K.; Xu, H.; Li, L.F.; Li, Y.H. Effects of nitrogen deficiency on photosynthetic traits of maize hybrids released in different years. Ann. Bot. 2005, 96, 925–930. [Google Scholar] [CrossRef]
- Zheng, L.W.; Ma, S.J.; Zhou, T.; Yue, C.P.; Hua, Y.P.; Huang, J.Y. Genome-wide identification of Brassicaceae B-BOX genes and molecular characterization of their transcriptional responses to various nutrient stresses in allotetraploid rapeseed. BMC Plant Biol. 2021, 21, 288. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.; Jiao, Y.; Jia, J.; Wang, X.; Li, H.; Shi, W.; Peng, C.; Polle, A.; Luo, Z.B. Phosphorus and nitrogen physiology of two contrasting poplar genotypes when exposed to phosphorus and/or nitrogen starvation. Tree Physiol. 2016, 36, 22–38. [Google Scholar] [CrossRef]
- Amrutha, R.N.; Sekhar, P.N.; Varshney, R.K.; Kishor, P.B.K. Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 2007, 172, 708–721. [Google Scholar] [CrossRef]
- Ashley, M.K.; Grant, M.; Grabov, A. Plant responses to potassium deficiencies: A role for potassium transport proteins. J. Exp. Bot. 2006, 57, 425–436. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhuang, Y.; Li, C.; Sun, X.; Zhao, S.; Ma, C.; Lin, H.; Zhou, H. SIMP1 modulates salt tolerance by elevating ERAD efficiency through UMP1A-mediated proteasome maturation in plants. New Phytol. 2021, 232, 625–641. [Google Scholar] [CrossRef]
- Zhang, Y.; Chao, J.; Li, X.; Zhang, C.; Khan, R.; Du, S.; Xu, N.; Song, L.; Liu, H.; Shi, Y. Comparative transcriptome combined with biochemical and physiological analyses provide new insights toward cadmium accumulation with two contrasting Nicotiana species. Physiol. Plant. 2021, 173, 369–383. [Google Scholar]
- Zhao, H.; Ma, H.; Yu, L.; Wang, X.; Zhao, J. Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS ONE 2012, 7, e49210. [Google Scholar] [CrossRef]
- Cheng, L.; Yuan, H.Y.; Ren, R.; Zhao, S.Q.; Han, Y.P.; Zhou, Q.Y.; Ke, D.X.; Wang, Y.X.; Wang, L. Genome-Wide Identification, Classification, and Expression Analysis of Amino Acid Transporter Gene Family in Glycine Max. Front. Plant Sci. 2016, 7, 515. