Redefining Roles: A Paradigm Shift in Tryptophan–Kynurenine Metabolism for Innovative Clinical Applications
Abstract
1. Introduction
2. Traditional Paradigm
2.1. Pro-Oxidants and Antioxidants
2.2. Excitotoxicity and Neuroprotection
2.2.1. Excitotoxicity
2.2.2. Neuroprotection
3. Emerging Evidence
3.1. Pro-Oxidants or Antioxidants?
3.2. Receptor Agonists or Antagonists?
3.3. Immunomodulators
3.4. The Body–Brain Axes
3.4.1. The Gut–Brain Axis
The Indoxyl Sulfate Pathway
The Indole-3-Acetamide Pathway
The Tryptamine Pathway
The Indole-3-Propionic Acid Pathway
3.4.2. The Muscle–Brain Axis
3.4.3. Other Axes
4. Paradigm Shift
4.1. Molecule–Molecule Interactions
4.2. Molecule–Neural Transmission Interactions
4.3. Molecule–Immune System Interactions
4.4. Connecting to Systems Biology
4.5. Zero-Order Responses, Resilience Measurement, and Intolerance, Among Others
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | anthranilic acid |
| AAD | amino acid decarboxylase |
| AD | Alzheimer’s disease |
| AhR | aryl hydrocarbon receptor |
| AMPA | alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| ALS | amyotrophic lateral sclerosis |
| ArAT | aromatic amino acid aminotransferase |
| BBB | blood–brain barrier |
| BPF | Bisphenol F |
| CA | cinnabarinic acid |
| CKD | chronic kidney disease |
| CNS | central nervous system |
| CrPic | chromium picolinate |
| GPR35 | G-protein-coupled receptor 35 |
| 3-HAA | 3-hydroxyanthranilic acid |
| HD | Huntington’s disease |
| 3-HK | 3-hydroxykynurenine |
| IAA | indole-3-acetic acid |
| IaaH | indole-3-acetamide hydrolase |
| IAAld | indole-3-acetaldehyde |
| IAcA | 3-indole acrylic acid |
| IAld | indole-3-aldehyde |
| IAM | indole-3-acetamide |
| IDO | indoleamine 2,3-dioxygenase |
| ILA | indole-3-lactic acid |
| INS | indoxyl sulfate |
| IPA | indole-3-propionic acid |
| IPyA | indole-3-pyruvic acid |
| JAK/STAT | Janus kinase/signal transducers and activators of transcription |
| KATs | kynurenine aminotransferases |
| KYN | kynurenine |
| KYNA | kynurenic acid |
| LPS | lipopolysaccharide |
| MAPK | mitogen-activated protein kinase |
| mGluRs | metabotropic glutamate receptors |
| MS | multiple sclerosis |
| NAD | nicotinamide adenine dinucleotide |
| NMDA | N-methyl-D-aspartate |
| PA | picolinic acid |
| PD | Parkinson’s disease |
| QUIN | quinolinic acid |
| ROS | reactive oxygen species |
| TDO | tryptophan 2,3-dioxygenase |
| TEAC | Trolox equivalent antioxidant capacity |
| TMO | tryptophan-2-monooxygenase |
| TNA | tryptophanase |
| TrD | tryptophan decarboxylase |
| Tregs | regulatory T cells |
| Trp | tryptophan |
| XA | xanthurenic acid |
References
- Török, N.; Tanaka, M.; Vécsei, L. Searching for peripheral biomarkers in neurodegenerative diseases: The tryptophan-kynurenine metabolic pathway. Int. J. Mol. Sci. 2020, 21, 9338. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, A.; Ikeda, Y.; Yoshikawa, S.; Taniguchi, K.; Sawamura, H.; Morikawa, S.; Nakashima, M.; Asai, T.; Matsuda, S. The tryptophan and kynurenine pathway involved in the development of immune-related diseases. Int. J. Mol. Sci. 2023, 24, 5742. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.A.-B. Tryptophan metabolism and disposition in cancer biology and immunotherapy. Biosci. Rep. 2022, 42, BSR20221682. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Chang, R.; Zou, J.; Tan, S.; Huang, Z. The role and mechanism of tryptophan–kynurenine metabolic pathway in depression. Rev. Neurosci. 2023, 34, 313–324. [Google Scholar] [CrossRef]
- Tutakhail, A.; Boulet, L.; Khabil, S.; Nazari, Q.A.; Hamid, H.; Coudoré, F. Neuropathology of kynurenine pathway of tryptophan metabolism. Curr. Pharmacol. Rep. 2020, 6, 8–23. [Google Scholar] [CrossRef]
- Hestad, K.; Alexander, J.; Rootwelt, H.; Aaseth, J.O. The role of tryptophan dysmetabolism and quinolinic acid in depressive and neurodegenerative diseases. Biomolecules 2022, 12, 998. [Google Scholar] [CrossRef]
- Dolšak, A.; Gobec, S.; Sova, M. Indoleamine and tryptophan 2, 3-dioxygenases as important future therapeutic targets. Pharmacol. Ther. 2021, 221, 107746. [Google Scholar] [CrossRef]
- Tanaka, M.; Tóth, F.; Polyák, H.; Szabó, Á.; Mándi, Y.; Vécsei, L. Immune influencers in action: Metabolites and enzymes of the tryptophan-kynurenine metabolic pathway. Biomedicines 2021, 9, 734. [Google Scholar] [CrossRef]
- Gáspár, R.; Halmi, D.; Demján, V.; Berkecz, R.; Pipicz, M.; Csont, T. Kynurenine pathway metabolites as potential clinical biomarkers in coronary artery disease. Front. Immunol. 2022, 12, 768560. [Google Scholar] [CrossRef]
- Rebnord, E.W.; Strand, E.; Midttun, Ø.; Svingen, G.F.; Christensen, M.H.; Ueland, P.M.; Mellgren, G.; Njølstad, P.R.; Tell, G.S.; Nygård, O.K. The kynurenine: Tryptophan ratio as a predictor of incident type 2 diabetes mellitus in individuals with coronary artery disease. Diabetologia 2017, 60, 1712–1721. [Google Scholar] [CrossRef]
- Liu, J.-J.; Ching, J.; Wee, H.N.; Liu, S.; Gurung, R.L.; Lee, J.; Zheng, H.; Lee, L.S.; Ang, K.; Shao, Y.M. Plasma tryptophan-kynurenine pathway metabolites and risk for progression to end-stage kidney disease in patients with type 2 diabetes. Diabetes Care 2023, 46, 2223–2231. [Google Scholar] [CrossRef] [PubMed]
- Basson, C.; Serem, J.C.; Hlophe, Y.N.; Bipath, P. The tryptophan–kynurenine pathway in immunomodulation and cancer metastasis. Cancer Med. 2023, 12, 18691–18701. [Google Scholar] [CrossRef] [PubMed]
- Fiore, A.; Murray, P.J. Tryptophan and indole metabolism in immune regulation. Curr. Opin. Immunol. 2021, 70, 7–14. [Google Scholar] [CrossRef]
- Yan, J.; Kuzhiumparambil, U.; Bandodkar, A.; Bandodkar, S.; Dale, R.C.; Fu, S. Cerebrospinal fluid metabolites in tryptophan-kynurenine and nitric oxide pathways: Biomarkers for acute neuroinflammation. Dev. Med. Child Neurol. 2021, 63, 552–559. [Google Scholar] [CrossRef]
- Mithaiwala, M.N.; Santana-Coelho, D.; Porter, G.A.; O’connor, J.C. Neuroinflammation and the kynurenine pathway in CNS disease: Molecular mechanisms and therapeutic implications. Cells 2021, 10, 1548. [Google Scholar] [CrossRef]
- Martin, K.S.; Azzolini, M.; Ruas, J.L. The kynurenine connection: How exercise shifts muscle tryptophan metabolism and affects energy homeostasis, the immune system, and the brain. Am. J. Physiol.-Cell Physiol. 2020, 318, C818–C830. [Google Scholar] [CrossRef]
- Castro-Portuguez, R.; Sutphin, G.L. Kynurenine pathway, NAD+ synthesis, and mitochondrial function: Targeting tryptophan metabolism to promote longevity and healthspan. Exp. Gerontol. 2020, 132, 110841. [Google Scholar] [CrossRef]
- Liebig, J. Über Kynurensäure. Justus Liebig’s Ann. Chem 1853, 86, 125–126. [Google Scholar] [CrossRef]
- Hopkins, F.G.; Cole, S.W. A contribution to the chemistry of proteids: Part I. A preliminary study of a hitherto undescribed product of tryptic digestion. J. Physiol. 1901, 27, 418. [Google Scholar] [CrossRef]
- Sir Frederick Hopkins Nobel—Lecture. Available online: https://www.nobelprize.org/prizes/medicine/1929/hopkins/lecture/ (accessed on 7 November 2024).
- Ellinger, A. Die Entstehung der Kynurensäure. Biol. Chem. 1905, 43, 325–337. [Google Scholar] [CrossRef][Green Version]
- Matsuoka, Z.; Yoshimatsu, N. Über eine neue Substanz, die aus Tryptophan im Tierkörper gebildet wird. Biol. Chem. 1925, 143. [Google Scholar] [CrossRef]
- Kotake, Y. The mechanism of kynurenic acid formation in the organism. Z. Physiol. Chem 1931, 195, 158–166. [Google Scholar]
- Kotake, Y.; Iwao, J. Studies on the intermediatary metabolism of tryptophan. I. Kynurenine, an intermediary metabolic product of tryptophan. Hoppe-Seylers Z Physiol. Chem. 1931, 195, 139. [Google Scholar] [CrossRef]
- Heidelberger, C.; Gullberg, M.E.; Morgan, A.F.; Lepkovsky, S. Tryptophan metabolism; concerning the mechanism of the mammalian conversion of tryptophan into kynurenine, kynurenic acid, and nicotinic acid. J. Biol. Chem. 1949, 179, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Kido, R. Kynurenate forming enzymes in liver, kidney and brain. Kynurenine Serotonin Pathw. Prog. Tryptophan Res. 1991, 294, 201–205. [Google Scholar]
- Saito, Y.; Hayashi, O.; Rothberg, S.; Senoh, S. L-Kynurenine hydroxylase. In Proceedings of the Fed Proc; 1957; p. 240. [Google Scholar]
- Soda, K.; Tanizawa, K. Kynureninases: Enzymological properties and regulation mechanism. Adv. Enzym. Relat. Areas Mol. Biol. 1979, 49, 1–40. [Google Scholar]
- Long, C.; Hill, H.N.; Weinstock, I.; Henderson, L. Studies of the enzymatic of transformation of 3-hydroxyanthranilate to quinolinate. J. Biol. Chem. 1954, 211, 405–417. [Google Scholar] [CrossRef]
- Nishizuka, Y.; Hayaishi, O. Studies on the biosynthesis of nicotinamide adenine dinucleotide: I. Enzymic synthesis of niacin ribonucleotides from 3-hydroxyanthranilic acid in mammalian tissues. J. Biol. Chem. 1963, 238, 3369–3377. [Google Scholar] [CrossRef]
- Katoh, A.; Hashimoto, T. Molecular biology of pyridine nucleotide and nicotine biosynthesis. Front. Biosci. 2004, 9, 1577–1586. [Google Scholar] [CrossRef]
- Krupa, A.; Kowalska, I. The kynurenine pathway—New linkage between innate and adaptive immunity in autoimmune endocrinopathies. Int. J. Mol. Sci. 2021, 22, 9879. [Google Scholar] [CrossRef]
- Schwarcz, R.; Stone, T.W. The kynurenine pathway and the brain: Challenges, controversies and promises. Neuropharmacology 2017, 112, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Savitz, J. The kynurenine pathway: A finger in every pie. Mol. Psychiatry 2020, 25, 131–147. [Google Scholar] [CrossRef]
- Török, N.; Maszlag-Török, R.; Molnár, K.; Szolnoki, Z.; Somogyvári, F.; Boda, K.; Tanaka, M.; Klivényi, P.; Vécsei, L. Single Nucleotide Polymorphisms of Indoleamine 2, 3-Dioxygenase1 Influenced the Age Onset of Parkinson’s Disease. Front. Biosci. -Landmark 2022, 27, 265. [Google Scholar] [CrossRef]
- Mor, A.; Tankiewicz-Kwedlo, A.; Krupa, A.; Pawlak, D. Role of kynurenine pathway in oxidative stress during neurodegenerative disorders. Cells 2021, 10, 1603. [Google Scholar] [CrossRef] [PubMed]
- Ala, M. The footprint of kynurenine pathway in every cancer: A new target for chemotherapy. Eur. J. Pharmacol. 2021, 896, 173921. [Google Scholar] [CrossRef]
- Marx, W.; McGuinness, A.J.; Rocks, T.; Ruusunen, A.; Cleminson, J.; Walker, A.J.; Gomes-da-Costa, S.; Lane, M.; Sanches, M.; Diaz, A.P. The kynurenine pathway in major depressive disorder, bipolar disorder, and schizophrenia: A meta-analysis of 101 studies. Mol. Psychiatry 2021, 26, 4158–4178. [Google Scholar] [CrossRef]
- Tanaka, M.; Vécsei, L. Monitoring the redox status in multiple sclerosis. Biomedicines 2020, 8, 406. [Google Scholar] [CrossRef]
- Lugo-Huitrón, R.; Blanco-Ayala, T.; Ugalde-Muñiz, P.; Carrillo-Mora, P.; Pedraza-Chaverrí, J.; Silva-Adaya, D.; Maldonado, P.; Torres, I.; Pinzón, E.; Ortiz-Islas, E. On the antioxidant properties of kynurenic acid: Free radical scavenging activity and inhibition of oxidative stress. Neurotoxicol. Teratol. 2011, 33, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Ostapiuk, A.; Urbanska, E.M. Kynurenic acid in neurodegenerative disorders—Unique neuroprotection or double-edged sword? CNS Neurosci. Ther. 2022, 28, 19–35. [Google Scholar] [CrossRef]
- Salvati, P.; Ukmar, G.; Dho, L.; Rosa, B.; Cini, M.; Marconi, M.; Molinari, A.; Post, C. Brain concentrations of kynurenic acid after a systemic neuroprotective dose in the gerbil model of global ischemia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 1999, 23, 741–752. [Google Scholar] [CrossRef]
- Leipnitz, G.; Schumacher, C.; Scussiato, K.; Dalcin, K.B.; Wannmacher, C.M.; Wyse, A.T.; Dutra-Filho, C.S.; Wajner, M.; Latini, A. Quinolinic acid reduces the antioxidant defenses in cerebral cortex of young rats. Int. J. Dev. Neurosci. 2005, 23, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A.; Laurie, C.; Mosley, R.L.; Gendelman, H.E. Oxidative stress and the pathogenesis of neurodegenerative disorders. Int. Rev. Neurobiol. 2007, 82, 297–325. [Google Scholar] [PubMed]
- Tanaka, M.; Bohár, Z.; Vécsei, L. Are kynurenines accomplices or principal villains in dementia? Maintenance of kynurenine metabolism. Molecules 2020, 25, 564. [Google Scholar] [CrossRef] [PubMed]
- Birner, A.; Platzer, M.; Bengesser, S.A.; Dalkner, N.; Fellendorf, F.T.; Queissner, R.; Pilz, R.; Rauch, P.; Maget, A.; Hamm, C. Increased breakdown of kynurenine towards its neurotoxic branch in bipolar disorder. PLoS ONE 2017, 12, e0172699. [Google Scholar] [CrossRef]
- Stone, T.W.; Darlington, L.G.; Badawy, A.A.-B.; Williams, R.O. The Complex World of Kynurenic Acid: Reflections on Biological Issues and Therapeutic Strategy. Int. J. Mol. Sci. 2024, 25, 9040. [Google Scholar] [CrossRef]
- Colín-González, A.L.; Maldonado, P.D.; Santamaría, A. 3-Hydroxykynurenine: An intriguing molecule exerting dual actions in the central nervous system. Neurotoxicology 2013, 34, 189–204. [Google Scholar] [CrossRef]
- Rózsa, E.; Robotka, H.; Vécsei, L.; Toldi, J. The Janus-face kynurenic acid. J. Neural Transm. 2008, 115, 1087–1091. [Google Scholar] [CrossRef]
- Pérez-González, A.; Alvarez-Idaboy, J.R.; Galano, A. Dual antioxidant/pro-oxidant behavior of the tryptophan metabolite 3-hydroxyanthranilic acid: A theoretical investigation of reaction mechanisms and kinetics. New J. Chem. 2017, 41, 3829–3845. [Google Scholar] [CrossRef]
- Hopkins, F.G. Feeding experiments illustrating the importance of accessory factors in normal dietaries. J. Physiol. 1912, 44, 425–460. [Google Scholar] [CrossRef]
- Klaessens, S.; Stroobant, V.; De Plaen, E.; Van den Eynde, B.J. Systemic tryptophan homeostasis. Front. Mol. Biosci. 2022, 9, 897929. [Google Scholar] [CrossRef]
- Leathwood, P.D. Tryptophan availability and serotonin synthesis. Proc. Nutr. Soc. 1987, 46, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Majewska, M.; Zajac, K.; Zemelka, M.; Szczepanik, M. Influence of melatonin and its precursor L-tryptophan on Th1 dependent contact hypersensitivity. J. Physiol. Pharmacol. 2007, 58, 125–132. [Google Scholar] [PubMed]
- Oxenkrug, G. Insulin resistance and dysregulation of tryptophan–kynurenine and kynurenine–nicotinamide adenine dinucleotide metabolic pathways. Mol. Neurobiol. 2013, 48, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Cetina Biefer, H.; Vasudevan, A.; Elkhal, A. Aspects of tryptophan and nicotinamide adenine dinucleotide in immunity: A new twist in an old tale. Int. J. Tryptophan Res. 2017, 10, 1178646917713491. [Google Scholar] [CrossRef] [PubMed]
- Mondanelli, G.; Volpi, C. The double life of serotonin metabolites: In the mood for joining neuronal and immune systems. Curr. Opin. Immunol. 2021, 70, 1–6. [Google Scholar] [CrossRef]
- Vaseghi, S.; Arjmandi-Rad, S.; Nasehi, M.; Zarrindast, M.-R. Cannabinoids and sleep-wake cycle: The potential role of serotonin. Behav. Brain Res. 2021, 412, 113440. [Google Scholar] [CrossRef]
- Bacqué-Cazenave, J.; Bharatiya, R.; Barrière, G.; Delbecque, J.-P.; Bouguiyoud, N.; Di Giovanni, G.; Cattaert, D.; De Deurwaerdère, P. Serotonin in animal cognition and behavior. Int. J. Mol. Sci. 2020, 21, 1649. [Google Scholar] [CrossRef]
- De Deurwaerdère, P.; Di Giovanni, G. Serotonin in health and disease. Int. J. Mol. Sci. 2020, 21, 3500. [Google Scholar] [CrossRef]
- Lin, J.; Liu, W.; Guan, J.; Cui, J.; Shi, R.; Wang, L.; Chen, D.; Liu, Y. Latest updates on the serotonergic system in depression and anxiety. Front. Synaptic Neurosci. 2023, 15, 1124112. [Google Scholar] [CrossRef]
