Precision Thermostability Predictions: Leveraging Machine Learning for Examining Laccases and Their Associated Genes
Abstract
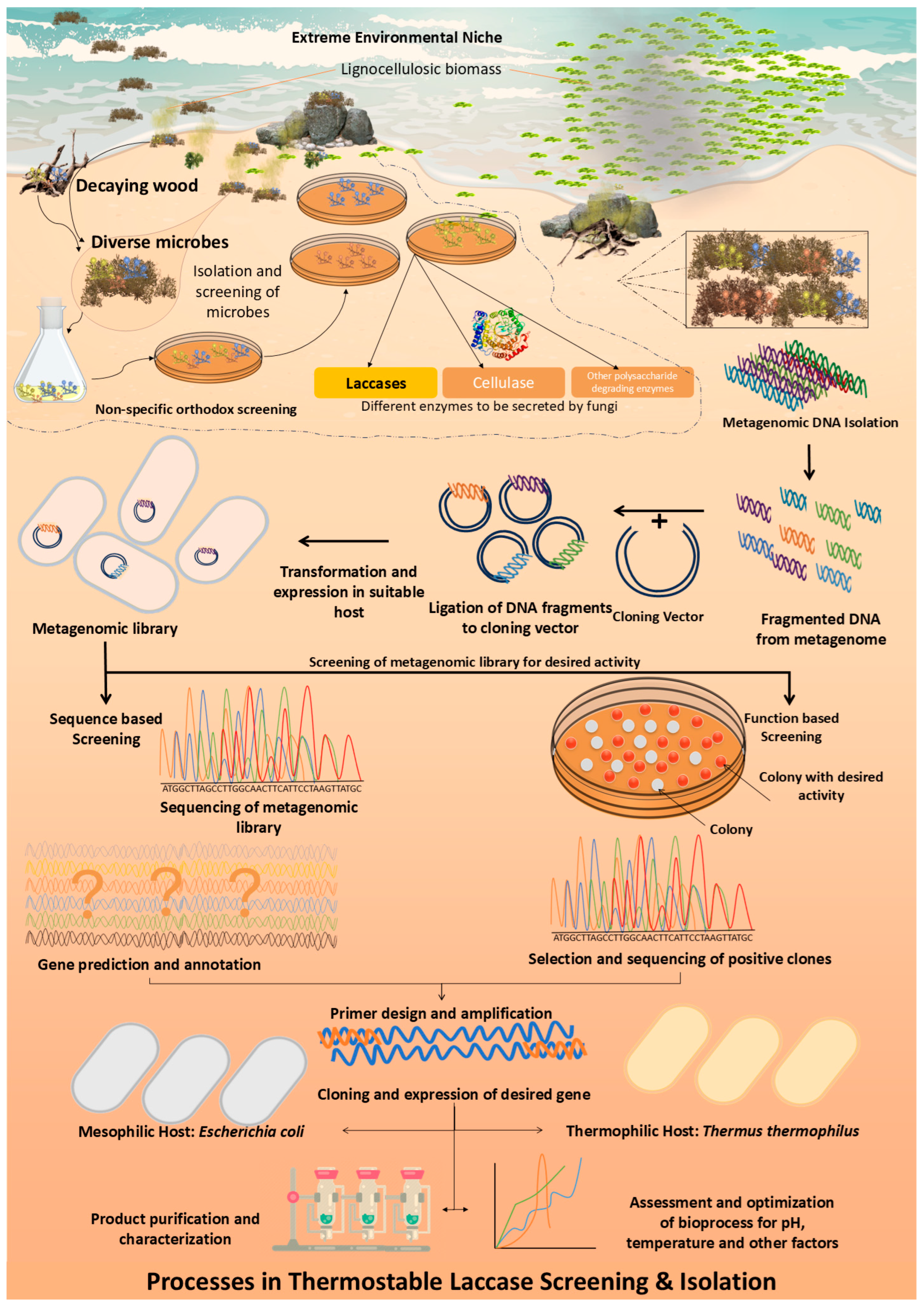
1. Introduction