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, M.; Zhu, D.; Pan, F.; Wang, Y.; Wang, Y.; Xiang, Y. Genome-Wide analysis of the AAAP gene family in moso bamboo (Phyllostachys edulis). BMC Plant Biol. 2017, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-B.; Liu, C.; Tang, D.-Y.; Yan, L.; Wang, D.; Yang, Y.-Z.; Gui, J.-S.; Zhao, X.-Y.; Li, L.-G.; Tang, X.-D.; et al. The Receptor-Like Cytoplasmic Kinase STRK1 Phosphorylates and Activates CatC, Thereby Regulating H2O2 Homeostasis and Improving Salt Tolerance in Rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.-Y.; Wang, P.; Chen, J.; Song, C.-P. Comprehensive Functional Analysis of the Catalase Gene Family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef]
- Su, T.; Wang, P.P.; Li, H.J.; Zhao, Y.W.; Lu, Y.; Dai, P.; Ren, T.Q.; Wang, X.F.; Li, X.Z.; Shao, Q.; et al. The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 2018, 60, 591–607. [Google Scholar] [CrossRef]
- Schmidt, R.; Mieulet, D.; Hubberten, H.-M.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fisahn, J.; Segundo, B.S.; Guiderdoni, E.; Schippers, J.H.M.; et al. SALT-RESPONSIVE ERF1 Regulates Reactive Oxygen Species-Dependent Signaling during the Initial Response to Salt Stress in Rice. Plant Cell 2013, 25, 2115–2131. [Google Scholar] [CrossRef]
- Willekens, H.; Villarroel, R.; Van Montagu, M.; Inzé, D.; Van Camp, W. Molecular identification of catalases from Nicotianaplumbaginifolia (L.). FEBS Lett. 1994, 352, 79–83. [Google Scholar] [CrossRef]
- Matsumura, T.; Tabayashi, N.; Kamagata, Y.; Souma, C.; Saruyama, H. Wheat catalase expressed in transgenic rice can improve tolerance against low temperature stress. Physiol. Plant. 2002, 116, 317–327. [Google Scholar] [CrossRef]
- Ma, H.; Cao, X.; Shi, S.; Li, S.; Gao, J.; Ma, Y.; Zhao, Q.; Chen, Q. Genome-wide survey and expression analysis of the amino acid transporter superfamily in potato (Solanum tuberosum L.). Plant Physiol. Biochem. 2016, 107, 164–177. [Google Scholar] [CrossRef]
- Wan, Y.; King, R.; Mitchell, R.A.C.; Hassani-Pak, K.; Hawkesford, M.J. Spatiotemporal expression patterns of wheat amino acid transporters reveal their putative roles in nitrogen transport and responses to abiotic stress. Sci. Rep. 2017, 7, 5461. [Google Scholar] [CrossRef]