- Xue, C.; Li, G.; Zheng, Q.; Gu, X.; Shi, Q.; Su, Y.; Chu, Q.; Yuan, X.; Bao, Z.; Lu, J. Tryptophan metabolism in health and disease. Cell Metab. 2023, 35, 1304–1326. [Google Scholar] [CrossRef]
- Strasser, B.; Becker, K.; Fuchs, D.; Gostner, J.M. Kynurenine pathway metabolism and immune activation: Peripheral measurements in psychiatric and co-morbid conditions. Neuropharmacology 2017, 112, 286–296. [Google Scholar] [CrossRef]
- Haroon, E.; Welle, J.R.; Woolwine, B.J.; Goldsmith, D.R.; Baer, W.; Patel, T.; Felger, J.C.; Miller, A.H. Associations among peripheral and central kynurenine pathway metabolites and inflammation in depression. Neuropsychopharmacology 2020, 45, 998–1007. [Google Scholar] [CrossRef]
- Pathak, S.; Nadar, R.; Kim, S.; Liu, K.; Govindarajulu, M.; Cook, P.; Watts Alexander, C.S.; Dhanasekaran, M.; Moore, T. The influence of kynurenine metabolites on neurodegenerative pathologies. Int. J. Mol. Sci. 2024, 25, 853. [Google Scholar] [CrossRef]
- Giil, L.M.; Midttun, Ø.; Refsum, H.; Ulvik, A.; Advani, R.; Smith, A.D.; Ueland, P.M. Kynurenine pathway metabolites in Alzheimer’s disease. J. Alzheimer’s Dis. 2017, 60, 495–504. [Google Scholar] [CrossRef]
- Fathi, M.; Vakili, K.; Yaghoobpoor, S.; Tavasol, A.; Jazi, K.; Hajibeygi, R.; Shool, S.; Sodeifian, F.; Klegeris, A.; McElhinney, A. Dynamic changes in metabolites of the kynurenine pathway in Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease: A systematic Review and meta-analysis. Front. Immunol. 2022, 13, 997240. [Google Scholar] [CrossRef]
- Chang, K.H.; Cheng, M.L.; Tang, H.Y.; Huang, C.Y.; Wu, Y.R.; Chen, C.M. Alternations of Metabolic Profile and Kynurenine Metabolism in the Plasma of Parkinson’s Disease. Mol. Neurobiol. 2018, 55, 6319–6328. [Google Scholar] [CrossRef]
- Heilman, P.L.; Wang, E.W.; Lewis, M.M.; Krzyzanowski, S.; Capan, C.D.; Burmeister, A.R.; Du, G.; Escobar Galvis, M.L.; Brundin, P.; Huang, X.; et al. Tryptophan Metabolites Are Associated With Symptoms and Nigral Pathology in Parkinson’s Disease. Mov. Disord. 2020, 35, 2028–2037. [Google Scholar] [CrossRef]
- Schwarcz, R.; Foo, A.; Sathyasaikumar, K.V.; Notarangelo, F.M. The Probiotic Lactobacillus reuteri Preferentially Synthesizes Kynurenic Acid from Kynurenine. Int. J. Mol. Sci. 2024, 25, 3679. [Google Scholar] [CrossRef]
- Espi, M.; Koppe, L.; Fouque, D.; Thaunat, O. Chronic kidney disease-associated immune dysfunctions: Impact of protein-bound uremic retention solutes on immune cells. Toxins 2020, 12, 300. [Google Scholar] [CrossRef]
- Ye, X.; Li, H.; Anjum, K.; Zhong, X.; Miao, S.; Zheng, G.; Liu, W.; Li, L. Dual role of indoles derived from intestinal microbiota on human health. Front. Immunol. 2022, 13, 903526. [Google Scholar] [CrossRef]
- Mishra, P.; Kaur, S.; Sharma, A.N.; Jolly, R.S. Characterization of an indole-3-acetamide hydrolase from Alcaligenes faecalis subsp. parafaecalis and its application in efficient preparation of both enantiomers of chiral building block 2, 3-dihydro-1, 4-benzodioxin-2-carboxylic acid. PLoS ONE 2016, 11, e0159009. [Google Scholar] [CrossRef] [PubMed]
- Mousseau, D.D. Tryptamine: A metabolite of tryptophan implicated in various neuropsychiatric disorders. Metab. Brain Dis. 1993, 8, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Brydges, C.R.; Fiehn, O.; Mayberg, H.S.; Schreiber, H.; Dehkordi, S.M.; Bhattacharyya, S.; Cha, J.; Choi, K.S.; Craighead, W.E.; Krishnan, R.R. Indoxyl sulfate, a gut microbiome-derived uremic toxin, is associated with psychic anxiety and its functional magnetic resonance imaging-based neurologic signature. Sci. Rep. 2021, 11, 21011. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, B.; Hu, Y.; Zhao, Y. New insights into gut-bacteria-derived indole and its derivatives in intestinal and liver diseases. Front. Pharmacol. 2021, 12, 769501. [Google Scholar] [CrossRef]
- Hou, Y.; Li, J.; Ying, S. Tryptophan metabolism and gut microbiota: A novel regulatory axis integrating the microbiome, immunity, and cancer. Metabolites 2023, 13, 1166. [Google Scholar] [CrossRef]
- Hajsl, M.; Hlavackova, A.; Broulikova, K.; Sramek, M.; Maly, M.; Dyr, J.E.; Suttnar, J. Tryptophan Metabolism, Inflammation, and Oxidative Stress in Patients with Neurovascular Disease. Metabolites 2020, 10, 208. [Google Scholar] [CrossRef]
- Iwaoka, K.; Otsuka, C.; Maeda, T.; Yamahara, K.; Kato, K.; Takahashi, K.; Takahashi, K.; Terayama, Y. Impaired metabolism of kynurenine and its metabolites in CSF of parkinson’s disease. Neurosci. Lett. 2020, 714, 134576. [Google Scholar] [CrossRef]
- Huang, Y.; Zhao, M.; Chen, X.; Zhang, R.; Le, A.; Hong, M.; Zhang, Y.; Jia, L.; Zang, W.; Jiang, C.; et al. Tryptophan Metabolism in Central Nervous System Diseases: Pathophysiology and Potential Therapeutic Strategies. Aging. Dis. 2023, 14, 858–878. [Google Scholar] [CrossRef]
- Castellano-González, G.; Jacobs, K.; Don, E.; Cole, N.; Adams, S.; Lim, C.; Lovejoy, D.; Guillemin, G. Kynurenine 3-Monooxygenase Activity in Human Primary Neurons and Effect on Cellular Bioenergetics Identifies New Neurotoxic Mechanisms. Neurotox. Res. 2019, 35, 530–541. [Google Scholar] [CrossRef]
- Tanaka, M.; Toldi, J.; Vécsei, L. Exploring the Etiological Links behind Neurodegenerative Diseases: Inflammatory Cytokines and Bioactive Kynurenines. Int. J. Mol. Sci. 2020, 21, 2431. [Google Scholar] [CrossRef]
- Stone, T.W.; Addae, J.I. The pharmacological manipulation of glutamate receptors and neuroprotection. Eur. J. Pharmacol. 2002, 447, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Goda, K.; Kishimoto, R.; Shimizu, S.; Hamane, Y.; Ueda, M. Quinolinic acid and active oxygens: Possible contribution of active oxygens during cell death in the brain. Recent Adv. Tryptophan Res. Tryptophan Serotonin Pathw. 1996, 398, 247–254. [Google Scholar]
- Hardeland, R.; Zsizsik, B. Kynurenic acid as a free radical scavenger: Measurements of educt and product fluorescence and of light emission from an excited intermediate state. Biol. Rhythm. Antioxidative Prot. 1997, 153–160. [Google Scholar]
- Eastman, C.L.; Guilarte, T.R. Cytotoxicity of 3-hydroxykynurenine in a neuronal hybrid cell line. Brain. Res. 1989, 495, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Stípek, S.; Stastný, F.; Pláteník, J.; Crkovská, J.; Zima, T. The effect of quinolinate on rat brain lipid peroxidation is dependent on iron. Neurochem. Int. 1997, 30, 233–237. [Google Scholar] [CrossRef]
- Lugo-Huitrón, R.; Ugalde Muñiz, P.; Pineda, B.; Pedraza-Chaverrí, J.; Ríos, C.; Pérez-de la Cruz, V. Quinolinic acid: An endogenous neurotoxin with multiple targets. Oxid. Med. Cell. Longev. 2013, 2013, 104024. [Google Scholar] [CrossRef]
- Behan, W.M.; McDonald, M.; Darlington, L.G.; Stone, T.W. Oxidative stress as a mechanism for quinolinic acid-induced hippocampal damage: Protection by melatonin and deprenyl. Br. J. Pharmacol. 1999, 128, 1754–1760. [Google Scholar] [CrossRef]
- Pérez-De La Cruz, V.; Carrillo-Mora, P.; Santamaría, A. Quinolinic Acid, an endogenous molecule combining excitotoxicity, oxidative stress and other toxic mechanisms. Int. J. Tryptophan. Res. 2012, 5, IJTR-S8158. [Google Scholar] [CrossRef]
- Guillemin, G.J. Quinolinic acid, the inescapable neurotoxin. FEBS J. 2012, 279, 1356–1365. [Google Scholar] [CrossRef]
- Okuda, S.; Nishiyama, N.; Saito, H.; Katsuki, H. Hydrogen peroxide-mediated neuronal cell death induced by an endogenous neurotoxin, 3-hydroxykynurenine. Proc. Natl. Acad. Sci. USA 1996, 93, 12553–12558. [Google Scholar] [CrossRef]
- Okuda, S.; Nishiyama, N.; Saito, H.; Katsuki, H. 3-Hydroxykynurenine, an endogenous oxidative stress generator, causes neuronal cell death with apoptotic features and region selectivity. J. Neurochem. 1998, 70, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.R.; Stocker, R. Redox reactions related to indoleamine 2,3-dioxygenase and tryptophan metabolism along the kynurenine pathway. Redox Rep. 1999, 4, 199–220. [Google Scholar] [CrossRef] [PubMed]
- Barbalho, S.M.; Sloan, L.A.; Araujo, A.C.; Laurindo, L.F.; Sloan, K.P. Vitamin D and Its Role on Inflammation, Oxidative Stress and Cardiovascular Disease. In Lipophilic Vitamins in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2024; pp. 291–311. [Google Scholar]
- Reyes-Ocampo, J.; Ramírez-Ortega, D.; Cervantes, G.I.; Pineda, B.; Balderas, P.M.; González-Esquivel, D.; Sánchez-Chapul, L.; Lugo-Huitrón, R.; Silva-Adaya, D.; Ríos, C.; et al. Mitochondrial dysfunction related to cell damage induced by 3-hydroxykynurenine and 3-hydroxyanthranilic acid: Non-dependent-effect of early reactive oxygen species production. Neurotoxicology 2015, 50, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Cerstiaens, A.; Huybrechts, J.; Kotanen, S.; Lebeau, I.; Meylaers, K.; De Loof, A.; Schoofs, L. Neurotoxic and neurobehavioral effects of kynurenines in adult insects. Biochem. Biophys. Res. Commun. 2003, 312, 1171–1177. [Google Scholar] [CrossRef]
- Ogawa, T.; Matson, W.R.; Beal, M.F.; Myers, R.H.; Bird, E.D.; Milbury, P.; Saso, S. Kynurenine pathway abnormalities in Parkinson’s disease. Neurology 1992, 42, 1702–1706. [Google Scholar] [CrossRef]
- Capucciati, A.; Galliano, M.; Bubacco, L.; Zecca, L.; Casella, L.; Monzani, E.; Nicolis, S. Neuronal Proteins as Targets of 3-Hydroxykynurenine: Implications in Neurodegenerative Diseases. ACS Chem. Neurosci. 2019, 10, 3731–3739. [Google Scholar] [CrossRef]
- Guidetti, P.; Schwarcz, R. 3-Hydroxykynurenine and quinolinate: Pathogenic synergism in early grade Huntington’s disease? Adv. Exp. Med. Biol. 2003, 527, 137–145. [Google Scholar] [CrossRef]
- Forrest, C.M.; Mackay, G.M.; Stoy, N.; Egerton, M.; Christofides, J.; Stone, T.W.; Darlington, L.G. Tryptophan loading induces oxidative stress. Free Radic. Res. 2004, 38, 1167–1171. [Google Scholar] [CrossRef]
- Tanaka, M.; Szabó, Á.; Spekker, E.; Polyák, H.; Tóth, F.; Vécsei, L. Mitochondrial Impairment: A Common Motif in Neuropsychiatric Presentation? The Link to the Tryptophan-Kynurenine Metabolic System. Cells 2022, 11, 2607. [Google Scholar] [CrossRef]
- Mishra, J.; Kumar, A. Improvement of mitochondrial function by paliperidone attenuates quinolinic acid-induced behavioural and neurochemical alterations in rats: Implications in Huntington’s disease. Neurotox. Res. 2014, 26, 363–381. [Google Scholar] [CrossRef]
- Fão, L.; Rego, A.C. Mitochondrial and Redox-Based Therapeutic Strategies in Huntington’s Disease. Antioxid. Redox Signal. 2021, 34, 650–673. [Google Scholar] [CrossRef] [PubMed]
- D’Egidio, F.; Castelli, V.; Cimini, A.; d’Angelo, M. Cell Rearrangement and Oxidant/Antioxidant Imbalance in Huntington’s Disease. Antioxidants 2023, 12, 571. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Singh, T.G.; Prabhakar, N.K.; Mannan, A. Kynurenine metabolism and alzheimer’s disease: The potential targets and approaches. Neurochem. Res. 2022, 47, 1459–1476. [Google Scholar] [CrossRef] [PubMed]
- Kubicova, L.; Chobot, V. Potential of kynurenine metabolites in drug development against neurodegenerative diseases. Neural Regen. Res. 2021, 16, 308–309. [Google Scholar]
- Tóth, F.; Cseh, E.K.; Vécsei, L. Natural molecules and neuroprotection: Kynurenic acid, pantethine and α-lipoic acid. Int. J. Mol. Sci. 2021, 22, 403. [Google Scholar] [CrossRef]
- Nunes, Y.C.; Mendes, N.M.; Pereira de Lima, E.; Chehadi, A.C.; Lamas, C.B.; Haber, J.F.; dos Santos Bueno, M.; Araújo, A.C.; Catharin, V.C.S.; Detregiachi, C.R.P. Curcumin: A Golden Approach to Healthy Aging: A Systematic Review of the Evidence. Nutrients 2024, 16, 2721. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, A. Ueber die Constitution der Indolgruppe im Eiweiss (Synthese der sogen. Skatolcarbonsäure) und die Quelle der Kynurensäure. Ber. Dtsch. Chem. Ges. (Rep. Ger. Chem. Soc.) 1904, 37, 1801–1808. [Google Scholar] [CrossRef]
- Giles, G.I.; Collins, C.A.; Stone, T.W.; Jacob, C. Electrochemical and in vitro evaluation of the redox-properties of kynurenine species. Biochem. Biophys. Res. Commun. 2003, 300, 719–724. [Google Scholar] [CrossRef]
- Németh, H.; Robotka, H.; Toldi, J.; Vécsei, L. Kynurenines in the central nervous system: Recent developments. Cent. Nerv. Syst. Agents Med. Chem. 2007, 7, 45–56. [Google Scholar] [CrossRef]
- Pérez-De La Cruz, V.; Königsberg, M.; Santamaría, A. Kynurenine pathway and disease: An overview. CNS Neurol. Disord. Drug Targets 2007, 6, 398–410. [Google Scholar]
- Stone, T.; Forrest, C.; Mackay, G.; Stoy, N.; Darlington, L. Tryptophan, adenosine, neurodegeneration and neuroprotection. Metab. Brain Dis. 2007, 22, 337–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guillemin, G.J. Kynurenine pathway metabolites in humans: Disease and healthy states. Int. J. Tryptophan Res. 2009, 2, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ramírez Ortega, D.; Ugalde Muñiz, P.E.; Blanco Ayala, T.; Vázquez Cervantes, G.I.; Lugo Huitrón, R.; Pineda, B.; González Esquivel, D.F.; Pérez de la Cruz, G.; Pedraza Chaverrí, J.; Sánchez Chapul, L. On the antioxidant properties of L-kynurenine: An efficient ROS scavenger and enhancer of rat brain antioxidant defense. Antioxidants 2021, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Guo, X.; Zhou, Y.; Du, J.; Lu, C.; Zhang, L.; Sun, S.; Wang, S.; Li, Y. Kynurenic acid inhibits macrophage pyroptosis by suppressing ROS production via activation of the NRF2 pathway. Mol. Med. Rep. 2023, 28, 211. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.S.; Schmitz, F.; Marques, E.P.; Siebert, C.; Wyse, A.T. Intrastriatal quinolinic acid administration impairs redox homeostasis and induces inflammatory changes: Prevention by kynurenic acid. Neurotox. Res. 2020, 38, 50–58. [Google Scholar] [CrossRef]
- Martos, D.; Lőrinczi, B.; Szatmári, I.; Vécsei, L.; Tanaka, M. The Impact of C-3 Side Chain Modifications on Kynurenic Acid: A Behavioral Analysis of Its Analogs in the Motor Domain. Int. J. Mol. Sci. 2024, 25, 3394. [Google Scholar] [CrossRef]
- Kearns, R. The Kynurenine Pathway in Gut Permeability and Inflammation. Inflammation 2024, 1–15. [Google Scholar] [CrossRef]
- Nemeth, H.; Toldi, J.; Vecsei, L. Role of kynurenines in the central and peripherial nervous systems. Curr. Neurovasc. Res. 2005, 2, 249–260. [Google Scholar] [CrossRef]
- Mándi, Y.; Vécsei, L. The kynurenine system and immunoregulation. J. Neural. Transm. 2012, 119, 197–209. [Google Scholar] [CrossRef]
- Rajda, C.; Majláth, Z.; Pukoli, D.; Vécsei, L. Kynurenines and Multiple Sclerosis: The Dialogue between the Immune System and the Central Nervous System. Int. J. Mol. Sci. 2015, 16, 18270–18282. [Google Scholar] [CrossRef]
- Tanaka, M.; Tuka, B.; Vécsei, L. Navigating the Neurobiology of Migraine: From pathways to potential therapies. Cells 2024, 13, 1098. [Google Scholar] [CrossRef] [PubMed]
- Barone, P. The ‘Yin’ and the ‘Yang’ of the kynurenine pathway: Excitotoxicity and neuroprotection imbalance in stress-induced disorders. Behav. Pharmacol. 2019, 30, 163–186. [Google Scholar] [CrossRef]
- Kincses, Z.T.; Toldi, J.; Vécsei, L. Kynurenines, neurodegeneration and Alzheimer’s disease. J. Cell. Mol. Med. 2010, 14, 2045–2054. [Google Scholar] [CrossRef] [PubMed]
- Zádori, D.; Klivényi, P.; Szalárdy, L.; Fülöp, F.; Toldi, J.; Vécsei, L. Mitochondrial disturbances, excitotoxicity, neuroinflammation and kynurenines: Novel therapeutic strategies for neurodegenerative disorders. J. Neurol. Sci. 2012, 322, 187–191. [Google Scholar] [CrossRef]
- Tanaka, M.; Török, N.; Vécsei, L. Novel Pharmaceutical Approaches in Dementia. In NeuroPsychopharmacotherapy; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–18. [Google Scholar]
- Vamos, E.; Pardutz, A.; Klivenyi, P.; Toldi, J.; Vecsei, L. The role of kynurenines in disorders of the central nervous system: Possibilities for neuroprotection. J. Neurol. Sci. 2009, 283, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Yu, J.T.; Tan, L. The kynurenine pathway in neurodegenerative diseases: Mechanistic and therapeutic considerations. J. Neurol. Sci. 2012, 323, 1–8. [Google Scholar] [CrossRef]
- Sas, K.; Szabó, E.; Vécsei, L. Mitochondria, Oxidative Stress and the Kynurenine System, with a Focus on Ageing and Neuroprotection. Molecules 2018, 23, 191. [Google Scholar] [CrossRef]
- Baumgartner, R.; Forteza, M.J.; Ketelhuth, D.F.J. The interplay between cytokines and the Kynurenine pathway in inflammation and atherosclerosis. Cytokine 2019, 122, 154148. [Google Scholar] [CrossRef]
- Gutiérrez-Vázquez, C.; Quintana, F.J. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, D.; Song, P.; Zou, M.H. Tryptophan-kynurenine pathway is dysregulated in inflammation, and immune activation. Front. Biosci. (Landmark Ed.) 2015, 20, 1116–1143. [Google Scholar] [CrossRef]
- Jászberényi, M.; Thurzó, B.; Bagosi, Z.; Vécsei, L.; Tanaka, M. The Orexin/Hypocretin System, the Peptidergic Regulator of Vigilance, Orchestrates Adaptation to Stress. Biomedicines 2024, 12, 448. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, G.J.; Meininger, V.; Brew, B.J. Implications for the kynurenine pathway and quinolinic acid in amyotrophic lateral sclerosis. Neurodegener. Dis. 2005, 2, 166–176. [Google Scholar] [CrossRef]
- Maddison, D.C.; Giorgini, F. The kynurenine pathway and neurodegenerative disease. Semin. Cell. Dev. Biol. 2015, 40, 134–141. [Google Scholar] [CrossRef]
- Stone, T.W. Endogenous neurotoxins from tryptophan. Toxicon 2001, 39, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Badawy, A.A. Hypothesis kynurenic and quinolinic acids: The main players of the kynurenine pathway and opponents in inflammatory disease. Med. Hypotheses 2018, 118, 129–138. [Google Scholar] [CrossRef]
- Kita, T.; Morrison, P.F.; Heyes, M.P.; Markey, S.P. Effects of systemic and central nervous system localized inflammation on the contributions of metabolic precursors to the L-kynurenine and quinolinic acid pools in brain. J. Neurochem. 2002, 82, 258–268. [Google Scholar] [CrossRef]
- Skorobogatov, K.; De Picker, L.; Verkerk, R.; Coppens, V.; Leboyer, M.; Müller, N.; Morrens, M. Brain Versus Blood: A Systematic Review on the Concordance Between Peripheral and Central Kynurenine Pathway Measures in Psychiatric Disorders. Front. Immunol. 2021, 12, 716980. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yu, S.; Long, Y.; Shi, A.; Deng, J.; Ma, Y.; Wen, J.; Li, X.; Liu, S.; Zhang, Y.; et al. Tryptophan metabolism: Mechanism-oriented therapy for neurological and psychiatric disorders. Front. Immunol. 2022, 13, 985378. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, A.G.; Mingoti, M.E.D.; Ignácio, Z.M. Neurobiological mechanisms in the kynurenine pathway and major depressive disorder. Rev. Neurosci. 2024. [Google Scholar] [CrossRef]
- Sharp, C.; Houghton, J.; Elrod, J.; Warren, A.; Jackson, T.; Jawahar, A.; Nanda, A.; Minagar, A.; Alexander, J. N-methyl-D-aspartate receptor activation in human cerebral endothelium promotes intracellular oxidant stress. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H1893–H1899. [Google Scholar] [CrossRef]
- Olivares, D.; Deshpande, V.K.; Shi, Y.; Lahiri, D.K.; Greig, N.H.; Rogers, J.T.; Huang, X. N-methyl D-aspartate (NMDA) receptor antagonists and memantine treatment for Alzheimer’s disease, vascular dementia and Parkinson’s disease. Curr. Alzheimer Res. 2012, 9, 746–758. [Google Scholar] [CrossRef] [PubMed]
- Bano, D.; Zanetti, F.; Mende, Y.; Nicotera, P. Neurodegenerative processes in Huntington’s disease. Cell Death Dis. 2011, 2, e228. [Google Scholar] [CrossRef] [PubMed]
- Gonsette, R.E. Neurodegeneration in multiple sclerosis: The role of oxidative stress and excitotoxicity. J. Neurol. Sci. 2008, 274, 48–53. [Google Scholar] [CrossRef]
- Beltramino, C.A.; de Olmos, J.S.; Gallyas, F.; Heimer, L.; Záborszky, L. Silver staining as a tool for neurotoxic assessment. NIDA Res. Monogr. 1993, 136, 101–126; discussion 126–132. [Google Scholar] [CrossRef]
- Foster, A.C.; Collins, J.F.; Schwarcz, R. On the excitotoxic properties of quinolinic acid, 2,3-piperidine dicarboxylic acids and structurally related compounds. Neuropharmacology 1983, 22, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Moroni, F.; Lombardi, G.; Carlà, V.; Moneti, G. The excitotoxin quinolinic acid is present and unevenly distributed in the rat brain. Brain Res. 1984, 295, 352–355. [Google Scholar] [CrossRef]
- Bruyn, R.P.; Stoof, J.C. The quinolinic acid hypothesis in Huntington’s chorea. J. Neurol. Sci. 1990, 95, 29–38. [Google Scholar] [CrossRef]
- Hunt, D.L.; Castillo, P.E. Synaptic plasticity of NMDA receptors: Mechanisms and functional implications. Curr. Opin. Neurobiol. 2012, 22, 496–508. [Google Scholar] [CrossRef]
- Li, F.; Tsien, J.Z. Memory and the NMDA receptors. N. Engl. J. Med. 2009, 361, 302–303. [Google Scholar] [CrossRef]
- Heyes, M.P.; Rubinow, D.; Lane, C.; Markey, S.P. Cerebrospinal fluid quinolinic acid concentrations are increased in acquired immune deficiency syndrome. Ann. Neurol. 1989, 26, 275–277. [Google Scholar] [CrossRef]
- Ogawa, M.; Araki, M.; Nagatsu, I.; Yoshida, M. Astroglial cell alteration caused by neurotoxins: Immunohistochemical observations with antibodies to glial fibrillary acidic protein, laminin, and tyrosine hydroxylase. Exp. Neurol. 1989, 106, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Eriksdotter-Nilsson, M.; Jonsson, G.; Dahl, D.; Björklund, H. Astroglial development in microencephalic rat brain after fetal methylazoxymethanol treatment. Int. J. Dev. Neurosci. 1986, 4, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Bordelon, Y.M.; Chesselet, M.F.; Nelson, D.; Welsh, F.; Erecińska, M. Energetic dysfunction in quinolinic acid-lesioned rat striatum. J. Neurochem. 1997, 69, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.A.; Grando, V.; Dutra Filho, C.S.; Wannmacher, C.M.; Wajner, M. Evidence that quinolinic acid severely impairs energy metabolism through activation of NMDA receptors in striatum from developing rats. J. Neurochem. 2006, 99, 1531–1542. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.S.Y.; Francis, H.M.; Lim, C.K. Exploring the roles of tryptophan metabolism in MS beyond neuroinflammation and neurodegeneration: A paradigm shift to neuropsychiatric symptoms. Brain Behav. Immun. Health 2021, 12, 100201. [Google Scholar] [CrossRef]
- Qin, W.; Shi, Y.; Chen, W.; Jia, X.; Asakawa, T. Can kynurenine pathway be considered as a next-generation therapeutic target for Parkinson’s disease? An update information. Biosci. Trends 2022, 16, 249–256. [Google Scholar] [CrossRef]
- Lee, J.M.; Tan, V.; Lovejoy, D.; Braidy, N.; Rowe, D.B.; Brew, B.J.; Guillemin, G.J. Involvement of quinolinic acid in the neuropathogenesis of amyotrophic lateral sclerosis. Neuropharmacology 2017, 112, 346–364. [Google Scholar] [CrossRef]
- Ghosh, C.; Marchi, N.; Hossain, M.; Rasmussen, P.; Alexopoulos, A.V.; Gonzalez-Martinez, J.; Yang, H.; Janigro, D. A pro-convulsive carbamazepine metabolite: Quinolinic acid in drug resistant epileptic human brain. Neurobiol. Dis. 2012, 46, 692–700. [Google Scholar] [CrossRef]
- Birch, P.J.; Grossman, C.J.; Hayes, A.G. Kynurenic acid antagonises responses to NMDA via an action at the strychnine-insensitive glycine receptor. Eur. J. Pharmacol. 1988, 154, 85–87. [Google Scholar] [CrossRef]
- Kumar, A. NMDA Receptor Function During Senescence: Implication on Cognitive Performance. Front. Neurosci. 2015, 9, 473. [Google Scholar] [CrossRef]
- Chen, Q.Y.; Li, X.H.; Zhuo, M. NMDA receptors and synaptic plasticity in the anterior cingulate cortex. Neuropharmacology 2021, 197, 108749. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.R.; Di Fazio, C.; Battaglia, S. Activated Tryptophan-Kynurenine metabolic system in the human brain is associated with learned fear. Front. Mol. Neurosci. 2023, 16, 1217090. [Google Scholar] [CrossRef]
- Miranda, A.F.; Boegman, R.J.; Beninger, R.J.; Jhamandas, K. Protection against quinolinic acid-mediated excitotoxicity in nigrostriatal dopaminergic neurons by endogenous kynurenic acid. Neuroscience 1997, 78, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Q.; Salituro, F.G.; Schwarcz, R. Enzyme-catalyzed production of the neuroprotective NMDA receptor antagonist 7-chlorokynurenic acid in the rat brain in vivo. Eur. J. Pharmacol. 1997, 319, 13–20. [Google Scholar] [CrossRef]
- Russi, P.; Alesiani, M.; Lombardi, G.; Davolio, P.; Pellicciari, R.; Moroni, F. Nicotinylalanine increases the formation of kynurenic acid in the brain and antagonizes convulsions. J. Neurochem. 1992, 59, 2076–2080. [Google Scholar] [CrossRef]
- Vécsei, L.; Beal, M.F. Comparative behavioral and pharmacological studies with centrally administered kynurenine and kynurenic acid in rats. Eur. J. Pharmacol. 1991, 196, 239–246. [Google Scholar] [CrossRef]
- Beal, M.F.; Matson, W.R.; Swartz, K.J.; Gamache, P.H.; Bird, E.D. Kynurenine pathway measurements in Huntington’s disease striatum: Evidence for reduced formation of kynurenic acid. J. Neurochem. 1990, 55, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Spekker, E.; Szabó, Á.; Polyák, H.; Vécsei, L. Modelling the neurodevelopmental pathogenesis in neuropsychiatric disorders. Bioactive kynurenines and their analogues as neuroprotective agents—In celebration of 80th birthday of Professor Peter Riederer. J. Neural Transm. 2022, 129, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Moroni, F. Tryptophan metabolism and brain function: Focus on kynurenine and other indole metabolites. Eur. J. Pharmacol. 1999, 375, 87–100. [Google Scholar] [CrossRef]
- Boegman, R.J.; Jhamandas, K.; Beninger, R.J. Neurotoxicity of tryptophan metabolites. Ann. N. Y. Acad. Sci. 1990, 585, 261–273. [Google Scholar] [CrossRef]
- Schwarcz, R. Metabolism and function of brain kynurenines. Biochem. Soc. Trans. 1993, 21, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Q.; Guidetti, P.; Goodman, J.H.; Varasi, M.; Ceresoli-Borroni, G.; Speciale, C.; Scharfman, H.E.; Schwarcz, R. Kynurenergic manipulations influence excitatory synaptic function and excitotoxic vulnerability in the rat hippocampus in vivo. Neuroscience 2000, 97, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Guillemin, G.J.; Kerr, S.J.; Smythe, G.A.; Smith, D.G.; Kapoor, V.; Armati, P.J.; Croitoru, J.; Brew, B.J. Kynurenine pathway metabolism in human astrocytes: A paradox for neuronal protection. J. Neurochem. 2001, 78, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Thevandavakkam, M.A.; Schwarcz, R.; Muchowski, P.J.; Giorgini, F. Targeting kynurenine 3-monooxygenase (KMO): Implications for therapy in Huntington’s disease. CNS Neurol. Disord. Drug Targets 2010, 9, 791–800. [Google Scholar] [CrossRef]
- Kocki, T.; Wnuk, S.; Kloc, R.; Kocki, J.; Owe-Larsson, B.; Urbanska, E.M. New insight into the antidepressants action: Modulation of kynurenine pathway by increasing the kynurenic acid/3-hydroxykynurenine ratio. J. Neural. Transm. 2012, 119, 235–243. [Google Scholar] [CrossRef]
- Ye, Z.; Yang, S.; Lu, L.; Zong, M.; Fan, L.; Kang, C. Unlocking the potential of the 3-hydroxykynurenine/kynurenic acid ratio: A promising biomarker in adolescent major depressive disorder. Eur. Arch. Psychiatry Clin. Neurosci. 2024, 1–13. [Google Scholar] [CrossRef]
- Tanaka, M.; Török, N.; Tóth, F.; Szabó, Á.; Vécsei, L. Co-players in chronic pain: Neuroinflammation and the tryptophan-kynurenine metabolic pathway. Biomedicines 2021, 9, 897. [Google Scholar] [CrossRef]
- Liloia, D.; Zamfira, D.A.; Tanaka, M.; Manuello, J.; Crocetta, A.; Keller, R.; Cozzolino, M.; Duca, S.; Cauda, F.; Costa, T. Disentangling the role of gray matter volume and concentration in autism spectrum disorder: A meta-analytic investigation of 25 years of voxel-based morphometry research. Neurosci. Biobehav. Rev. 2024, 164, 105791. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, S.; Avenanti, A.; Vécsei, L.; Tanaka, M. Neural correlates and molecular mechanisms of memory and learning. Int. J. Mol. Sci. 2024, 25, 2724. [Google Scholar] [CrossRef]
- Tanaka, M.; Vécsei, L. A Decade of Dedication: Pioneering Perspectives on Neurological Diseases and Mental Illnesses. Biomedicines 2024, 12, 1083. [Google Scholar] [CrossRef]
- Valotto Neto, L.J.; Reverete de Araujo, M.; Moretti Junior, R.C.; Mendes Machado, N.; Joshi, R.K.; dos Santos Buglio, D.; Barbalho Lamas, C.; Direito, R.; Fornari Laurindo, L.; Tanaka, M. Investigating the Neuroprotective and Cognitive-Enhancing Effects of Bacopa monnieri: A Systematic Review Focused on Inflammation, Oxidative Stress, Mitochondrial Dysfunction, and Apoptosis. Antioxidants 2024, 13, 393. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, S.; Avenanti, A.; Vécsei, L.; Tanaka, M. Neurodegeneration in cognitive impairment and mood disorders for experimental, clinical and translational neuropsychiatry. Biomedicines 2024, 12, 574. [Google Scholar] [CrossRef] [PubMed]
- Balogh, L.; Tanaka, M.; Török, N.; Vécsei, L.; Taguchi, S. Crosstalk between existential phenomenological psychotherapy and neurological sciences in mood and anxiety disorders. Biomedicines 2021, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Battaglia, S.; Giménez-Llort, L.; Chen, C.; Hepsomali, P.; Avenanti, A.; Vécsei, L. Innovation at the intersection: Emerging translational research in neurology and psychiatry. Cells 2024, 13, 790. [Google Scholar] [CrossRef] [PubMed]
- Juhász, L.; Szolnoki, B.Z.; Nászai, A.; Szabó, Á.; Rutai, A.; Tallósy, S.P.; Toldi, J.; Tanaka, M.; Ono, E.; Vécsei, L. Electron transport disturbances in kynurenine aminotransferase knockout mice. Biochim. Biophys. Acta-Bioenerg. 2024, 1865, 149389. [Google Scholar]
- Hiraku, Y.; Inoue, S.; Oikawa, S.; Yamamoto, K.; Tada, S.; Nishino, K.; Kawanishi, S. Metal-mediated oxidative damage to cellular and isolated DNA by certain tryptophan metabolites. Carcinogenesis 1995, 16, 349–356. [Google Scholar] [CrossRef]
- Thomas, S.R.; Witting, P.K.; Stocker, R. 3-Hydroxyanthranilic acid is an efficient, cell-derived co-antioxidant for α-tocopherol, inhibiting human low density lipoprotein and plasma lipid peroxidation. J. Biol. Chem. 1996, 271, 32714–32721. [Google Scholar] [CrossRef]
- Esaki, H.; Onozaki, H.; Kawakishi, S.; Osawa, T. New antioxidant isolated from tempeh. J. Agric. Food Chem. 1996, 44, 696–700. [Google Scholar] [CrossRef]
- Christen, S.; Peterhans, E.; Stocker, R. Antioxidant activities of some tryptophan metabolites: Possible implication for inflammatory diseases. Proc. Natl. Acad. Sci. USA 1990, 87, 2506–2510. [Google Scholar] [CrossRef]
- Krause, D.; Suh, H.-S.; Tarassishin, L.; Cui, Q.L.; Durafourt, B.A.; Choi, N.; Bauman, A.; Cosenza-Nashat, M.; Antel, J.P.; Zhao, M.-L. The tryptophan metabolite 3-hydroxyanthranilic acid plays anti-inflammatory and neuroprotective roles during inflammation: Role of hemeoxygenase-1. Am. J. Pathol. 2011, 179, 1360–1372. [Google Scholar] [CrossRef]
- Gawel, K. A Review on the Role and Function of Cinnabarinic Acid, a “Forgotten” Metabolite of the Kynurenine Pathway. Cells 2024, 13, 453. [Google Scholar] [CrossRef] [PubMed]
- Parker, N.R.; Jamie, J.F.; Davies, M.J.; Truscott, R.J. Protein-bound kynurenine is a photosensitizer of oxidative damage. Free Radic. Biol. Med. 2004, 37, 1479–1489. [Google Scholar] [CrossRef]