- Wu, X.M.; Kou, S.J.; Liu, Y.L.; Fang, Y.N.; Xu, Q.; Guo, W.W. Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA- and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol. J. 2015, 13, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Aan den Toorn, M.; Albrecht, C.; de Vries, S. On the Origin of SERKs: Bioinformatics Analysis of the Somatic Embryogenesis Receptor Kinases. Mol. Plant 2015, 8, 762–782. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.R.; Zhang, Q.; Bae, D.W.; Kim, T.H. Pod removal responsive change in phytohormones and its impact on protein degradation and amino acid transport in source leaves of Brassica napus. Plant Physiol. Biochem. 2016, 106, 159–164. [Google Scholar] [CrossRef]
- Shin, K.; Lee, S.; Song, W.Y.; Lee, R.A.; Lee, I.; Ha, K.; Koo, J.C.; Park, S.K.; Nam, H.G.; Lee, Y.; et al. Genetic identification of ACC-RESISTANT2 reveals involvement of LYSINE HISTIDINE TRANSPORTER1 in the uptake of 1-aminocyclopropane-1-carboxylic acid in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 572–582. [Google Scholar] [CrossRef]
- Perchlik, M.; Tegeder, M. Improving Plant Nitrogen Use Efficiency through Alteration of Amino Acid Transport Processes. Plant Physiol. 2017, 175, 235–247. [Google Scholar] [CrossRef]
- Pratelli, R.; Pilot, G. Regulation of amino acid metabolic enzymes and transporters in plants. J. Exp. Bot. 2014, 65, 5535–5556. [Google Scholar] [CrossRef]
- Avice, J.C.; Etienne, P. Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.). J. Exp. Bot. 2014, 65, 3813–3824. [Google Scholar] [CrossRef]
- Santiago, J.P.; Tegeder, M. Connecting Source with Sink: The Role of Arabidopsis AAP8 in Phloem Loading of Amino Acids. Plant Physiol. 2016, 171, 508–521. [Google Scholar] [CrossRef]
- Couturier, J.; Doidy, J.; Guinet, F.; Wipf, D.; Blaudez, D.; Chalot, M. Glutamine, arginine and the amino acid transporter Pt-CAT11 play important roles during senescence in poplar. Ann. Bot. 2010, 105, 1159–1169. [Google Scholar] [PubMed]
- Liu, Y.; von Wirén, N. Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Bot. 2017, 68, 2581–2592. [Google Scholar]
- Sun, S.K.; Xu, X.; Tang, Z.; Tang, Z.; Huang, X.Y.; Wirtz, M.; Hell, R.; Zhao, F.J. A molecular switch in sulfur metabolism to reduce arsenic and enrich selenium in rice grain. Nat. Commun. 2021, 12, 1392. [Google Scholar]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar]