- Song, H.; Park, H.; Kim, Y.-S.; Kim, K.D.; Lee, H.-K.; Cho, D.-H.; Yang, J.-W.; Hur, D.Y. L-kynurenine-induced apoptosis in human NK cells is mediated by reactive oxygen species. Int. Immunopharmacol. 2011, 11, 932–938. [Google Scholar] [CrossRef]
- Palzkill, V.R.; Thome, T.; Murillo, A.L.; Khattri, R.B.; Ryan, T.E. Increasing plasma L-kynurenine impairs mitochondrial oxidative phosphorylation prior to the development of atrophy in murine skeletal muscle: A pilot study. Front. Physiol. 2022, 13, 992413. [Google Scholar] [CrossRef]
- Bratek-Gerej, E.; Ziembowicz, A.; Godlewski, J.; Salinska, E. The mechanism of the neuroprotective effect of kynurenic acid in the experimental model of neonatal hypoxia–ischemia: The link to oxidative stress. Antioxidants 2021, 10, 1775. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.W. Does kynurenic acid act on nicotinic receptors? An assessment of the evidence. J. Neurochem. 2020, 152, 627–649. [Google Scholar] [CrossRef]
- Cosi, C.; Mannaioni, G.; Cozzi, A.; Carlà, V.; Sili, M.; Cavone, L.; Maratea, D.; Moroni, F. G-protein coupled receptor 35 (GPR35) activation and inflammatory pain: Studies on the antinociceptive effects of kynurenic acid and zaprinast. Neuropharmacology 2011, 60, 1227–1231. [Google Scholar] [CrossRef] [PubMed]
- Fallarini, S.; Magliulo, L.; Paoletti, T.; de Lalla, C.; Lombardi, G. Expression of functional GPR35 in human iNKT cells. Biochem. Biophys. Res. Commun. 2010, 398, 420–425. [Google Scholar] [CrossRef]
- Guo, J.; Williams, D.J.; Puhl, H.L.; Ikeda, S.R. Inhibition of N-type calcium channels by activation of GPR35, an orphan receptor, heterologously expressed in rat sympathetic neurons. J. Pharmacol. Exp. Ther. 2008, 324, 342–351. [Google Scholar] [CrossRef]
- Korlimbinis, A.; Hains, P.G.; Truscott, R.J.; Aquilina, J.A. 3-Hydroxykynurenine oxidizes alpha-crystallin: Potential role in cataractogenesis. Biochemistry 2006, 45, 1852–1860. [Google Scholar] [CrossRef]
- Leipnitz, G.; Schumacher, C.; Dalcin, K.B.; Scussiato, K.; Solano, A.; Funchal, C.; Dutra-filho, C.; Wyse, A.; Wannmacher, C.; Latini, A.; et al. In vitro evidence for an antioxidant role of 3-hydroxykynurenine and 3-hydroxyanthranilic acid in the brain. Neurochem. Int. 2007, 50, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Haneda, M.; Yoshino, M. Prooxidant action of xanthurenic acid and quinoline compounds: Role of transition metals in the generation of reactive oxygen species and enhanced formation of 8-hydroxy-2’-deoxyguanosine in DNA. Biometals 2006, 19, 429–435. [Google Scholar] [CrossRef] [PubMed]
- González Esquivel, D.; Ramírez-Ortega, D.; Pineda, B.; Castro, N.; Ríos, C.; Pérez de la Cruz, V. Kynurenine pathway metabolites and enzymes involved in redox reactions. Neuropharmacology 2017, 112, 331–345. [Google Scholar] [CrossRef]
- Murakami, K.; Ito, M.; Yoshino, M. Xanthurenic acid inhibits metal ion-induced lipid peroxidation and protects NADP-isocitrate dehydrogenase from oxidative inactivation. J. Nutr. Sci. Vitaminol. 2001, 47, 306–310. [Google Scholar] [CrossRef]
- Malina, H.Z.; Hess, O.M. Xanthurenic acid translocates proapoptotic Bcl-2 family proteins into mitochondria and impairs mitochondrial function. BMC Cell Biol. 2004, 5, 14. [Google Scholar] [CrossRef][Green Version]
- Lai, Q.; Wu, L.; Dong, S.; Zhu, X.; Fan, Z.; Kou, J.; Liu, F.; Yu, B.; Li, F. Inhibition of KMO Ameliorates Myocardial Ischemia Injury via Maintaining Mitochondrial Fusion and Fission Balance. Int. J. Biol. Sci. 2023, 19, 3077–3098. [Google Scholar] [CrossRef] [PubMed]
- Neale, S.A.; Copeland, C.S.; Uebele, V.N.; Thomson, F.J.; Salt, T.E. Modulation of hippocampal synaptic transmission by the kynurenine pathway member xanthurenic acid and other VGLUT inhibitors. Neuropsychopharmacology 2013, 38, 1060–1067. [Google Scholar] [CrossRef]
- Copeland, C.S.; Neale, S.A.; Salt, T.E. Actions of Xanthurenic acid, a putative endogenous Group II metabotropic glutamate receptor agonist, on sensory transmission in the thalamus. Neuropharmacology 2013, 66, 133–142. [Google Scholar] [CrossRef]
- Chakraborty, P.; Dey, A.; Gopalakrishnan, A.V.; Swati, K.; Ojha, S.; Prakash, A.; Kumar, D.; Ambasta, R.K.; Jha, N.K.; Jha, S.K.; et al. Glutamatergic neurotransmission: A potential pharmacotherapeutic target for the treatment of cognitive disorders. Ageing Res. Rev. 2023, 85, 101838. [Google Scholar] [CrossRef]
- Hiramatsu, R.; Hara, T.; Akimoto, H.; Takikawa, O.; Kawabe, T.; Isobe, K.; Nagase, F. Cinnabarinic acid generated from 3-hydroxyanthranilic acid strongly induces apoptosis in thymocytes through the generation of reactive oxygen species and the induction of caspase. J. Cell. Biochem. 2008, 103, 42–53. [Google Scholar] [CrossRef]
- Hu, S.; Hu, C.; Luo, L.; Zhang, H.; Zhao, S.; Liu, Z.; Zeng, L. Pu-erh tea increases the metabolite Cinnabarinic acid to improve circadian rhythm disorder-induced obesity. Food Chem. 2022, 394, 133500. [Google Scholar] [CrossRef] [PubMed]
- Natella, F.; Nardini, M.; Di Felice, M.; Scaccini, C. Benzoic and cinnamic acid derivatives as antioxidants: Structure− activity relation. J. Agric. Food Chem. 1999, 47, 1453–1459. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Shan, C.; Pan, B.; Pignatello, J.J. The Fenton reaction in water assisted by picolinic acid: Accelerated iron cycling and co-generation of a selective Fe-based oxidant. Environ. Sci. Technol. 2021, 55, 8299–8308. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Wang, J.; Ashley, D.C.; Sharma, V.K.; Huang, C.-H. Enhanced degradation of micropollutants in a peracetic acid–Fe (III) system with picolinic acid. Environ. Sci. Technol. 2022, 56, 4437–4446. [Google Scholar] [CrossRef]
- Subramaniam, P.; Selvi, N.T. Picolinic acid promoted oxidative decarboxylation of phenylsulfinylacetic acid by Cr (VI). Bull. Chem. Soc. Ethiop. 2016, 30, 137–146. [Google Scholar] [CrossRef][Green Version]
- Tan, G.-Y.; Zheng, S.-S.; Zhang, M.-H.; Feng, J.-H.; Xie, P.; Bi, J.-M. Study of Oxidative Damage in Growing–Finishing Pigs with Continuous Excess Dietary Chromium Picolinate Intake. Biol. Trace Elem. Res. 2008, 126, 129–140. [Google Scholar] [CrossRef]
- Sundaram, B.; Aggarwal, A.; Sandhir, R. Chromium picolinate attenuates hyperglycemia-induced oxidative stress in streptozotocin-induced diabetic rats. J. Trace Elem. Med. Biol. 2013, 27, 117–121. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Watanabe, H.; Otagiri, M.; Maruyama, T. New Insight Into the Redox Properties of Uremic Solute Indoxyl Sulfate as a Pro-and Anti-Oxidant. Ther. Apher. Dial. 2011, 15, 129–131. [Google Scholar] [CrossRef]
- Rapa, S.F.; Prisco, F.; Popolo, A.; Iovane, V.; Autore, G.; Di Iorio, B.R.; Dal Piaz, F.; Paciello, O.; Nishijima, F.; Marzocco, S. Pro-Inflammatory Effects of Indoxyl Sulfate in Mice: Impairment of Intestinal Homeostasis and Immune Response. Int. J. Mol. Sci. 2021, 22, 1135. [Google Scholar] [CrossRef]
- Tan, Y.-Q.; Wang, Y.-N.; Feng, H.-Y.; Guo, Z.-Y.; Li, X.; Nie, X.-L.; Zhao, Y.-Y. Host/microbiota interactions-derived tryptophan metabolites modulate oxidative stress and inflammation via aryl hydrocarbon receptor signaling. Free Radic. Biol. Med. 2022, 184, 30–41. [Google Scholar] [CrossRef]
- Praschberger, M.; Hermann, M.; Laggner, C.; Jirovetz, L.; Exner, M.; Kapiotis, S.; Gmeiner, B.M.; Laggner, H. Carbamoylation abrogates the antioxidant potential of hydrogen sulfide. Biochimie 2013, 95, 2069–2075. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.C.; DiNatale, B.C.; Murray, I.A.; Flaveny, C.A.; Liu, Q.; Laurenzana, E.M.; Lin, J.M.; Strom, S.C.; Omiecinski, C.J.; Amin, S. The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry 2010, 49, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, I.; Tatebe, J.; Namba, S.; Koizumi, M.; Yamazaki, J.; Morita, T. Activation of aryl hydrocarbon receptor mediates indoxyl sulfate-induced monocyte chemoattractant protein-1 expression in human umbilical vein endothelial cells. Circ. J. 2013, 77, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.; Edgley, A.J.; Kelly, D.J.; Kompa, A.R. Aryl hydrocarbon receptor inhibition restores indoxyl sulfate-mediated endothelial dysfunction in rat aortic rings. Toxins 2022, 14, 100. [Google Scholar] [CrossRef]
- Ölgen, S.; Çoban, T. Synthesis and Antioxidant Properties of Novel N-Substituted Indole-2-carboxamide and Indole-3-acetamide Derivatives. Arch. Pharm. Int. J. Pharm. Med. Chem. 2002, 335, 331–338. [Google Scholar] [CrossRef]
- Kanwal; Khan, K.M.; Chigurupati, S.; Ali, F.; Younus, M.; Aldubayan, M.; Wadood, A.; Khan, H.; Taha, M.; Perveen, S. Indole-3-acetamides: As potential antihyperglycemic and antioxidant agents; synthesis, in vitro α-amylase inhibitory activity, structure–activity relationship, and in silico studies. ACS Omega 2021, 6, 2264–2275. [Google Scholar] [CrossRef]
- Ölgen, S.; Bakar, F.; Aydin, S.; Nebioğlu, D.; Nebioğlu, S. Synthesis of new indole-2-carboxamide and 3-acetamide derivatives and evaluation their antioxidant properties. J. Enzym. Inhib. Med. Chem. 2013, 28, 58–64. [Google Scholar] [CrossRef]
- Vyhlídalová, B.; Krasulová, K.; Pečinková, P.; Marcalíková, A.; Vrzal, R.; Zemánková, L.; Vančo, J.; Trávníček, Z.; Vondráček, J.; Karasová, M. Gut microbial catabolites of tryptophan are ligands and agonists of the aryl hydrocarbon receptor: A detailed characterization. Int. J. Mol. Sci. 2020, 21, 2614. [Google Scholar] [CrossRef]
- Ng, T.; Liu, F.; Zhao, L. Antioxidative and free radical scavenging activities of pineal indoles. J. Neural Transm. 2000, 107, 1243–1251. [Google Scholar] [CrossRef]
- Choi, S.; Kim, Y.; Oh, S.; Oh, S.; Chun, T.; Kim, S. Inhibitory effect of skatole (3-methylindole) on enterohemorrhagic Escherichia coli O157: H7 ATCC 43894 biofilm formation mediated by elevated endogenous oxidative stress. Lett. Appl. Microbiol. 2014, 58, 454–461. [Google Scholar] [CrossRef]
- Hong, S.-H.; Hong, Y.; Lee, M.; Keum, B.-R.; Kim, G.-H. Natural product skatole ameliorates lipotoxicity-induced multiple hepatic damage under hyperlipidemic conditions in hepatocytes. Nutrients 2023, 15, 1490. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.K.; Balaguer, P.; Ekstrand, B.; Daujat-Chavanieu, M.; Gerbal-Chaloin, S. Skatole (3-methylindole) is a partial aryl hydrocarbon receptor agonist and induces CYP1A1/2 and CYP1B1 expression in primary human hepatocytes. PLoS ONE 2016, 11, e0154629. [Google Scholar] [CrossRef] [PubMed]
- Churro, C.; Fernandes, A.; Alverca, E.; Sam-Bento, F.; Paulino, S.; Figueira, V.; Bento, A.; Prabhakar, S.; Lobo, A.; Martins, L. Effects of tryptamine on growth, ultrastructure, and oxidative stress of cyanobacteria and microalgae cultures. Hydrobiologia 2010, 649, 195–206. [Google Scholar] [CrossRef]
- Bentz, E.N.; Lobayan, R.M.; Martinez, H.; Redondo, P.; Largo, A. Intrinsic antioxidant potential of the aminoindole structure: A computational kinetics study of tryptamine. J. Phys. Chem. B 2018, 122, 6386–6395. [Google Scholar] [CrossRef] [PubMed]
- Estevão, M.S.; Carvalho, L.C.; Ribeiro, D.; Couto, D.; Freitas, M.; Gomes, A.; Ferreira, L.; Fernandes, E.; Marques, M.M.B. Antioxidant activity of unexplored indole derivatives: Synthesis and screening. Eur. J. Med. Chem. 2010, 45, 4869–4878. [Google Scholar] [CrossRef]
- Asghar, S.; Mushtaq, N.; Ahmad, A.; Munawwar, R.; Ansari, S.; Rizvi, S.A. Design, Synthesis and Therapeutic investigation of Tryptamine derivatives as Potential Antioxidant and Amyloid inhibitor/disaggregator. Res. J. Pharm. Technol. 2023, 16, 3622–3632. [Google Scholar] [CrossRef]
- Cheng, Y.; Jin, U.-H.; Allred, C.D.; Jayaraman, A.; Chapkin, R.S.; Safe, S. Aryl hydrocarbon receptor activity of tryptophan metabolites in young adult mouse colonocytes. Drug Metab. Dispos. 2015, 43, 1536–1543. [Google Scholar] [CrossRef]
- Jin, U.-H.; Lee, S.-O.; Sridharan, G.; Lee, K.; Davidson, L.A.; Jayaraman, A.; Chapkin, R.S.; Alaniz, R.; Safe, S. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol. Pharmacol. 2014, 85, 777–788. [Google Scholar] [CrossRef]
- Dopkins, N.; Becker, W.; Miranda, K.; Walla, M.; Nagarkatti, P.; Nagarkatti, M. Tryptamine attenuates experimental multiple sclerosis through activation of aryl hydrocarbon receptor. Front. Pharmacol. 2021, 11, 619265. [Google Scholar] [CrossRef]
- Smirnova, A.; Wincent, E.; Vikström Bergander, L.; Alsberg, T.; Bergman, J.; Rannug, A.; Rannug, U. Evidence for new light-independent pathways for generation of the endogenous aryl hydrocarbon receptor agonist FICZ. Chem. Res. Toxicol. 2016, 29, 75–86. [Google Scholar] [CrossRef]
- Herraiz, T.; Galisteo, J. Endogenous and dietary indoles: A class of antioxidants and radical scavengers in the ABTS assay. Free Radic. Res. 2004, 38, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Aoki, R.; Aoki-Yoshida, A.; Suzuki, C.; Takayama, Y. Indole-3-pyruvic acid, an aryl hydrocarbon receptor activator, suppresses experimental colitis in mice. J. Immunol. 2018, 201, 3683–3693. [Google Scholar] [CrossRef] [PubMed]
- Kamata, K.; Hara, A.; Minaga, K.; Yoshikawa, T.; Kurimoto, M.; Sekai, I.; Okai, N.; Omaru, N.; Masuta, Y.; Otsuka, Y. Activation of the aryl hydrocarbon receptor inhibits the development of experimental autoimmune pancreatitis through IL-22-mediated signaling pathways. Clin. Exp. Immunol. 2023, 212, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Kosaka, M.; Shindo, K.; Kawasumi, T.; Kimoto-Nira, H.; Suzuki, C. Identification of antioxidants produced by Lactobacillus plantarum. Biosci. Biotechnol. Biochem. 2013, 77, 1299–1302. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 2017, 22, 25–37. [Google Scholar] [CrossRef]
- Chyan, Y.-J.; Poeggeler, B.; Omar, R.A.; Chain, D.G.; Frangione, B.; Ghiso, J.; Pappolla, M.A. Potent neuroprotective properties against the Alzheimer β-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid. J. Biol. Chem. 1999, 274, 21937–21942. [Google Scholar] [CrossRef]
- Rynkowska, A.; Stępniak, J.; Karbownik-Lewińska, M. Melatonin and indole-3-propionic acid reduce oxidative damage to membrane lipids induced by high iron concentrations in porcine skin. Membranes 2021, 11, 571. [Google Scholar] [CrossRef]
- Bendheim, P.E.; Poeggeler, B.; Neria, E.; Ziv, V.; Pappolla, M.A.; Chain, D.G. Development of indole-3-propionic acid (OXIGON™) for alzheimer’s disease. J. Mol. Neurosci. 2002, 19, 213–217. [Google Scholar] [CrossRef]
- Motta, J.-P.; Zhu, K.; Liu, W.; Liu, K.-X.; Huang, Z.-B.; Hu, Z.; Lu, C.-X.; Luo, S.-D.; Chen, Y.; Zhou, Z.-P.; et al. Gut microbiota-derived indole 3-propionic acid partially activates aryl hydrocarbon receptor to promote macrophage phagocytosis and attenuate septic injury. Front. Cell. Infect. Microbiol. 2022, 12, 1015386. [Google Scholar]
- Chobot, V.; Hadacek, F.; Weckwerth, W.; Kubicova, L. Iron chelation and redox chemistry of anthranilic acid and 3-hydroxyanthranilic acid: A comparison of two structurally related kynurenine pathway metabolites to obtain improved insights into their potential role in neurological disease development. J. Organomet. Chem. 2015, 782, 103–110. [Google Scholar] [CrossRef]
- Francisco-Marquez, M.; Fernández, M.A.; Galano, A. Anthranilic acid as a secondary antioxidant. Comput. Theor. Chem. 2015, 1077, 18–24. [Google Scholar] [CrossRef]
- Ramírez-Ortega, D.; Ramiro-Salazar, A.; González-Esquivel, D.; Ríos, C.; Pineda, B.; De La Cruz, P. 3-Hydroxykynurenine and 3-Hydroxyanthranilic Acid Enhance the Toxicity Induced by Copper in Rat Astrocyte Culture. Oxid. Med. Cell Longev. 2017, 2017, 2371895. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Piñeiro, R.J.; Dali, M.; Mansuy, D.; Boucher, J.-L. Unstability of cinnabarinic acid, an endogenous metabolite of tryptophan, under situations mimicking physiological conditions. Biochimie 2022, 199, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Zollner, H. Effects of cinnabarinic acid on mitochondrial respiration. Biochem. Pharmacol. 1976, 25, 643–648. [Google Scholar] [CrossRef]
- Nagamura, Y.; Uesugi, K.; Naito, J.; Ishiguro, I. Cinnabarinic acid was formed in damaged mitochondria and its effect on mitochondrial respiration. Adv. Exp. Med. Biol. 1999, 467, 419–423. [Google Scholar] [CrossRef]
- Jovanovic, F.; Candido, K.D.; Knezevic, N.N. The Role of the Kynurenine Signaling Pathway in Different Chronic Pain Conditions and Potential Use of Therapeutic Agents. Int. J. Mol. Sci. 2020, 21, 6045. [Google Scholar] [CrossRef]
- Jacobs, K.; Lim, C.; Blennow, K.; Zetterberg, H.; Chatterjee, P.; Martins, R.; Brew, B.; Guillemin, G.; Lovejoy, D. Correlation between plasma and CSF concentrations of kynurenine pathway metabolites in Alzheimer’s disease and relationship to amyloid-β and tau. Neurobiol. Aging 2019, 80, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mezrich, J.D.; Fechner, J.H.; Zhang, X.; Johnson, B.P.; Burlingham, W.J.; Bradfield, C.A. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 2010, 185, 3190–3198. [Google Scholar] [CrossRef]
- Kawasaki, H.; Chang, H.W.; Tseng, H.C.; Hsu, S.C.; Yang, S.J.; Hung, C.H.; Zhou, Y.; Huang, S.K. A tryptophan metabolite, kynurenine, promotes mast cell activation through aryl hydrocarbon receptor. Allergy 2014, 69, 445–452. [Google Scholar] [CrossRef]
- Kaiser, H.; Parker, E.; Hamrick, M.W. Kynurenine signaling through the aryl hydrocarbon receptor: Implications for aging and healthspan. Exp. Gerontol. 2020, 130, 110797. [Google Scholar] [CrossRef]
- Badawy, A.A.; Dawood, S. Molecular Insights into the Interaction of Tryptophan Metabolites with the Human Aryl Hydrocarbon Receptor in Silico: Tryptophan as Antagonist and no Direct Involvement of Kynurenine. Front. Biosci. -Landmark 2024, 29, 333. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.W. Kynurenic acid antagonists and kynurenine pathway inhibitors. Expert. Opin. Investig. Drugs 2001, 10, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Tuboly, G.; Tar, L.; Bohar, Z.; Safrany-Fark, A.; Petrovszki, Z.; Kekesi, G.; Vecsei, L.; Pardutz, A.; Horvath, G. The inimitable kynurenic acid: The roles of different ionotropic receptors in the action of kynurenic acid at a spinal level. Brain Res. Bull. 2015, 112, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Patte-Mensah, C.; Taleb, O.; Bourguignon, J.; Schmitt, M.; Bihel, F.; Maitre, M.; Mensah-Nyagan, A. The neuroprotector kynurenic acid increases neuronal cell survival through neprilysin induction. Neuropharmacology 2013, 70, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Dietrich, D.; Gräsel, I.; Reuter, G.; Seifert, G.; Steinhäuser, C. 6-Hydroxykynurenic acid and kynurenic acid differently antagonise AMPA and NMDA receptors in hippocampal neurones. J. Neurochem. 2001, 77, 1108–1115. [Google Scholar] [CrossRef]
- Prescott, C.; Weeks, A.M.; Staley, K.J.; Partin, K.M. Kynurenic acid has a dual action on AMPA receptor responses. Neurosci. Lett. 2006, 402, 108–112. [Google Scholar] [CrossRef]
- Foley, C.M.; Moffitt, J.A.; Hay, M.; Hasser, E.M. Glutamate in the nucleus of the solitary tract activates both ionotropic and metabotropic glutamate receptors. Am. J. Physiol. 1998, 275, R1858–R1866. [Google Scholar] [CrossRef]
- Deora, G.S.; Kantham, S.; Chan, S.; Dighe, S.N.; Veliyath, S.K.; McColl, G.; Parat, M.O.; McGeary, R.P.; Ross, B.P. Multifunctional Analogs of Kynurenic Acid for the Treatment of Alzheimer’s Disease: Synthesis, Pharmacology, and Molecular Modeling Studies. ACS Chem. Neurosci. 2017, 8, 2667–2675. [Google Scholar] [CrossRef]
- Wang, Y.T.; Wang, X.L.; Feng, S.T.; Chen, N.H.; Wang, Z.Z.; Zhang, Y. Novel rapid-acting glutamatergic modulators: Targeting the synaptic plasticity in depression. Pharmacol. Res. 2021, 171, 105761. [Google Scholar] [CrossRef]
- DiNatale, B.C.; Murray, I.A.; Schroeder, J.C.; Flaveny, C.A.; Lahoti, T.S.; Laurenzana, E.M.; Omiecinski, C.J.; Perdew, G.H. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol. Sci. 2010, 115, 89–97. [Google Scholar] [CrossRef]
- Moroni, F.; Cozzi, A.; Sili, M.; Mannaioni, G. Kynurenic acid: A metabolite with multiple actions and multiple targets in brain and periphery. J. Neural. Transm. 2012, 119, 133–139. [Google Scholar] [CrossRef] [PubMed]
- García-Lara, L.; Pérez-Severiano, F.; González-Esquivel, D.; Elizondo, G.; Segovia, J. Absence of aryl hydrocarbon receptors increases endogenous kynurenic acid levels and protects mouse brain against excitotoxic insult and oxidative stress. J. Neurosci. Res. 2015, 93, 1423–1433. [Google Scholar] [CrossRef] [PubMed]
- Cortés Malagón, E.M.; López Ornelas, A.; Olvera Gómez, I.; Bonilla Delgado, J. The Kynurenine Pathway, Aryl Hydrocarbon Receptor, and Alzheimer’s Disease. Brain Sci. 2024, 14, 950. [Google Scholar] [CrossRef]
- Fazio, F.; Lionetto, L.; Curto, M.; Iacovelli, L.; Copeland, C.S.; Neale, S.A.; Bruno, V.; Battaglia, G.; Salt, T.E.; Nicoletti, F. Cinnabarinic acid and xanthurenic acid: Two kynurenine metabolites that interact with metabotropic glutamate receptors. Neuropharmacology 2017, 112, 365–372. [Google Scholar] [CrossRef]
- Sun, M.; Ma, N.; He, T.; Johnston, L.J.; Ma, X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit. Rev. Food Sci. Nutr. 2020, 60, 1760–1768. [Google Scholar] [CrossRef]
- Lin, J.; Sun-Waterhouse, D.; Cui, C. The therapeutic potential of diet on immune-related diseases: Based on the regulation on tryptophan metabolism. Crit. Rev. Food Sci. Nutr. 2022, 62, 8793–8811. [Google Scholar] [CrossRef]
- Ambrosio, L.F.; Insfran, C.; Volpini, X.; Acosta Rodriguez, E.; Serra, H.M.; Quintana, F.J.; Cervi, L.; Motrán, C.C. Role of Aryl Hydrocarbon Receptor (AhR) in the Regulation of Immunity and Immunopathology During Trypanosoma cruzi Infection. Front. Immunol. 2019, 10, 631. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.K.; Kwon, B. Immune regulation through tryptophan metabolism. Exp. Mol. Med. 2023, 55, 1371–1379. [Google Scholar] [CrossRef]
- Dong, F.; Perdew, G.H. The aryl hydrocarbon receptor as a mediator of host-microbiota interplay. Gut. Microbes. 2020, 12, 1859812. [Google Scholar] [CrossRef]
- Fazio, F.; Lionetto, L.; Molinaro, G.; Bertrand, H.O.; Acher, F.; Ngomba, R.T.; Notartomaso, S.; Curini, M.; Rosati, O.; Scarselli, P.; et al. Cinnabarinic acid, an endogenous metabolite of the kynurenine pathway, activates type 4 metabotropic glutamate receptors. Mol. Pharmacol. 2012, 81, 643–656. [Google Scholar] [CrossRef]
- Lowe, M.M.; Mold, J.E.; Kanwar, B.; Huang, Y.; Louie, A.; Pollastri, M.P.; Wang, C.; Patel, G.; Franks, D.G.; Schlezinger, J.; et al. Identification of cinnabarinic acid as a novel endogenous aryl hydrocarbon receptor ligand that drives IL-22 production. PLoS ONE 2014, 9, e87877. [Google Scholar] [CrossRef] [PubMed]
- Seok, S.-H.; Ma, Z.-X.; Feltenberger, J.B.; Chen, H.; Chen, H.; Scarlett, C.; Lin, Z.; Satyshur, K.A.; Cortopassi, M.; Jefcoate, C.R. Trace derivatives of kynurenine potently activate the aryl hydrocarbon receptor (AHR). J. Biol. Chem. 2018, 293, 1994–2005. [Google Scholar] [CrossRef] [PubMed]
- Plattén, M.; Litzenburger, U.; Wick, W. The aryl hydrocarbon receptor in tumor immunity. Oncoimmunology 2012, 1, 396–397. [Google Scholar] [CrossRef]
- Rojas, I.Y.; Moyer, B.J.; Ringelberg, C.S.; Wilkins, O.M.; Pooler, D.B.; Ness, D.B.; Coker, S.; Tosteson, T.D.; Lewis, L.D.; Chamberlin, M.D. Kynurenine-Induced Aryl Hydrocarbon Receptor Signaling in Mice Causes Body Mass Gain, Liver Steatosis, and Hyperglycemia. Obesity 2021, 29, 337–349. [Google Scholar] [CrossRef]
- Duarte, J.H.; Di Meglio, P.; Hirota, K.; Ahlfors, H.; Stockinger, B. Differential influences of the aryl hydrocarbon receptor on Th17 mediated responses in vitro and in vivo. PLoS ONE 2013, 8, e79819. [Google Scholar] [CrossRef]
- Haruki, H.; Hovius, R.; Pedersen, M.G.; Johnsson, K. Tetrahydrobiopterin biosynthesis as a potential target of the kynurenine pathway metabolite xanthurenic acid. J. Biol. Chem. 2016, 291, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Kwon, I.-S.; Kwak, J.H.; Pyo, S.; Lee, H.-W.; Kim, A.; Schmitz, F.J. Oscarellin, an anthranilic acid derivative from a Philippine sponge, Oscarella stillans, as an inhibitor of inflammatory cytokines in macrophages. J. Nat. Prod. 2017, 80, 149–155. [Google Scholar] [CrossRef]
- Xue, C.; Gu, X.; Zheng, Q.; Shi, Q.; Yuan, X.; Chu, Q.; Jia, J.; Su, Y.; Bao, Z.; Lu, J. Effects of 3-HAA on HCC by Regulating the Heterogeneous Macrophages—A scRNA-Seq Analysis. Adv. Sci. 2023, 10, 2207074. [Google Scholar] [CrossRef]
- Kennedy, P.J.; Cryan, J.F.; Dinan, T.G.; Clarke, G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology 2017, 112, 399–412. [Google Scholar] [CrossRef]
- Copeland, E.; Whitley, K.; Cleverdon, R.; Baranowski, B.; Marko, D.; MacPherson, R.; Allison, D.; Fajardo, V. Kynurenine Metabolism in the D2 mdx Mouse: A Muscle-to-Brain Connection. FASEB J. 2021, 35. [Google Scholar] [CrossRef]
- Parrott, J.M.; Redus, L.; O’Connor, J.C. Kynurenine metabolic balance is disrupted in the hippocampus following peripheral lipopolysaccharide challenge. J. Neuroinflamm. 2016, 13, 124. [Google Scholar] [CrossRef] [PubMed]
- Topczewska-Bruns, J.; Pawlak, D.; Chabielska, E.; Tankiewicz, A.; Buczko, W. Increased levels of 3-hydroxykynurenine in different brain regions of rats with chronic renal insufficiency. Brain Res. Bull. 2002, 58, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Fuertig, R.; Azzinnari, D.; Bergamini, G.; Cathomas, F.; Sigrist, H.; Seifritz, E.; Vavassori, S.; Luippold, A.; Hengerer, B.; Ceci, A. Mouse chronic social stress increases blood and brain kynurenine pathway activity and fear behaviour: Both effects are reversed by inhibition of indoleamine 2, 3-dioxygenase. Brain Behav. Immun. 2016, 54, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Rea, K.; Dinan, T.G.; Cryan, J.F. The microbiome: A key regulator of stress and neuroinflammation. Neurobiol. Stress 2016, 4, 23–33. [Google Scholar] [CrossRef]
- Mörkl, S.; Butler, M.I.; Lackner, S. Advances in the gut microbiome and mood disorders. Curr. Opin. Psychiatry 2023, 36, 1–7. [Google Scholar] [CrossRef]
- Su, C.-H.; Chuang, H.-C.; Hong, C.-J. Physical exercise prevents mice from L-Kynurenine-induced depression-like behavior. Asian J. Psychiatry 2020, 48, 101894. [Google Scholar] [CrossRef]
- Vegas-Suárez, S.; Simón, J.; Martínez-Chantar, M.L.; Moratalla, R. Metabolic diffusion in Neuropathologies: The relevance of Brain-Liver Axis. Front. Physiol. 2022, 13, 864263. [Google Scholar] [CrossRef]
- Capuron, L.; Miller, A.H. Immune system to brain signaling: Neuropsychopharmacological implications. Pharmacol. Ther. 2011, 130, 226–238. [Google Scholar] [CrossRef]
- Osadchiy, V.; Martin, C.R.; Mayer, E.A. The gut–brain axis and the microbiome: Mechanisms and clinical implications. Clin. Gastroenterol. Hepatol. 2019, 17, 322–332. [Google Scholar] [CrossRef]
- Martin, C.R.; Mayer, E.A. Gut-brain axis and behavior. Intest. Microbiome Funct. Asp. Health Dis. 2017, 88, 45–54. [Google Scholar]
- Gershon, M.D.; Margolis, K.G. The gut, its microbiome, and the brain: Connections and communications. J. Clin. Investig. 2021, 131, e143768. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Inslicht, S.S.; Bhargava, A. Gut-Brain Axis: Role of Microbiome, Metabolomics, Hormones, and Stress in Mental Health Disorders. Cells 2024, 13, 1436. [Google Scholar] [CrossRef] [PubMed]
- Undieh, U.A.U. Dysfunction of the Gut Microbiome-Brain Axis in Neurodegenerative Disease: Role of Indole and Its Metabolites. Master’s Thesis, Drexel University, Philadelphia, PA, USA, May 2023. [Google Scholar]
- Młynarska, E.; Gadzinowska, J.; Tokarek, J.; Forycka, J.; Szuman, A.; Franczyk, B.; Rysz, J. The role of the microbiome-brain-gut axis in the pathogenesis of depressive disorder. Nutrients 2022, 14, 1921. [Google Scholar] [CrossRef] [PubMed]
- Margolis, K.G.; Cryan, J.F.; Mayer, E.A. The microbiota-gut-brain axis: From motility to mood. Gastroenterology 2021, 160, 1486–1501. [Google Scholar] [CrossRef]
- Kern, L.; Mastandrea, I.; Melekhova, A.; Elinav, E. Mechanisms by which microbiome-derived metabolites exert their impacts on neurodegeneration. Cell Chem. Biol. 2024, S2451-9456(24)00363-5. [Google Scholar] [CrossRef] [PubMed]
- Spivak, I.; Fluhr, L.; Elinav, E. Local and systemic effects of microbiome-derived metabolites. EMBO Rep. 2022, 23, e55664. [Google Scholar] [CrossRef]
- Shapiro, H.; Thaiss, C.A.; Levy, M.; Elinav, E. The cross talk between microbiota and the immune system: Metabolites take center stage. Curr. Opin. Immunol. 2014, 30, 54–62. [Google Scholar] [CrossRef]
- Yusufu, I.; Ding, K.; Smith, K.; Wankhade, U.D.; Sahay, B.; Patterson, G.T.; Pacholczyk, R.; Adusumilli, S.; Hamrick, M.W.; Hill, W.D. A tryptophan-deficient diet induces gut microbiota dysbiosis and increases systemic inflammation in aged mice. Int. J. Mol. Sci. 2021, 22, 5005. [Google Scholar] [CrossRef]
- Lin, P.; Li, D.; Shi, Y.; Li, Q.; Guo, X.; Dong, K.; Chen, Q.; Lou, X.; Li, Z.; Li, P. Dysbiosis of the gut microbiota and kynurenine (Kyn) pathway activity as potential biomarkers in patients with major depressive disorder. Nutrients 2023, 15, 1752. [Google Scholar] [CrossRef]
- Capuco, A.; Urits, I.; Hasoon, J.; Chun, R.; Gerald, B.; Wang, J.K.; Kassem, H.; Ngo, A.L.; Abd-Elsayed, A.; Simopoulos, T. Current perspectives on gut microbiome dysbiosis and depression. Adv. Ther. 2020, 37, 1328–1346. [Google Scholar] [CrossRef]
- Tao, Y.; Zhou, H.; Li, Z.; Wu, H.; Wu, F.; Miao, Z.; Shi, H.; Huang, F.; Wu, X. TGR5 deficiency-induced anxiety and depression-like behaviors: The role of gut microbiota dysbiosis. J. Affect. Disord. 2024, 344, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Intili, G.; Paladino, L.; Rappa, F.; Alberti, G.; Plicato, A.; Calabrò, F.; Fucarino, A.; Cappello, F.; Bucchieri, F.; Tomasello, G. From Dysbiosis to neurodegenerative diseases through different communication pathways: An overview. Biology 2023, 12, 195. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.; Dinan, T.G.; Cryan, J. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef]
- Delzenne, N.M.; Cani, P.D.; Everard, A.; Neyrinck, A.M.; Bindels, L.B. Gut microorganisms as promising targets for the management of type 2 diabetes. Diabetologia 2015, 58, 2206–2217. [Google Scholar] [CrossRef]
- Leclercq, S.; Schwarz, M.; Delzenne, N.M.; Stärkel, P.; de Timary, P. Alterations of kynurenine pathway in alcohol use disorder and abstinence: A link with gut microbiota, peripheral inflammation and psychological symptoms. Transl. Psychiatry 2021, 11, 503. [Google Scholar] [CrossRef]
- Bercik, P.; Collins, S.M. The effects of inflammation, infection and antibiotics on the microbiota-gut-brain axis. Microb. Endocrinol. Microbiota-Gut-Brain Axis Health Dis. 2014, 817, 279–289. [Google Scholar]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Hubková, B.; Valko-Rokytovská, M.; Čižmárová, B.; Zábavníková, M.; Mareková, M.; Birková, A. Tryptophan: Its metabolism along the kynurenine, serotonin, and indole pathway in malignant melanoma. Int. J. Mol. Sci. 2022, 23, 9160. [Google Scholar] [CrossRef]
- Herderich, M.; Gutsche, B. Tryptophan-derived bioactive compounds in food. Food Rev. Int. 1997, 13, 103–135. [Google Scholar] [CrossRef]