- Wang, X.; Wu, J.; Liang, J.; Cheng, F.; Wang, X. Brassica database (BRAD) version 2.0: Integrating and mining Brassicaceae species genomic resources. Database 2015, 2015, bav093. [Google Scholar]
- Ostergaard, L.; King, G.J. Standardized gene nomenclature for the Brassica genus. Plant Methods 2008, 4, 10. [Google Scholar]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2009, 26, 136–138. [Google Scholar] [CrossRef]



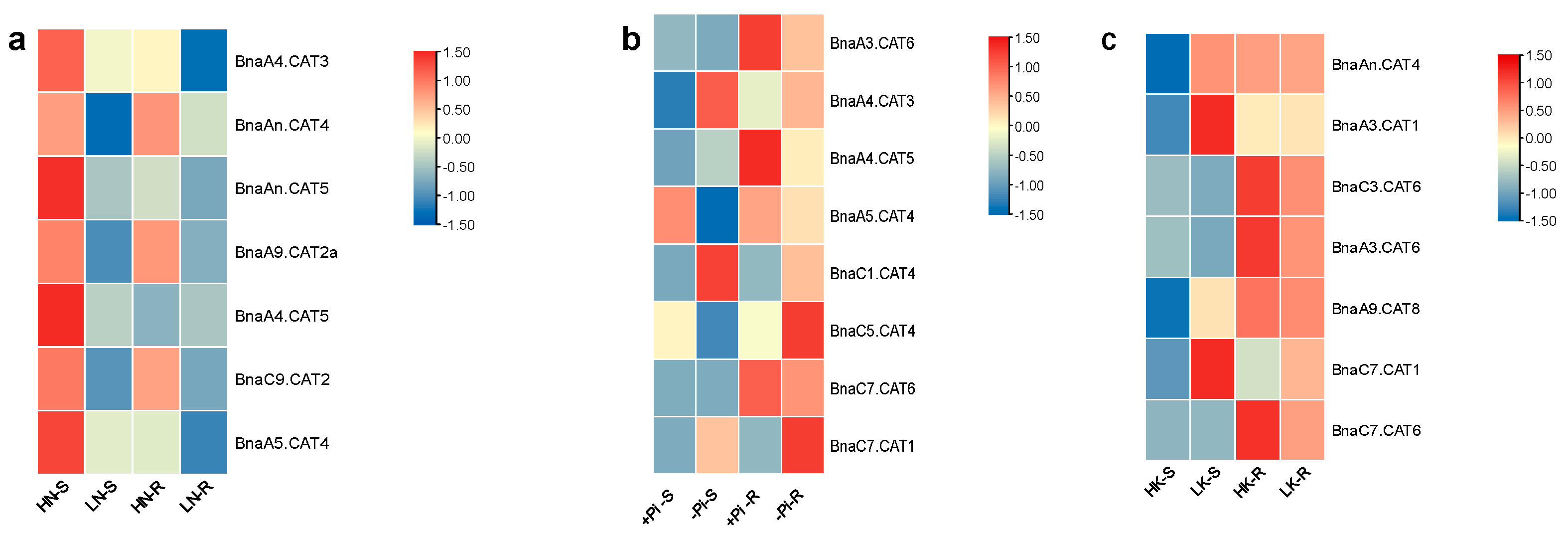
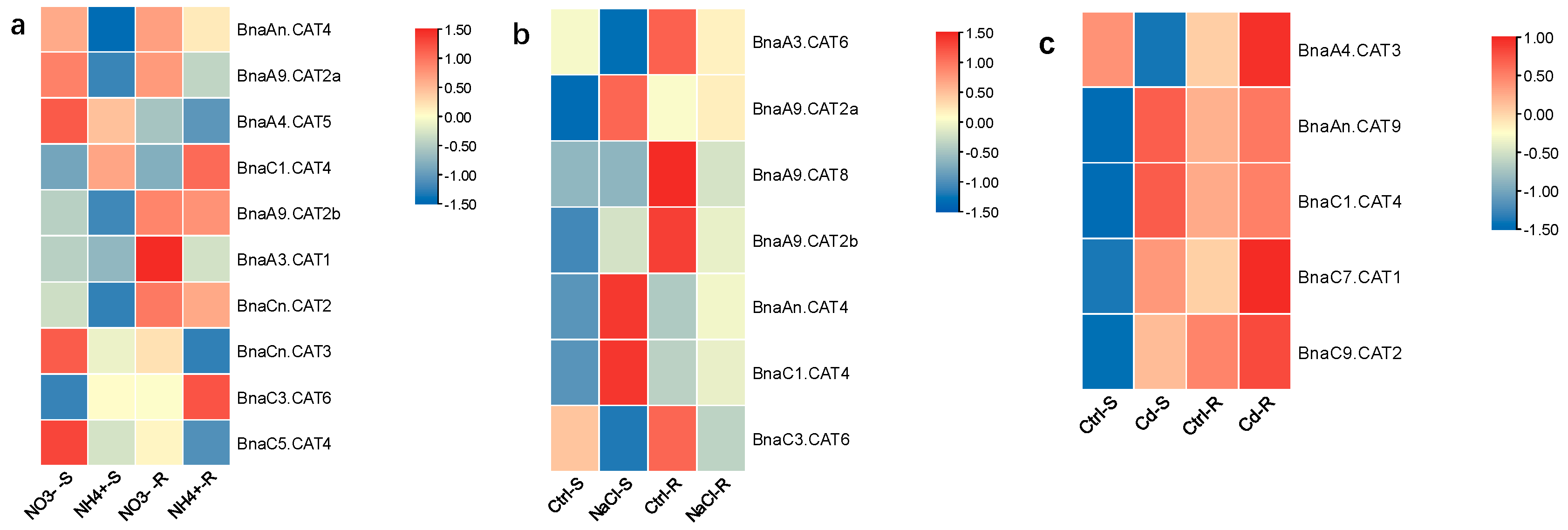
| Gene Name | Arabidopsis thaliana (125 Mb) | Brassica rapa (465 Mb) | Brassica oleracea (485 Mb) | Brassica napus (1130 Mb) |
|---|---|---|---|---|
| CAT1 | 1 | 2 | 2 | 4 |
| CAT2 | 1 | 2 | 2 | 4 |
| CAT3 | 1 | 1 | 1 | 2 |
| CAT4 | 1 | 2 | 2 | 4 |
| CAT5 | 1 | 1 | 1 | 2 |
| CAT6 | 1 | 2 | 2 | 3 |
| CAT7 | 1 | 1 | 1 | 0 |
| CAT8 | 1 | 1 | 1 | 2 |
| CAT9 | 1 | 1 | 1 | 1 |
| Total | 9 | 13 | 13 | 22 |
| Gene ID | Gene Name | Block | Amino Acids (aa) | CDS (bp) | Ka | Ks | Ka/Ks | Divergent Times (Mya) |
|---|---|---|---|---|---|---|---|---|
| BnaC07g36580D | BnaC7.CAT1 | U | 595 | 1788 | 0.065 | 0.49 | 0.13 | 16.56 |
| BnaC03g64380D | BnaC3.CAT1 | U | 598 | 1797 | 0.076 | 0.50 | 0.15 | 16.91 |
| BnaA03g58530D | BnaA3.CAT1 | U | 596 | 1791 | 0.063 | 0.48 | 0.13 | 16.07 |
| AT1G58030.1 | AtCAT2 | D | 635 | 1908 | ||||
| BnaA09g12110D | BnaA9.CAT2a | D | 638 | 1917 | 0.056 | 0.46 | 0.12 | 15.37 |
| BnaA09g53040D | BnaA9.CAT2b | D | 634 | 1905 | 0.056 | 0.43 | 0.12 | 14.46 |
| BnaCnng25140D | BnaCn.CAT2 | D | 634 | 1905 | 0.056 | 0.43 | 0.13 | 14.42 |
| BnaC09g12080D | BnaC9.CAT2 | D | 639 | 1920 | 0.057 | 0.43 | 0.13 | 14.64 |
| AT5G36940.1 | AtCAT3 | S | 609 | 1830 | ||||
| BnaCnng50730D | BnaCn.CAT3 | S | 636 | 1911 | 0.064 | 0.36 | 0.17 | 12.07 |
| BnaA04g07800D | BnaA4.CAT3 | S | 632 | 1899 | 0.067 | 0.36 | 0.18 | 12.05 |
| AT3G03720.2 | AtCAT4 | F | 600 | 1803 | ||||
| BnaC01g40540D | BnaC1.CAT4 | F | 604 | 1815 | 0.046 | 0.34 | 0.13 | 11.51 |
| BnaA05g32770D | BnaA5.CAT4 | F | 614 | 1845 | 0.046 | 0.33 | 0.14 | 11.12 |
| BnaC05g48070D | BnaC5.CAT4 | F | 613 | 1842 | 0.046 | 0.33 | 0.14 | 11.02 |
| BnaAnng23630D | BnaAn.CAT4 | F | 604 | 1815 | 0.042 | 0.34 | 0.12 | 11.52 |
| AT2G34960.1 | AtCAT5 | J | 569 | 1710 | ||||