- Badawy, A.A.-B. Tryptophan metabolism in alcoholism. Nutr. Res. Rev. 2002, 15, 123–152. [Google Scholar] [CrossRef]
- Salyers, Z.R.; Coleman, M.; Balestrieri, N.P.; Ryan, T.E. Indoxyl sulfate impairs angiogenesis via chronic aryl hydrocarbon receptor activation. Am. J. Physiol. -Cell Physiol. 2021, 320, C240–C249. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-C.; Shyu, J.-F.; Lim, P.S.; Fang, T.-C.; Lu, C.-L.; Zheng, C.-M.; Hou, Y.-C.; Wu, C.-C.; Lin, Y.-F.; Lu, K.-C. Concentration and duration of indoxyl sulfate exposure affects osteoclastogenesis by regulating NFATc1 via aryl hydrocarbon receptor. Int. J. Mol. Sci. 2020, 21, 3486. [Google Scholar] [CrossRef] [PubMed]
- Shivanna, S.; Kolandaivelu, K.; Shashar, M.; Belghasim, M.; Al-Rabadi, L.; Balcells, M.; Zhang, A.; Weinberg, J.; Francis, J.; Pollastri, M.P. The aryl hydrocarbon receptor is a critical regulator of tissue factor stability and an antithrombotic target in uremia. J. Am. Soc. Nephrol. 2016, 27, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Lee, J.H.; Lee, J. Diverse roles of microbial indole compounds in eukaryotic systems. Biol. Rev. 2021, 96, 2522–2545. [Google Scholar] [CrossRef]
- Zgarbová, E.; Vrzal, R. Skatole: A thin red line between its benefits and toxicity. Biochimie 2023, 208, 1–12. [Google Scholar] [CrossRef]
- Tennoune, N.; Andriamihaja, M.; Blachier, F. Production of indole and indole-related compounds by the intestinal microbiota and consequences for the host: The good, the bad, and the ugly. Microorganisms 2022, 10, 930. [Google Scholar] [CrossRef]
- Tang, J.; Li, Y.; Zhang, L.; Mu, J.; Jiang, Y.; Fu, H.; Zhang, Y.; Cui, H.; Yu, X.; Ye, Z. Biosynthetic pathways and functions of indole-3-acetic acid in microorganisms. Microorganisms 2023, 11, 2077. [Google Scholar] [CrossRef] [PubMed]
- Shatova, O.; Shestopalov, A. Tryptophan Metabolism: A New Look at the Role of Tryptophan Derivatives in the Human Body. Biol. Bull. Rev. 2023, 13, 81–91. [Google Scholar] [CrossRef]
- Hwang, I.K.; Yoo, K.Y.; Li, H.; Park, O.K.; Lee, C.H.; Choi, J.H.; Jeong, Y.G.; Lee, Y.L.; Kim, Y.M.; Kwon, Y.G. Indole-3-propionic acid attenuates neuronal damage and oxidative stress in the ischemic hippocampus. J. Neurosci. Res. 2009, 87, 2126–2137. [Google Scholar] [CrossRef]
- Konopelski, P.; Mogilnicka, I. Biological effects of indole-3-propionic acid, a gut microbiota-derived metabolite, and its precursor tryptophan in mammals’ health and disease. Int. J. Mol. Sci. 2022, 23, 1222. [Google Scholar] [CrossRef]
- Arosio, B.; Calvani, R.; Ferri, E.; Coelho-Junior, H.J.; Carandina, A.; Campanelli, F.; Ghiglieri, V.; Marzetti, E.; Picca, A. Sarcopenia and Cognitive decline in older adults: Targeting the muscle–brain Axis. Nutrients 2023, 15, 1853. [Google Scholar] [CrossRef] [PubMed]
- de Lima, E.P.; Tanaka, M.; Lamas, C.B.; Quesada, K.; Detregiachi, C.R.P.; Araújo, A.C.; Guiguer, E.L.; Catharin, V.M.C.S.; de Castro, M.V.M.; Junior, E.B. Vascular Impairment, Muscle Atrophy, and Cognitive Decline: Critical Age-Related Conditions. Biomedicines 2024, 12, 2096. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.K. Physical activity and muscle–brain crosstalk. Nat. Rev. Endocrinol. 2019, 15, 383–392. [Google Scholar] [CrossRef]
- Saran, T.; Turska, M.; Kocki, T.; Zawadka, M.; Zieliński, G.; Turski, W.A.; Gawda, P. Effect of 4-week physical exercises on tryptophan, kynurenine and kynurenic acid content in human sweat. Sci. Rep. 2021, 11, 11092. [Google Scholar] [CrossRef]
- Dugué, P.-A.; Li, S.; Marinis, S.; Hodge, A. Combined association of diet and physical activity with plasma concentrations of markers of the kynurenine pathway. Proc. Nutr. Soc. 2023, 82, E201. [Google Scholar] [CrossRef]
- Lim, A.; Harijanto, C.; Vogrin, S.; Guillemin, G.; Duque, G. Does exercise influence kynurenine/tryptophan metabolism and psychological outcomes in persons with age-related diseases? A systematic review. Int. J. Tryptophan Res. 2021, 14, 1178646921991119. [Google Scholar] [CrossRef]
- Cervenka, I.; Agudelo, L.Z.; Ruas, J.L. Kynurenines: Tryptophan’s metabolites in exercise, inflammation, and mental health. Science 2017, 357, eaaf9794. [Google Scholar] [CrossRef]
- Valente-Silva, P.; Ruas, J.L. Tryptophan-Kynurenine Metabolites in Exercise and Mental Health. In Hormones, Metabolism and the Benefits of Exercise; Spiegelman, B., Ed.; Springer: Cham, Switzerland, 2017; pp. 83–91. [Google Scholar]
- Valente-Silva, P.; Cervenka, I.; Ferreira, D.M.; Correia, J.C.; Edman, S.; Horwath, O.; Heng, B.; Chow, S.; Jacobs, K.R.; Guillemin, G.J. Effects of tryptophan supplementation and exercise on the fate of kynurenine metabolites in mice and humans. Metabolites 2021, 11, 508. [Google Scholar] [CrossRef]
- Agudelo, L.Z.; Femenía, T.; Orhan, F.; Porsmyr-Palmertz, M.; Goiny, M.; Martinez-Redondo, V.; Correia, J.C.; Izadi, M.; Bhat, M.; Schuppe-Koistinen, I. Skeletal muscle PGC-1α1 modulates kynurenine metabolism and mediates resilience to stress-induced depression. Cell 2014, 159, 33–45. [Google Scholar] [CrossRef]
- Holzer, P.; Farzi, A.; Hassan, A.M.; Zenz, G.; Jačan, A.; Reichmann, F. Visceral inflammation and immune activation stress the brain. Front. Immunol. 2017, 8, 1613. [Google Scholar] [CrossRef]
- Yu, Z.; Lin, Y.; Wu, L.; Wang, L.; Fan, Y.; Xu, L.; Zhang, L.; Wu, W.; Tao, J.; Huan, F. Bisphenol F exposure induces depression-like changes: Roles of the kynurenine metabolic pathway along the “liver-brain” axis ☆. Environ. Pollut. 2024, 346, 123356. [Google Scholar] [CrossRef] [PubMed]
- Lucchetti, J.; Fumagalli, F.; Olivari, D.; Affatato, R.; Fracasso, C.; De Giorgio, D.; Perego, C.; Motta, F.; Passoni, A.; Staszewsky, L. Brain kynurenine pathway and functional outcome of rats resuscitated from cardiac arrest. J. Am. Heart Assoc. 2021, 10, e021071. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.Y.; van Praag, H. On the run for hippocampal plasticity. Cold Spring Harb. Perspect. Med. 2018, 8, a029736. [Google Scholar]
- Kindler, J.; Lim, C.K.; Weickert, C.S.; Boerrigter, D.; Galletly, C.; Liu, D.; Jacobs, K.R.; Balzan, R.; Bruggemann, J.; O’Donnell, M. Dysregulation of kynurenine metabolism is related to proinflammatory cytokines, attention, and prefrontal cortex volume in schizophrenia. Mol. Psychiatry 2020, 25, 2860–2872. [Google Scholar] [CrossRef] [PubMed]
- Sheibani, M.; Shayan, M.; Khalilzadeh, M.; Soltani, Z.E.; Jafari-Sabet, M.; Ghasemi, M.; Dehpour, A.R. Kynurenine pathway and its role in neurologic, psychiatric, and inflammatory bowel diseases. Mol. Biol. Rep. 2023, 50, 10409–10425. [Google Scholar] [CrossRef]
- Jafari, R.M.; Shayesteh, S.; Ala, M.; Yousefi-Manesh, H.; Rashidian, A.; Hashemian, S.M.; Sorouri, M.; Dehpour, A.R. Dapsone ameliorates colitis through TLR4/NF-kB pathway in TNBS induced colitis model in rat. Arch. Med. Res. 2021, 52, 595–602. [Google Scholar] [CrossRef]
- Zlokovic, B.V. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat. Rev. Neurosci. 2011, 12, 723–738. [Google Scholar] [CrossRef]
- Mor, A.; Kalaska, B.; Pawlak, D. Kynurenine pathway in chronic kidney disease: What’s old, what’s new, and what’s next? Int. J. Tryptophan Res. 2020, 13, 1178646920954882. [Google Scholar] [CrossRef]
- Wee, H.N.; Liu, J.-J.; Ching, J.; Kovalik, J.-P.; Lim, S.C. The kynurenine pathway in acute kidney injury and chronic kidney disease. Am. J. Nephrol. 2021, 52, 771–787. [Google Scholar] [CrossRef]
- Tankiewicz, A.; Pawlak, D.; Topczewska-Bruns, J.; Buczko, W. Kidney and Liver Kynurenine Pathway Enzymes in Chronic Renal Failure. In Developments in Tryptophan and Serotonin Metabolism; Springer: Berlin/Heidelberg, Germany, 2003; pp. 409–414. [Google Scholar]
- Gorji, H.; Yang, J.W.; Hannan, M.; He, J.; Miller, L.M.; Mathew, A.V.; Seliger, S.L.; Tamura, M.; Mehta, R.; Kotanko, P. Kynurenine Metabolism and Neurocognition in CKD: CRIC Study: TH-PO1066. J. Am. Soc. Nephrol. 2023, 34, 390–391. [Google Scholar] [CrossRef]
- Pawlak, D.; Pawlak, K.; Malyszko, J.; Mysliwiec, M.; Buczko, W. Accumulation of toxic products degradation of kynurenine in hemodialyzed patients. Int. Urol. Nephrol. 2001, 33, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Banks, W.A. The blood–brain barrier as an endocrine tissue. Nat. Rev. Endocrinol. 2019, 15, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, B.; Babaeizad, A.; Banihashemian, S.Z.; Feyzabadi, Z.K.; Dadashpour, M.; Pahlevan, D.; Ghaffari, H.; Eslami, M. Gastrointestinal tract, microbiota and multiple sclerosis (MS) and the link between gut microbiota and CNS. Curr. Microbiol. 2023, 80, 38. [Google Scholar] [CrossRef] [PubMed]
- Quettier, T.; Ippolito, G.; Però, L.; Cardellicchio, P.; Battaglia, S.; Borgomaneri, S. Individual differences in intracortical inhibition predict action control when facing emotional stimuli. Front. Psychol. 2024, 15, 1391723. [Google Scholar] [CrossRef]
- Battaglia, S.; Nazzi, C.; Lonsdorf, T.B.; Thayer, J.F. Neuropsychobiology of fear-induced bradycardia in humans: Progress and pitfalls. Mol. Psychiatry 2024, 29, 3826–3840. [Google Scholar] [CrossRef]
- Nazzi, C.; Avenanti, A.; Battaglia, S. The Involvement of Antioxidants in Cognitive Decline and Neurodegeneration: Mens Sana in Corpore Sano. Antioxidants 2024, 13, 701. [Google Scholar] [CrossRef]
- Gregorio, F.D.; Battaglia, S. The intricate brain–body interaction in psychiatric and neurological diseases. Adv. Clin. Exp. Med. 2024, 33, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, S.; Nazzi, C.; Thayer, J.F. Genetic differences associated with dopamine and serotonin release mediate fear-induced bradycardia in the human brain. Transl. Psychiatry 2024, 14, 24. [Google Scholar] [CrossRef]
- Alcocer-Cuarón, C.; Rivera, A.L.; Castaño, V.M. Hierarchical structure of biological systems: A bioengineering approach. Bioengineered 2014, 5, 73–79. [Google Scholar] [CrossRef]
- Bick, C.; Gross, E.; Harrington, H.A.; Schaub, M.T. What are higher-order networks? SIAM Rev. 2023, 65, 686–731. [Google Scholar] [CrossRef]
- Xia, C.; Wang, J.; Perc, M.; Wang, Z. Reputation and reciprocity. Phys. Life Rev. 2023, 46, 8–45. [Google Scholar] [CrossRef] [PubMed]
- Neurock, M.; Stark, S.M.; Klein, M.T. Molecular Simulation of Kinetic Interactions in Complex Mixtures. In Computer-Aided Design of Catalysts; CRC Press: Boca Raton, FL, USA, 2020; pp. 55–88. [Google Scholar]
- Sánchez, W.N.; Robeson, L.; Carrasco, V.; Figueroa, N.L.; Burgos-Bravo, F.; Wilson, C.A.; Casanova-Morales, N. Determination of protein–protein interactions at the single-molecule level using optical tweezers. Q. Rev. Biophys. 2022, 55, e8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Liao, W.; Tong, Z.; Yuan, F.; Mao, L.; Liu, J.; Gao, Y. The concentration-, pH-and temperature-responsive self-assembly of undenatured type II collagen: Kinetics, thermodynamics, nanostructure and molecular mechanism. Food Hydrocoll. 2023, 137, 108424. [Google Scholar] [CrossRef]
- Ohi, Y.; Kimura, S.; Haji, A. Modulation of glutamatergic transmission by presynaptic N-methyl-D-aspartate mechanisms in second-order neurons of the rat nucleus tractus solitarius. Neurosci. Lett. 2015, 587, 62–67. [Google Scholar] [CrossRef]
- Todd, A.J. An historical perspective: The second order neuron in the pain pathway. Front. Pain Res. 2022, 3, 845211. [Google Scholar] [CrossRef]
- Yamada, D.; Bushey, D.; Li, F.; Hibbard, K.L.; Sammons, M.; Funke, J.; Litwin-Kumar, A.; Hige, T.; Aso, Y. Hierarchical architecture of dopaminergic circuits enables second-order conditioning in Drosophila. eLife 2023, 12, e79042. [Google Scholar] [CrossRef]
- Subramanian, N.; Torabi-Parizi, P.; Gottschalk, R.A.; Germain, R.N.; Dutta, B. Network representations of immune system complexity. Wiley Interdiscip. Rev. Syst. Biol. Med. 2015, 7, 13–38. [Google Scholar] [CrossRef]
- You, X.; Zhang, M.; Ma, Y.; Tan, J.; Liu, Z. Impact of higher-order interactions and individual emotional heterogeneity on information-disease coupled dynamics in multiplex networks. Chaos Solitons Fractals 2023, 177, 114186. [Google Scholar] [CrossRef]
- Treur, J. On Structure, Dynamics, and Adaptivity for Biological and Mental Processes: A Higher-Order Adaptive Dynamical System Modeling Perspective. In Proceedings of the Annual Meeting of the Cognitive Science Society, Rotterdam, The Netherlands, 24–27 July 2024. [Google Scholar]
- Nijhout, H.F.; Best, J.A.; Reed, M.C. Systems biology of robustness and homeostatic mechanisms. Wiley Interdiscip. Rev. Syst. Biol. Med. 2019, 11, e1440. [Google Scholar] [CrossRef]
- Cohen, A.A.; Ferrucci, L.; Fülöp, T.; Gravel, D.; Hao, N.; Kriete, A.; Levine, M.E.; Lipsitz, L.A.; Olde Rikkert, M.G.; Rutenberg, A. A complex systems approach to aging biology. Nat. Aging 2022, 2, 580–591. [Google Scholar] [CrossRef]
- Bi, D.; Almpanis, A.; Noel, A.; Deng, Y.; Schober, R. A survey of molecular communication in cell biology: Establishing a new hierarchy for interdisciplinary applications. IEEE Commun. Surv. Tutor. 2021, 23, 1494–1545. [Google Scholar] [CrossRef]
- Karafyllis, I.; Krstic, M. Nonlinear stabilization under sampled and delayed measurements, and with inputs subject to delay and zero-order hold. IEEE Trans. Autom. Control 2011, 57, 1141–1154. [Google Scholar] [CrossRef]
- Laracuente, M.-L.; Marina, H.Y.; McHugh, K.J. Zero-order drug delivery: State of the art and future prospects. J. Control. Release 2020, 327, 834–856. [Google Scholar] [CrossRef]
- Domínguez-Hüttinger, E.; Christodoulides, P.; Miyauchi, K.; Irvine, A.D.; Okada-Hatakeyama, M.; Kubo, M.; Tanaka, R.J. Mathematical modeling of atopic dermatitis reveals “double-switch” mechanisms underlying 4 common disease phenotypes. J. Allergy Clin. Immunol. 2017, 139, 1861–1872.e7. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Vijayasree, V.; Das, S.; Sivamani, Y.; Elayaperumal, S. Overview of Drugs and drug Targets. In Biochemical and Molecular Pharmacology in Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2024; pp. 45–69. [Google Scholar]
- Fischer, E. Einfluss der Configuration auf die Wirkung der Enzyme. Berichte Dtsch. Chem. Ges. 1894, 27, 2985–2993. [Google Scholar] [CrossRef]
- Cramer, F. Emil Fischer’s Lock-and-Key Hypothesis after 100 years-Towards a Supracellular Chemistry’. Perspect. Supramol. Chem. Lock Key Princ. 1994, 1, 1–23. [Google Scholar]
- Brunori, M. Allostery turns 50: Is the vintage yet attractive? Protein Sci. A Publ. Protein Soc. 2011, 20, 1097. [Google Scholar] [CrossRef]
- Hill, A.V. The possible effects of the aggregation of the molecules of hemoglobin on its dissociation curves. J. Physiol. 1910, 40, iv–vii. [Google Scholar]
- Cattoni, D.I.; Chara, O.; Kaufman, S.B.; González Flecha, F.L. Cooperativity in binding processes: New insights from phenomenological modeling. PLoS ONE 2015, 10, e0146043. [Google Scholar] [CrossRef]
- Sykes, D.A.; Stoddart, L.A.; Kilpatrick, L.E.; Hill, S.J. Binding kinetics of ligands acting at GPCRs. Mol. Cell. Endocrinol. 2019, 485, 9–19. [Google Scholar] [CrossRef]
- Tummino, P.J.; Copeland, R.A. Residence time of receptor− ligand complexes and its effect on biological function. Biochemistry 2008, 47, 5481–5492. [Google Scholar] [CrossRef] [PubMed]
- Peletier, L.A. An Extended Model Including Target Turnover, Ligand–Target Complex Kinetics, and Binding Properties to Describe Drug–Receptor Interactions. Comput. Methods Estim. Kinet. Parameters Biol. Syst. 2021, 2385, 19–46. [Google Scholar]
- Borisov, D.; Veselovsky, A. Ligand–receptor binding kinetics in drug design. Biochem. Suppl. Ser. B Biomed. Chem. 2020, 14, 228–240. [Google Scholar]
- Srinivasan, B. A guide to enzyme kinetics in early drug discovery. FEBS J. 2023, 290, 2292–2305. [Google Scholar] [CrossRef]
- Gibson, Q.H. The reaction of oxygen with hemoglobin and the kinetic basis of the effect of salt on binding of oxygen. J. Biol. Chem. 1970, 245, 3285–3288. [Google Scholar] [CrossRef]
- Gibson, Q.H. Kinetics of oxygen binding to hemoglobin A. Biochemistry 1999, 38, 5191–5199. [Google Scholar] [CrossRef] [PubMed]
- Berger, R.L.; Antonini, E.; Brunori, M.; Wyman, J.; Rossi-Fanelli, A. Observations on the kinetics of the reaction of hemoglobin with oxygen. J. Biol. Chem. 1967, 242, 4841–4843. [Google Scholar] [CrossRef]
- Zomaya, A.Y.; Pan, Y. Biomolecular Networks: Methods and Applications in Systems Biology; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Lü, J.; Wang, P. Modeling and Analysis of Bio-Molecular Networks; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Hampel, H.; Nisticò, R.; Seyfried, N.T.; Levey, A.I.; Modeste, E.; Lemercier, P.; Baldacci, F.; Toschi, N.; Garaci, F.; Perry, G. Omics sciences for systems biology in Alzheimer’s disease: State-of-the-art of the evidence. Ageing Res. Rev. 2021, 69, 101346. [Google Scholar] [CrossRef]
- Azer, K.; Kaddi, C.D.; Barrett, J.S.; Bai, J.P.; McQuade, S.T.; Merrill, N.J.; Piccoli, B.; Neves-Zaph, S.; Marchetti, L.; Lombardo, R. History and future perspectives on the discipline of quantitative systems pharmacology modeling and its applications. Front. Physiol. 2021, 12, 637999. [Google Scholar] [CrossRef]
- Li, L.; Ji, J.; Song, F.; Hu, J. Intercellular receptor-ligand binding: Effect of protein-membrane interaction. J. Mol. Biol. 2023, 435, 167787. [Google Scholar] [CrossRef]
- Markin, C.; Mokhtari, D.; Sunden, F.; Appel, M.; Akiva, E.; Longwell, S.; Sabatti, C.; Herschlag, D.; Fordyce, P. Revealing enzyme functional architecture via high-throughput microfluidic enzyme kinetics. Science 2021, 373, eabf8761. [Google Scholar] [CrossRef] [PubMed]
- Loisios-Konstantinidis, I.; Mavroudis, P.D.; Macheras, P. Dynamical Aspects of Pharmacokinetic/Pharmacodynamic & Quantitative Systems Pharmacology Models. In Approaching Complex Diseases: Network-Based Pharmacology and Systems Approach in Bio-Medicine; Springer: Berlin/Heidelberg, Germany, 2020; pp. 35–61. [Google Scholar]
- Foster, M. A Textbook of Physiology, Book Three: The Central Nervous System and Its Instruments; Macmillan: New York, NY, USA, 1897. [Google Scholar]
- Bennett, M.R. The early history of the synapse: From Plato to Sherrington. Brain Res. Bull. 1999, 50, 95–118. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, G.M.; Erulkar, S.D. Centenary of the synapse: From Sherrington to the molecular biology of the synapse and beyond. Trends Neurosci. 1997, 20, 385–392. [Google Scholar] [CrossRef]
- Loewi, O. Über humorale übertragbarkeit der Herznervenwirkung. Pflügers Arch. Eur. J. Physiol. 1921, 189, 239–242. [Google Scholar] [CrossRef]
- Borges, R.; García, A.G. One hundred years from Otto Loewi experiment, a dream that revolutionized our view of neurotransmission. Pflügers Arch. -Eur. J. Physiol. 2021, 473, 977–981. [Google Scholar] [CrossRef]
- Valenstein, E.S. The discovery of chemical neurotransmitters. Brain Cogn. 2002, 49, 73–95. [Google Scholar] [CrossRef] [PubMed]
- Ince, R.A.; Montani, F.; Arabzadeh, E.; Diamond, M.E.; Panzeri, S. On the Presence of High-Order Interactions Among Somatosensory Neurons and Their Effect on Information Transmission. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2009; p. 012013. [Google Scholar]
- Wang, B.; Dudko, O.K. A theory of synaptic transmission. eLife 2021, 10, e73585. [Google Scholar] [CrossRef]
- Destexhe, A.; Mainen, Z.; Sejnowski, T. An E cient Method for Computing Synaptic Conductances Based on a Kinetic Model of Receptor Binding. Neural Comput. 1994, 6, 14–18. [Google Scholar] [CrossRef]
- Norman, C.A.; Krishnakumar, S.S.; Timofeeva, Y.; Volynski, K.E. The release of inhibition model reproduces kinetics and plasticity of neurotransmitter release in central synapses. Commun. Biol. 2023, 6, 1091. [Google Scholar] [CrossRef]
- Levine, M.S.; Li, S.; Cepeda, C.; Cromwell, H.C.; Altemus, K.L. Neuromodulatory actions of dopamine on synaptically-evoked neostriatal responses in slices. Synapse 1996, 24, 65–78. [Google Scholar] [CrossRef]
- Trevathan, J.K.; Yousefi, A.; Park, H.O.; Bartoletta, J.J.; Ludwig, K.A.; Lee, K.H.; Lujan, J.L. Computational modeling of neurotransmitter release evoked by electrical stimulation: Nonlinear approaches to predicting stimulation-evoked dopamine release. ACS Chem. Neurosci. 2017, 8, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Seeholzer, A.; Deger, M.; Gerstner, W. Stability of working memory in continuous attractor networks under the control of short-term plasticity. PLoS Comput. Biol. 2019, 15, e1006928. [Google Scholar] [CrossRef]
- Tompa, P.; Töth-Boconádi, R.; Friedrich, P. Frequency decoding of fast calcium oscillations by calpain. Cell Calcium 2001, 29, 161–170. [Google Scholar] [CrossRef]
- Schild, J.; Clark, J.; Canavier, C.; Kunze, D.; Andresen, M. Afferent synaptic drive of rat medial nucleus tractus solitarius neurons: Dynamic simulation of graded vesicular mobilization, release, and non-NMDA receptor kinetics. J. Neurophysiol. 1995, 74, 1529–1548. [Google Scholar] [CrossRef]
- FallahBagheri, N.; Akan, O.B. A Molecular Communication Perspective of Alzheimer’s Disease: Impact of Amyloid Beta Oligomers on Glutamate Diffusion in the Synaptic Cleft. arXiv 2024, arXiv:2409.03396. [Google Scholar]
- Bosl, W.J. Systems biology by the rules: Hybrid intelligent systems for pathway modeling and discovery. BMC Syst. Biol. 2007, 1, 1–25. [Google Scholar] [CrossRef][Green Version]
- Singel, D.J.; Stamler, J.S. Chemical physiology of blood flow regulation by red blood cells: The role of nitric oxide and S-nitrosohemoglobin. Annu. Rev. Physiol. 2005, 67, 99–145. [Google Scholar] [CrossRef]
- Araki, T.; Tanji, H.; Kato, H.; Mizugaki, M.; Itoyama, Y. Alterations of second messenger systems in the rat brain after 6-hydroxydopamine lesions of the medial forebrain bundle. Eur. J. Pharm. Sci. 1999, 8, 261–267. [Google Scholar] [CrossRef]
- Frade, J.G.L.d.O. Real-time change of nitric oxide in rat hippocampal slices and astrocytic glutathione release via glutamate-dependent pathways. Master’s Thesis, Universidade de Coimbra, Coimbra, Portugal, 2008. [Google Scholar]
- Gosak, M.; Markovič, R.; Dolenšek, J.; Rupnik, M.S.; Marhl, M.; Stožer, A.; Perc, M. Network science of biological systems at different scales: A review. Phys. Life Rev. 2018, 24, 118–135. [Google Scholar] [CrossRef]
- De Luca, C.; Colangelo, A.M.; Virtuoso, A.; Alberghina, L.; Papa, M. Neurons, glia, extracellular matrix and neurovascular unit: A systems biology approach to the complexity of synaptic plasticity in health and disease. Int. J. Mol. Sci. 2020, 21, 1539. [Google Scholar] [CrossRef]
- Hodgkin, A.L.; Huxley, A.F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952, 117, 500. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Collado, A.; Rueda, C. A simple parametric representation of the Hodgkin-Huxley model. PLoS ONE 2021, 16, e0254152. [Google Scholar] [CrossRef] [PubMed]
- Bickle, J. Hodgkin’s and Huxley’s own assessments of their “quantitative description” of nerve membrane current. Hist. Philos. Life Sci. 2023, 45, 25. [Google Scholar] [CrossRef] [PubMed]
- Behring, E.V. Ueber das zustandekommen der diphtherie-immunität und der tetanus-immunität bei thieren. Drucke 19. Jh. 1890. [Google Scholar]
- Winau, F.; Winau, R. Emil von Behring and serum therapy. Microbes Infect. 2002, 4, 185–188. [Google Scholar] [CrossRef]
- Lahaie, Y.M.; Watier, H. From the Gorgon’s blood to Behring’s Blutserumtherapie: A long path towards serum therapy. Immunol. Rev. 2024. ahead of print. [Google Scholar] [CrossRef]
- Jiang, P.; Zhang, Y.; Ru, B.; Yang, Y.; Vu, T.; Paul, R.; Mirza, A.; Altan-Bonnet, G.; Liu, L.; Ruppin, E. Systematic investigation of cytokine signaling activity at the tissue and single-cell levels. Nat. Methods 2021, 18, 1181–1191. [Google Scholar] [CrossRef]
- Hwang, J.-R.; Byeon, Y.; Kim, D.; Park, S.-G. Recent insights of T cell receptor-mediated signaling pathways for T cell activation and development. Exp. Mol. Med. 2020, 52, 750–761. [Google Scholar] [CrossRef]
- Wu, L.; Wei, Q.; Brzostek, J.; Gascoigne, N.R. Signaling from T cell receptors (TCRs) and chimeric antigen receptors (CARs) on T cells. Cell. Mol. Immunol. 2020, 17, 600–612. [Google Scholar] [CrossRef]
- Toraño, A.; Moreno, I.; Infantes, J.A.; Domínguez, M. An Overview of ELISA-Based Initial Velocity Methods to Measure the Immunoreactive Fraction, Association Rate, And Equilibrium Constants of Monoclonal Antibodies. Asian J. Complement. Altern. Med. 2022, 10, 148. [Google Scholar] [CrossRef]
- Witt, S.N.; McConnell, H.M. A first-order reaction controls the binding of antigenic peptides to major histocompatibility complex class II molecules. Proc. Natl. Acad. Sci. USA 1991, 88, 8164–8168. [Google Scholar] [CrossRef] [PubMed]
- Šimánek, V.; Pecen, L.; Řezáčková, H.; Topolčan, O.; Fajfrlík, K.; Sedláček, D.; Šín, R.; Bludovská, M.; Pazdiora, P.; Slouka, D. Long-Term Monitoring of the Antibody Response to a SARS-CoV-2 Infection. Diagnostics 2021, 11, 1915. [Google Scholar] [CrossRef] [PubMed]
- Leslie, R. Macrophage handling of soluble immune complexes. Immunol. Today 1980, 1, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Klasse, P. Neutralization of virus infectivity by antibodies: Old problems in new perspectives. Adv. Biol. 2014, 2014, 157895. [Google Scholar] [CrossRef] [PubMed]
- Elveborg, S.; Monteil, V.M.; Mirazimi, A. Methods of inactivation of highly pathogenic viruses for molecular, serology or vaccine development purposes. Pathogens 2022, 11, 271. [Google Scholar] [CrossRef]
- Zheng, Y.; Balakrishnan, V.; Buzzard, G.; Geahlen, R.; Harrison, M.; Rundell, A. Modeling and analysis of early events in T-lymphocyte antigen-activated intracellular-signaling pathways. J. Comput. Appl. Math. 2005, 184, 320–341. [Google Scholar] [CrossRef]
- Kennedy, R.H.; Silver, R. Neuroimmune Signaling: Cytokines and the Central Nervous System. In Neuroscience in the 21st Century: From Basic to Clinical; Springer: Berlin/Heidelberg, Germany, 2022; pp. 883–922. [Google Scholar]
- Carreño, L.J.; Bueno, S.M.; Bull, P.; Nathenson, S.G.; Kalergis, A.M. The half-life of the T-cell receptor/peptide–major histocompatibility complex interaction can modulate T-cell activation in response to bacterial challenge. Immunology 2007, 121, 227–237. [Google Scholar] [CrossRef]
- Perley, J.P.; Mikolajczak, J.; Buzzard, G.T.; Harrison, M.L.; Rundell, A.E. Resolving early signaling events in T-cell activation leading to IL-2 and FOXP3 transcription. Processes 2014, 2, 867–900. [Google Scholar] [CrossRef]
- Slansky, J.E.; Nakayama, M. Peptide mimotopes alter T cell function in cancer and autoimmunity. Semin. Immunol. 2020, 47, 101395. [Google Scholar] [CrossRef]
- Glover, R.E.; Koshkin, V.; Dunford, H.B.; Mason, R.P. The reaction rates of NO with horseradish peroxidase compounds I and II. Nitric Oxide 1999, 3, 439–444. [Google Scholar] [CrossRef]
- Cai, C.; Fordsmann, J.C.; Jensen, S.H.; Gesslein, B.; Lønstrup, M.; Hald, B.O.; Zambach, S.A.; Brodin, B.; Lauritzen, M.J. Stimulation-induced increases in cerebral blood flow and local capillary vasoconstriction depend on conducted vascular responses. Proc. Natl. Acad. Sci. USA 2018, 115, E5796–E5804. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Li, D.; Li, Y.; Yang, P.; Zhang, Q.; Zhong, W.; Shan, H.; Dai, H.; Chen, L. Identifying function modules from protein–protein interaction networks based on Szemerédi’s Regularity Lemma. Int. J. Biomath. 2024, 2024, 2350104. [Google Scholar] [CrossRef]
- Yuan, S.; Jiang, S.-C.; Zhang, Z.-W.; Fu, Y.-F.; Hu, J.; Li, Z.-L. Quantification of cytokine storms during virus infections. Front. Immunol. 2021, 12, 659419. [Google Scholar] [CrossRef]
- Bocquet-Garçon, A. Impact of the SARS-CoV-2 Spike Protein on the Innate Immune System: A Review. Cureus 2024, 16, e57008. [Google Scholar] [CrossRef]
- Kang, S.; Kishimoto, T. Interplay between interleukin-6 signaling and the vascular endothelium in cytokine storms. Exp. Mol. Med. 2021, 53, 1116–1123. [Google Scholar] [CrossRef]
- Kwong, A.C.; Ordovas-Montanes, J. Deconstructing inflammatory memory across tissue setpoints using cell circuit motifs. J. Allergy Clin. Immunol. 2024, 154, 1095–1105. [Google Scholar] [CrossRef]
- Reddy, N. Synthetic Biology Approaches to Understand and Engineer Immune Cells; University of California: San Francisco, CA, USA, 2023. [Google Scholar]
- Bocharov, G. Understanding complex regulatory systems: Integrating molecular biology and systems analysis. Transfus. Med. Hemother. 2005, 32, 304–321. [Google Scholar] [CrossRef]
- Germain, R.N.; Meier-Schellersheim, M.; Nita-Lazar, A.; Fraser, I.D. Systems biology in immunology: A computational modeling perspective. Annu. Rev. Immunol. 2011, 29, 527–585. [Google Scholar] [CrossRef]
- Zhou, L.; Fu, F.; Wang, Y.; Yang, L. Interlocked feedback loops balance the adaptive immune response. Math. Biosci. Eng. 2022, 19, 4084–4100. [Google Scholar] [CrossRef]
- Madhavan, M.; Mustafa, S. Systems biology–the transformative approach to integrate sciences across disciplines: Systems Biology: Integrating Biological Sciences. Phys. Sci. Rev. 2023, 8, 2523–2545. [Google Scholar] [CrossRef]
- Kitano, H. Looking beyond the details: A rise in system-oriented approaches in genetics and molecular biology. Curr. Genet. 2002, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kitano, H. Nobel Turing Challenge: Creating the engine for scientific discovery. NPJ Syst. Biol. Appl. 2021, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Stelling, J. Mathematical models in microbial systems biology. Curr. Opin. Microbiol. 2004, 7, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Ingalls, B.P. Mathematical Modeling in Systems Biology: An Introduction; MIT Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Banerjee, S. Mathematical Modeling: Models, Analysis and Applications; Chapman and Hall/CRC: New York, NY, USA, 2021. [Google Scholar]
- Ricardo, H.J. A Modern Introduction to Differential Equations; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Nakada, T.; Mager, D.E. Systems model identifies baseline cytokine concentrations as potential predictors of rheumatoid arthritis inflammatory response to biologics. Br. J. Pharmacol. 2022, 179, 4063–4077. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Sohail, A.; Nutini, A.; Arif, R.; Tavares, J.M.R. Computational model to explore the endocrine response to trastuzumab action in HER-2/neu positive breast cancer. Saudi J. Biol. Sci. 2022, 29, 123–131. [Google Scholar] [CrossRef]
- Ernst, A.; Schütte, C.; Sigrist, S.J.; Winkelmann, S. Variance of filtered signals: Characterization for linear reaction networks and application to neurotransmission dynamics. Math. Biosci. 2022, 343, 108760. [Google Scholar] [CrossRef]
- Wang, M.; Xie, Y.; Liu, J.; Li, A.; Chen, L.; Stromberg, A.; Arnold, S.M.; Liu, C.; Wang, C. A Probabilistic Approach to Estimate the Temporal Order of Pathway Mutations Accounting for Intra-Tumor Heterogeneity. Cancers 2024, 16, 2488. [Google Scholar] [CrossRef]
- Simoni, G.; Reali, F.; Priami, C.; Marchetti, L. Stochastic simulation algorithms for computational systems biology: Exact, approximate, and hybrid methods. Wiley Interdiscip. Rev. Syst. Biol. Med. 2019, 11, e1459. [Google Scholar] [CrossRef]