| BnaAnng19530D | BnaAn.CAT5 | J | 570 | 1713 | 0.045 | 0.55 | 0.08 | 18.61 |
| BnaA04g20450D | BnaA4.CAT5 | J | 571 | 1716 | 0.046 | 0.55 | 0.08 | 18.35 |
| AT5G04770.1 | AtCAT6 | R | 583 | 1752 | ||||
| BnaA03g01440D | BnaA3.CAT6 | R | 529 | 1590 | 0.036 | 0.59 | 0.061 | 19.86 |
| BnaC03g01740D | BnaC3.CAT6 | R | 529 | 1590 | 0.036 | 0.53 | 0.067 | 17.98 |
| BnaC07g34120D | BnaC7.CAT6 | R | 580 | 1743 | 0.42 | 2.14 | 0.19 | 71.58 |
| AT1G17120.1 | AtCAT8 | A | 590 | 1773 | ||||
| BnaC08g37970D | BnaC8.CAT8 | A | 586 | 1761 | 0.041 | 0.75 | 0.054 | 25.29 |
| BnaA09g45150D | BnaA9.CAT8 | A | 586 | 1761 | 0.037 | 0.68 | 0.054 | 22.92 |
| AT1G05940.1 | AtCAT9 | A | 569 | 1710 | ||||
| BnaAnng20490D | BnaAn.CAT9 | A | 571 | 1716 | 0.037 | 0.27 | 0.13 | 9.23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.-Q.; Sun, S.-S.; Zhou, T.; Zhang, L.; Feng, Y.-N.; Zhang, K.-L.; Hua, Y.-P. Genome-Wide Identification of the CAT Genes and Molecular Characterization of Their Transcriptional Responses to Various Nutrient Stresses in Allotetraploid Rapeseed. Int. J. Mol. Sci. 2024, 25, 12658. https://doi.org/10.3390/ijms252312658
Du X-Q, Sun S-S, Zhou T, Zhang L, Feng Y-N, Zhang K-L, Hua Y-P. Genome-Wide Identification of the CAT Genes and Molecular Characterization of Their Transcriptional Responses to Various Nutrient Stresses in Allotetraploid Rapeseed. International Journal of Molecular Sciences. 2024; 25(23):12658. https://doi.org/10.3390/ijms252312658
Chicago/Turabian StyleDu, Xiao-Qian, Si-Si Sun, Ting Zhou, Lu Zhang, Ying-Na Feng, Kun-Long Zhang, and Ying-Peng Hua. 2024. "Genome-Wide Identification of the CAT Genes and Molecular Characterization of Their Transcriptional Responses to Various Nutrient Stresses in Allotetraploid Rapeseed" International Journal of Molecular Sciences 25, no. 23: 12658. https://doi.org/10.3390/ijms252312658
APA StyleDu, X.-Q., Sun, S.-S., Zhou, T., Zhang, L., Feng, Y.-N., Zhang, K.-L., & Hua, Y.-P. (2024). Genome-Wide Identification of the CAT Genes and Molecular Characterization of Their Transcriptional Responses to Various Nutrient Stresses in Allotetraploid Rapeseed. International Journal of Molecular Sciences, 25(23), 12658. https://doi.org/10.3390/ijms252312658