- Ayers, D.; Day, P.J. Systems medicine: The application of systems biology approaches for modern medical research and drug development. Mol. Biol. Int. 2015, 2015, 698169. [Google Scholar] [CrossRef]
- Soyer, O.S.; O’Malley, M.A. Evolutionary systems biology: What it is and why it matters. BioEssays 2013, 35, 696–705. [Google Scholar] [CrossRef]
- El-Samad, H. Biological feedback control—Respect the loops. Cell Syst. 2021, 12, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Cinquin, O.; Demongeot, J. Roles of positive and negative feedback in biological systems. Comptes Rendus. Biol. 2002, 325, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Paludan, S.R.; Pradeu, T.; Masters, S.L.; Mogensen, T.H. Constitutive immune mechanisms: Mediators of host defence and immune regulation. Nat. Rev. Immunol. 2021, 21, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Ranganath, V.A.; Maity, I. Artificial Homeostasis Systems Based on Feedback Reaction Networks: Design Principles and Future Promises. Angew. Chem. Int. Ed. 2024, 63, e202318134. [Google Scholar] [CrossRef]
- Ferraz de Arruda, G.; Aleta, A.; Moreno, Y. Contagion dynamics on higher-order networks. Nat. Rev. Phys. 2024, 6, 468–482. [Google Scholar] [CrossRef]
- Khammash, M.H. Perfect adaptation in biology. Cell Syst. 2021, 12, 509–521. [Google Scholar] [CrossRef]
- Angarita-Rodríguez, A.; González-Giraldo, Y.; Rubio-Mesa, J.J.; Aristizábal, A.F.; Pinzón, A.; González, J. Control Theory and Systems Biology: Potential Applications in Neurodegeneration and Search for Therapeutic Targets. Int. J. Mol. Sci. 2023, 25, 365. [Google Scholar] [CrossRef]
- Filo, M.; Chang, C.-H.; Khammash, M. Biomolecular feedback controllers: From theory to applications. Curr. Opin. Biotechnol. 2023, 79, 102882. [Google Scholar] [CrossRef]
- Graw, S.; Chappell, K.; Washam, C.L.; Gies, A.; Bird, J.; Robeson, M.S.; Byrum, S.D. Multi-omics data integration considerations and study design for biological systems and disease. Mol. Omics 2021, 17, 170–185. [Google Scholar] [CrossRef]
- Jung, G.T.; Kim, K.-P.; Kim, K. How to interpret and integrate multi-omics data at systems level. Anim. Cells Syst. 2020, 24, 1–7. [Google Scholar] [CrossRef]
- Jendoubi, T. Approaches to integrating metabolomics and multi-omics data: A primer. Metabolites 2021, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Gehlen, W. Wirkungsstärke intravenös verabreichter Arzneimittel als Zeitfunktion: Ein Beitrag zur mathematischen Behandlung pharmakologischer Probleme. Naunyn-Schmiedebergs Arch. Exp. Pathol. Pharmakol. 1933, 171, 541–554. [Google Scholar] [CrossRef]
- Gladtke, E. History of Pharmacokinetics. In Pharmacokinetics: Mathematical and Statistical Approaches to Metabolism and Distribution of Chemicals and Drugs; Springer: Berlin/Heidelberg, Germany, 1988; pp. 1–9. [Google Scholar]
- Talevi, A.; Ruiz, M.E. Zero-Order Drug Release. In The ADME Encyclopedia: A Comprehensive Guide on Biopharmacy and Pharmacokinetics; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–6. [Google Scholar]
- Widmark, E. The theoretical basis and practical application of medical-legal determination of alcohol. Berl. Urban Schwarz. 1932, 1–140. [Google Scholar]
- Himeoka, Y.; Furusawa, C. Perturbation-response analysis of in silico metabolic dynamics in nonlinear regime: Hard-coded responsiveness in the cofactors and network sparsity. eLife 2024, 13, RP98800. [Google Scholar]
- Filo, M.; Kumar, S.; Khammash, M. A hierarchy of biomolecular proportional-integral-derivative feedback controllers for robust perfect adaptation and dynamic performance. Nat. Commun. 2022, 13, 2119. [Google Scholar] [CrossRef]
- Jamshed, L.; Debnath, A.; Jamshed, S.; Wish, J.; Raine, J.; Tomy, G.; Thomas, P.; Holloway, A. An Emerging Cross-Species Marker for Organismal Health: Tryptophan-Kynurenine Pathway. Int. J. Mol. Sci. 2022, 23, 6300. [Google Scholar] [CrossRef]
- Kozieł, K.; Urbańska, E. Kynurenine Pathway in Diabetes Mellitus—Novel Pharmacological Target? Cells 2023, 12, 460. [Google Scholar] [CrossRef]
- Morris, G.; F Carvalho, A.; Anderson, G.; Galecki, P.; Maes, M. The many neuroprogressive actions of tryptophan catabolites (TRYCATs) that may be associated with the pathophysiology of neuro-immune disorders. Curr. Pharm. Des. 2016, 22, 963–977. [Google Scholar] [CrossRef]
- Riedel, W.J.; Klaassen, T.; Schmitt, J.A. Tryptophan, mood, and cognitive function. Brain Behav. Immun. 2002, 16, 581–589. [Google Scholar] [CrossRef]
- Almulla, A.F.; Supasitthumrong, T.; Amrapala, A.; Tunvirachaisakul, C.; Jaleel, A.-K.K.A.; Oxenkrug, G.; Al-Hakeim, H.K.; Maes, M. The tryptophan catabolite or kynurenine pathway in Alzheimer’s disease: A systematic review and meta-analysis. J. Alzheimer’s Dis. 2022, 88, 1325–1339. [Google Scholar] [CrossRef]
- Venkatesan, D.; Iyer, M.; Narayanasamy, A.; Siva, K.; Vellingiri, B. Kynurenine pathway in Parkinson’s disease—An update. Eneurologicalsci 2020, 21, 100270. [Google Scholar] [CrossRef] [PubMed]
- Bohár, Z.; Toldi, J.; Fülöp, F.; Vécsei, L. Changing the face of kynurenines and neurotoxicity: Therapeutic considerations. Int. J. Mol. Sci. 2015, 16, 9772–9793. [Google Scholar] [CrossRef] [PubMed]
- Varga, D.; Herédi, J.; Kánvási, Z.; Ruszka, M.; Kis, Z.; Ono, E.; Iwamori, N.; Iwamori, T.; Takakuwa, H.; Vécsei, L.; et al. Systemic L-Kynurenine sulfate administration disrupts object recognition memory, alters open field behavior and decreases c-Fos immunopositivity in C57Bl/6 mice. Front. Behav. Neurosci. 2015, 9, 157. [Google Scholar] [CrossRef] [PubMed]
- Zhuravlev, A.V.; Zakharov, G.A.; Shchegolev, B.F.; Savvateeva-Popova, E.V. Antioxidant properties of kynurenines: Density functional theory calculations. PLoS Comput. Biol. 2016, 12, e1005213. [Google Scholar] [CrossRef]
- Hiratsuka, C.; Fukuwatari, T.; Sano, M.; Saito, K.; Sasaki, S.; Shibata, K. Supplementing healthy women with up to 5.0 g/d of L-tryptophan has no adverse effects. J. Nutr. 2013, 143, 859–866. [Google Scholar] [CrossRef]
- Ghiboub, M.; Verburgt, C.M.; Sovran, B.; Benninga, M.A.; de Jonge, W.J.; Van Limbergen, J.E. Nutritional therapy to modulate tryptophan metabolism and aryl hydrocarbon-receptor signaling activation in human diseases. Nutrients 2020, 12, 2846. [Google Scholar] [CrossRef]
- Sadok, I.; Jędruchniewicz, K. Dietary Kynurenine pathway metabolites—Source, fate, and chromatographic determinations. Int. J. Mol. Sci. 2023, 24, 16304. [Google Scholar] [CrossRef]
- de Fátima Alves, L.; Moore, J.B.; Kell, D.B. The Biology and Biochemistry of Kynurenic Acid, a Potential Nutraceutical with Multiple Biological Effects. Int. J. Mol. Sci. 2024, 25, 9082. [Google Scholar] [CrossRef]
- Khoshnevisan, K.; Chehrehgosha, M.; Conant, M.; Meftah, A.M.; Baharifar, H.; Ejtahed, H.S.; Angoorani, P.; Gholami, M.; Sharifi, F.; Maleki, H. Interactive relationship between Trp metabolites and gut microbiota: The impact on human pathology of disease. J. Appl. Microbiol. 2022, 132, 4186–4207. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Sun, X.-Z.; Zhao, D.-Y.; Zhou, Y.-C.; Wang, Q.-Q.; Qin, G.; Yao, S.-K. Alteration of fecal tryptophan metabolism correlates with shifted microbiota and may be involved in pathogenesis of colorectal cancer. World J. Gastroenterol. 2020, 26, 7173. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Vécsei, L. From Lab to Life: Exploring Cutting-Edge Models for Neurological and Psychiatric Disorders. Biomedicines 2024, 12, 613. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Szabó, Á.; Vécsei, L.; Giménez-Llort, L. Emerging translational research in neurological and psychiatric diseases: From in vitro to in vivo models. Int. J. Mol. Sci. 2023, 24, 15739. [Google Scholar] [CrossRef]
- Tanaka, M.; Szabó, Á.; Körtési, T.; Szok, D.; Tajti, J.; Vécsei, L. From CGRP to PACAP, VIP, and beyond: Unraveling the next chapters in migraine treatment. Cells 2023, 12, 2649. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Chen, C. Towards a mechanistic understanding of depression, anxiety, and their comorbidity: Perspectives from cognitive neuroscience. Front. Behav. Neurosci. 2023, 17, 1268156. [Google Scholar] [CrossRef]
- Maoz, B.M. Brain-on-a-Chip: Characterizing the next generation of advanced in vitro platforms for modeling the central nervous system. APL Bioeng. 2021, 5, 030902. [Google Scholar] [CrossRef]
- Mofazzal Jahromi, M.A.; Abdoli, A.; Rahmanian, M.; Bardania, H.; Bayandori, M.; Moosavi Basri, S.M.; Kalbasi, A.; Aref, A.R.; Karimi, M.; Hamblin, M.R. Microfluidic brain-on-a-chip: Perspectives for mimicking neural system disorders. Mol. Neurobiol. 2019, 56, 8489–8512. [Google Scholar] [CrossRef] [PubMed]
- Brofiga, M.; Massobrio, P. Brain-on-a-chip: Dream or reality? Front. Neurosci. 2022, 16, 837623. [Google Scholar] [CrossRef]
- Subramanyam, M.; Goyal, J. Translational biomarkers: From discovery and development to clinical practice. Drug Discov. Today Technol. 2016, 21, 3–10. [Google Scholar] [CrossRef]
- Cook, N.; Jodrell, D.I.; Tuveson, D.A. Predictive in vivo animal models and translation to clinical trials. Drug Discov. Today 2012, 17, 253–260. [Google Scholar] [CrossRef]
- Serelli-Lee, V.; Ito, K.; Koibuchi, A.; Tanigawa, T.; Ueno, T.; Matsushima, N.; Imai, Y. A State-of-the-Art Roadmap for Biomarker-Driven Drug Development in the Era of Personalized Therapies. J. Pers. Med. 2022, 12, 669. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Vécsei, L. Revolutionizing our understanding of Parkinson’s disease: Dr. Heinz Reichmann’s pioneering research and future research direction. J. Neural Transm. 2024, 1–21, ahead of print. [Google Scholar] [CrossRef]
- Luke, J.J.; Fakih, M.; Schneider, C.; Chiorean, E.G.; Bendell, J.; Kristeleit, R.; Kurzrock, R.; Blagden, S.P.; Brana, I.; Goff, L.W. Phase I/II sequencing study of azacitidine, epacadostat, and pembrolizumab in advanced solid tumors. Br. J. Cancer 2023, 128, 2227–2235. [Google Scholar] [CrossRef]
- Long, G.V.; Dummer, R.; Hamid, O.; Gajewski, T.F.; Caglevic, C.; Dalle, S.; Arance, A.; Carlino, M.S.; Grob, J.-J.; Kim, T.M. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): A phase 3, randomised, double-blind study. Lancet Oncol. 2019, 20, 1083–1097. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Leonardi, G.C.; Puccetti, P.; Fallarino, F.; Bianconi, V.; Sahebkar, A.; Baglivo, S.; Chiari, R.; Pirro, M. Targeting indoleamine-2, 3-dioxygenase in cancer: Scientific rationale and clinical evidence. Pharmacol. Ther. 2019, 196, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Sun, L. Advances in the discovery and development of selective heme-displacing IDO1 inhibitors. Expert Opin. Drug Discov. 2020, 15, 1223–1232. [Google Scholar] [CrossRef]
- Kim, D.K.; Synn, C.-B.; Yang, S.M.; Kang, S.; Baek, S.; Oh, S.-W.; Lee, G.-J.; Kang, H.-W.; Lee, Y.-S.; Park, J.S. YH29407 with anti-PD-1 ameliorates anti-tumor effects via increased T cell functionality and antigen presenting machinery in the tumor microenvironment. Front. Chem. 2022, 10, 998013. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, K.; Eder, P.; Piha-Paul, S.A.; Gimmi, C.; Hussey, E.; Zhang, S.; Li, L.; Habermehl, C.; Naing, A. Abstract CT011: First-in-human Phase I study of M4112, the first dual inhibitor of indoleamine 2, 3-dioxygenase-1 and tryptophan 2, 3-dioxygenase 2, in patients with advanced solid malignancies. Cancer Res. 2019, 79, CT011. [Google Scholar] [CrossRef]
- Naing, A.; Eder, J.P.; Piha-Paul, S.A.; Gimmi, C.; Hussey, E.; Zhang, S.; Hildebrand, V.; Hosagrahara, V.; Habermehl, C.; Moisan, J. Preclinical investigations and a first-in-human phase I trial of M4112, the first dual inhibitor of indoleamine 2, 3-dioxygenase 1 and tryptophan 2, 3-dioxygenase 2, in patients with advanced solid tumors. J. Immunother. Cancer 2020, 8, e000870. [Google Scholar] [CrossRef]
- Rebai, R.; Carmena-Bargueño, M.; Toumi, M.E.; Derardja, I.; Jasmin, L.; Pérez-Sánchez, H.; Boudah, A. Identification of potent inhibitors of kynurenine-3-monooxygenase from natural products: In silico and in vitro approaches. Heliyon 2024, 10, e30287. [Google Scholar] [CrossRef]
- Pocivavsek, A.; Schwarcz, R.; Erhardt, S. Neuroactive Kynurenines as Pharmacological Targets: New Experimental Tools and Exciting Therapeutic Opportunities. Pharmacol. Rev. 2024, 76, 978–1008. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-S.; Ogbechi, J.; Clanchy, F.I.; Williams, R.O.; Stone, T.W. IDO and kynurenine metabolites in peripheral and CNS disorders. Front. Immunol. 2020, 11, 388. [Google Scholar] [CrossRef] [PubMed]
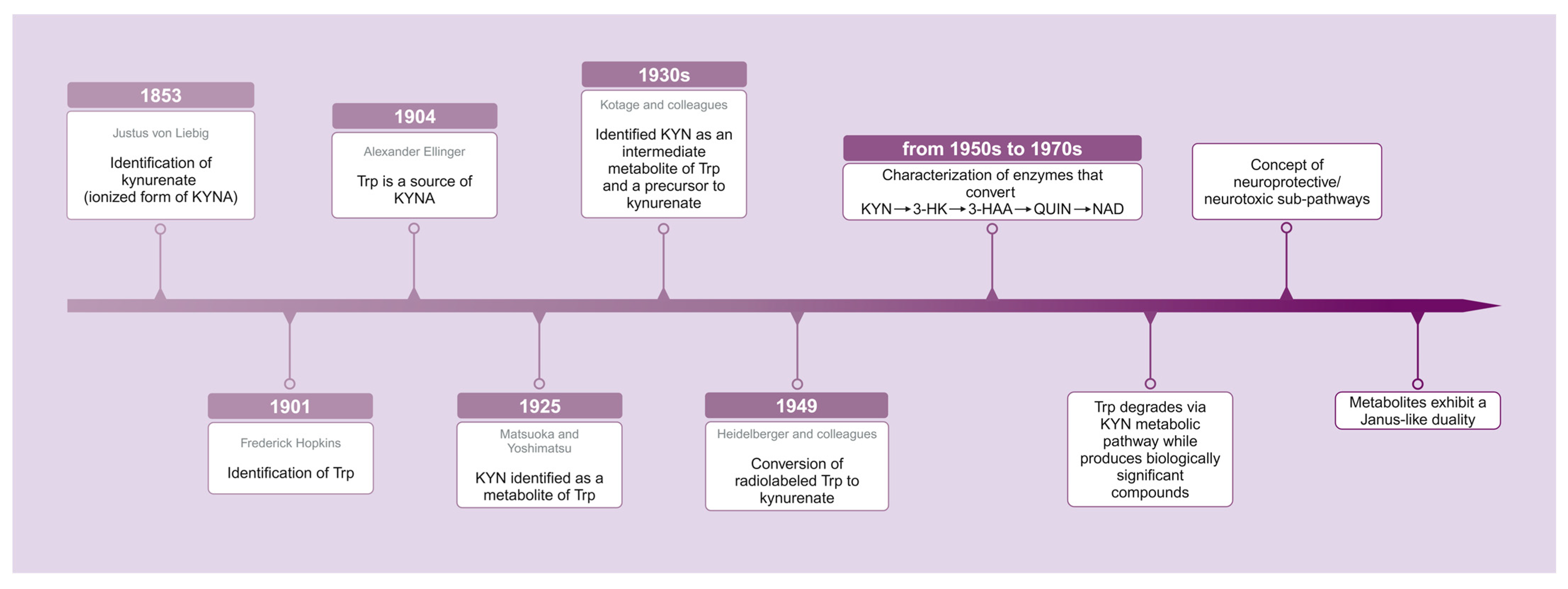

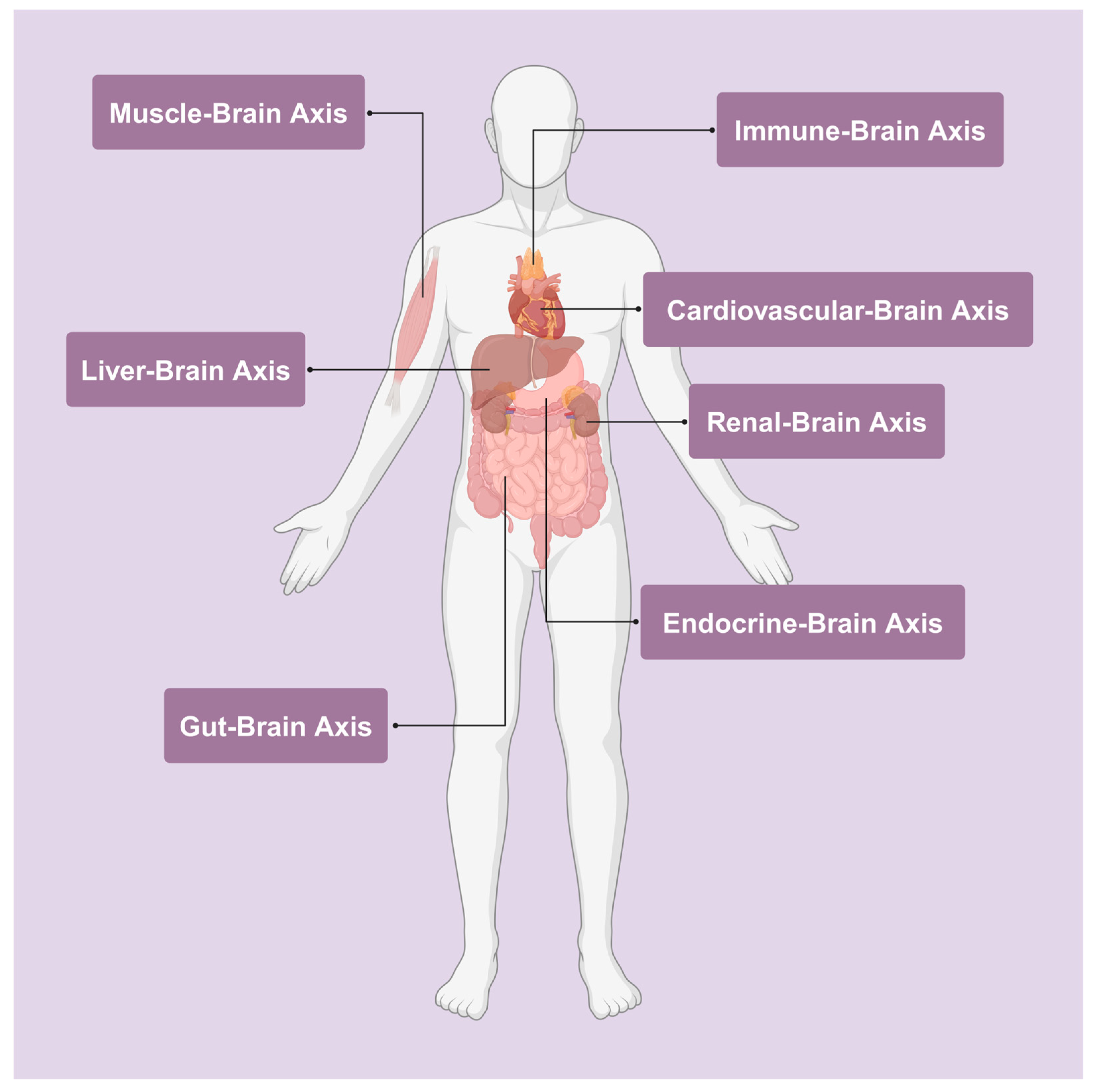
| Metabolites | Pro-Oxidant | Antioxidant | Receptors | Ref. | |
|---|---|---|---|---|---|
| Agonist | Antagonist | ||||
| Kynurenic acid (KYNA) | - | + | - | + 1, 2 | [40,83,84,85] |
| 3-Hydroxykynurenine (3-HK) | + | - | - | - | [86] |
| Quinolinic acid (QUIN) | + | - | + 1 | - | [87,88,89,90,91] |
| Metabolites | Pro- Oxidant | Anti- Oxidant | Receptors | AhR Agonist | Ref. | |
|---|---|---|---|---|---|---|
| Agonist | Antagonist | |||||
| Kynurenine (KYN) | + | + | - | - | + | [116,196,197,198] |
| Kynurenic acid (KYNA) | + | + | + 1, + 2 | + 3, ? 4 | + | [40,83,199,200,201,202,203] |
| Anthranilic acid (AA) | + | + | - | - | + | [190,191,192,193] |
| 3-Hydroxykynurenine (3-HK) | + | + | - | - | - | [67,193,204,205] |
| Xanthurenic acid (XA) | + | + | + 5 | - | + | [78,193,206,207,208,209,210,211,212,213] |
| Cinnabarinic acid (CA) | + | + | + 6 | - | + | [214,215,216] |
| 3-Hydroxyanthranilic acid (3-HAA) | + | + | - | - | - | [193,205] |
| Quinolinic acid (QUIN) | + | - | + | - | - | [87,88,89,90,91] |
| Picolinic acid (PA) | + | + | - | - | - | [217,218,219,220,221] |
| Indoxyl sulfate (INS) | + | + | - | - | + | [222,223,224,225,226,227,228] |
| Indole-3-acetamide (IAM) | - | + | - | - | + | [229,230,231,232] |
| Indole-3-acetic acid (IAA) | + | + | - | - | - | [230,233] |
| Indole-3-acetaldehyde (IAld) | - | - | - | - | + 7 | [232] |
| 3-methylindole (skatole) | + | + | - | - | + | [232,234,235,236] |
| Tryptamine | + | + | - | - | + 8 | [237,238,239,240,241,242,243] |
| Indole-3-acetaldehyde (IAAld) | - | - | - | - | + | [232,241,244] |
| Indole-3-ethanol (tryptophol) | - | + | - | - | + | [232,233,245] |
| Indole-3-pyruvic acid (IPyA) | - | - | - | - | + | [246,247] |
| Indole-3-lactic acid (ILA) | - | + | - | - | - | [248] |
| 3-indole acrylic acid (IAcA) | - | + | - | - | - | [249] |
| Indole-3-propionic acid (IPA) | - | + | - | - | + | [250,251,252,253] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, M.; Szabó, Á.; Vécsei, L. Redefining Roles: A Paradigm Shift in Tryptophan–Kynurenine Metabolism for Innovative Clinical Applications. Int. J. Mol. Sci. 2024, 25, 12767. https://doi.org/10.3390/ijms252312767
Tanaka M, Szabó Á, Vécsei L. Redefining Roles: A Paradigm Shift in Tryptophan–Kynurenine Metabolism for Innovative Clinical Applications. International Journal of Molecular Sciences. 2024; 25(23):12767. https://doi.org/10.3390/ijms252312767
Chicago/Turabian StyleTanaka, Masaru, Ágnes Szabó, and László Vécsei. 2024. "Redefining Roles: A Paradigm Shift in Tryptophan–Kynurenine Metabolism for Innovative Clinical Applications" International Journal of Molecular Sciences 25, no. 23: 12767. https://doi.org/10.3390/ijms252312767
APA StyleTanaka, M., Szabó, Á., & Vécsei, L. (2024). Redefining Roles: A Paradigm Shift in Tryptophan–Kynurenine Metabolism for Innovative Clinical Applications. International Journal of Molecular Sciences, 25(23), 12767. https://doi.org/10.3390/ijms252312767