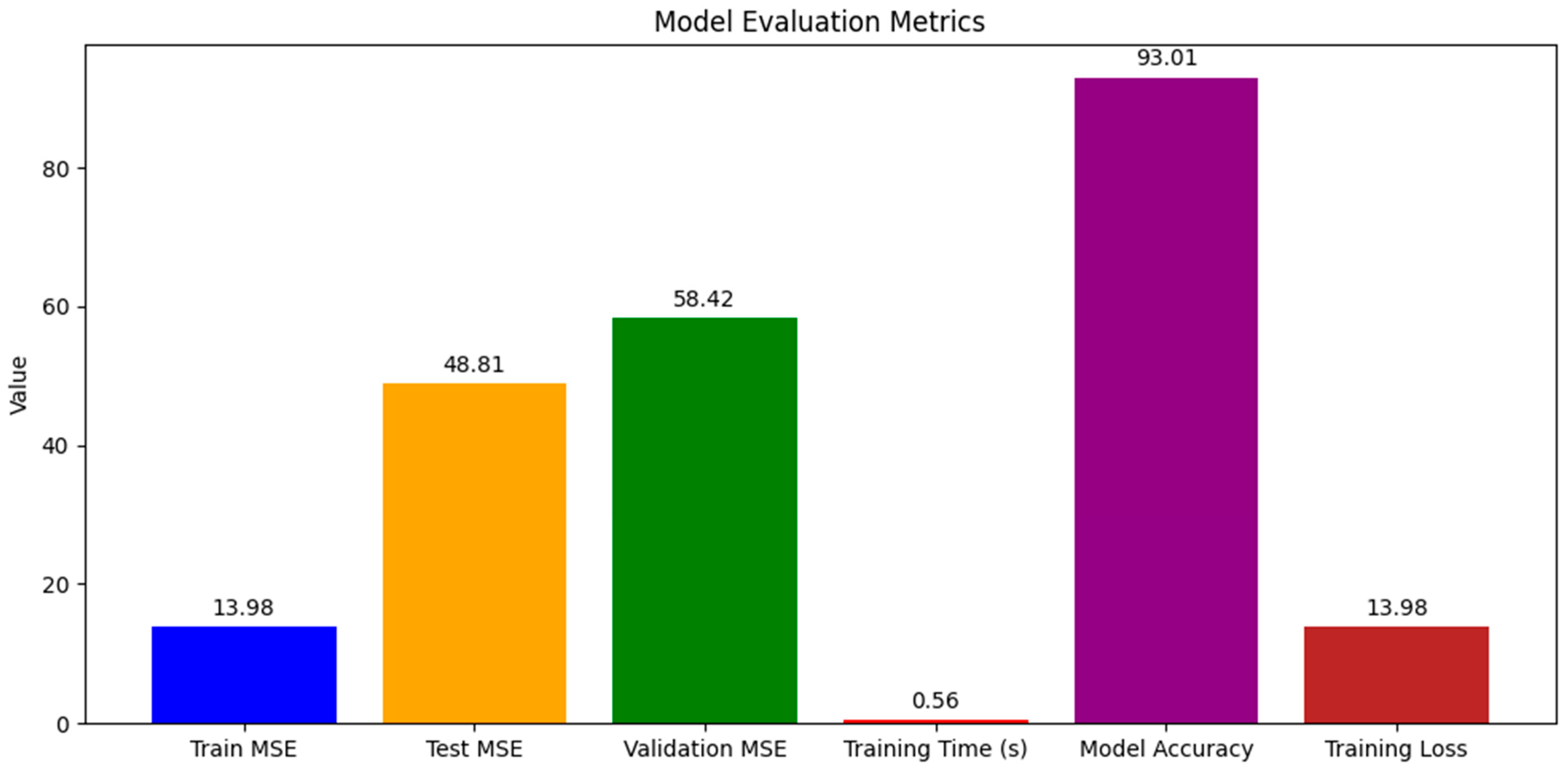
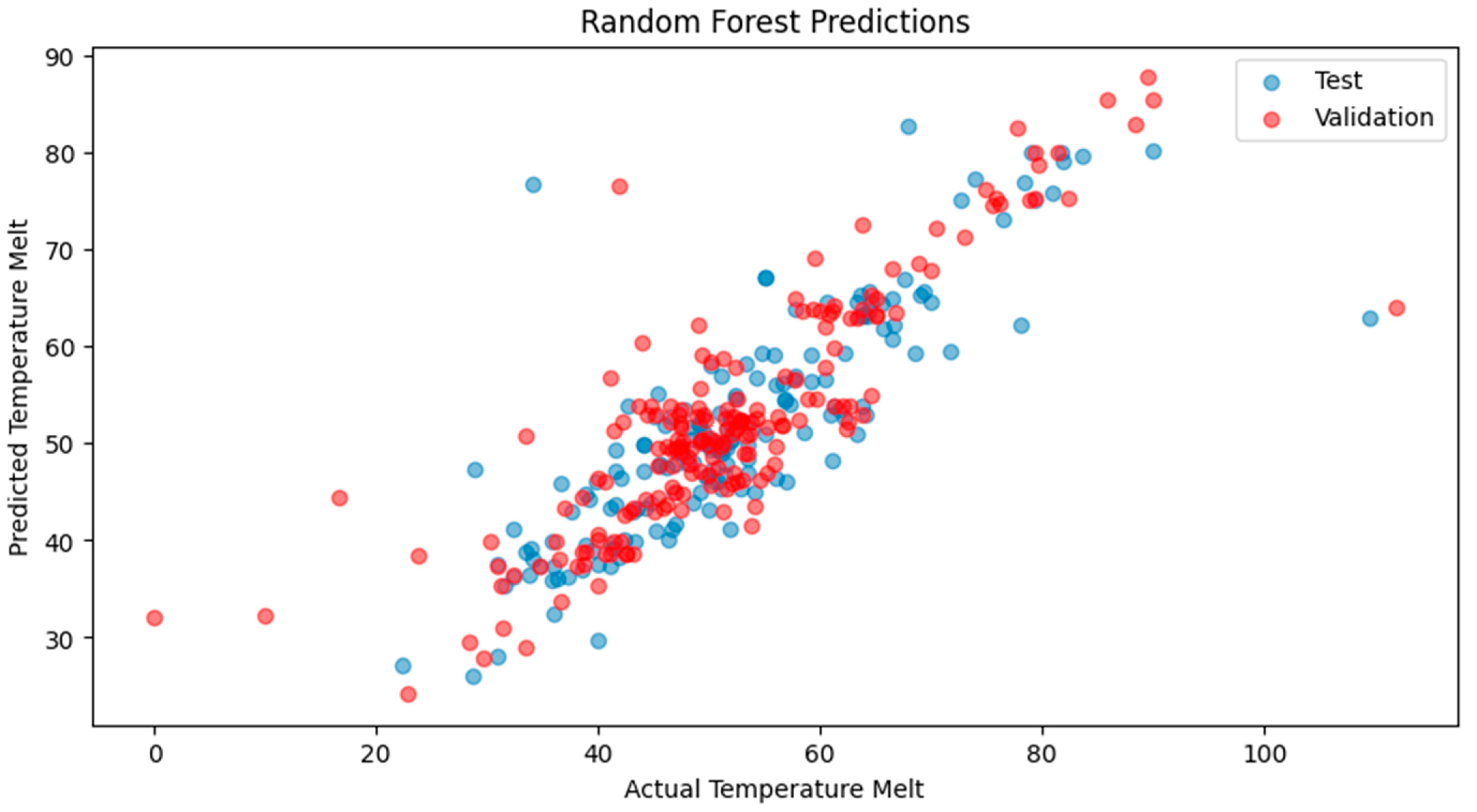
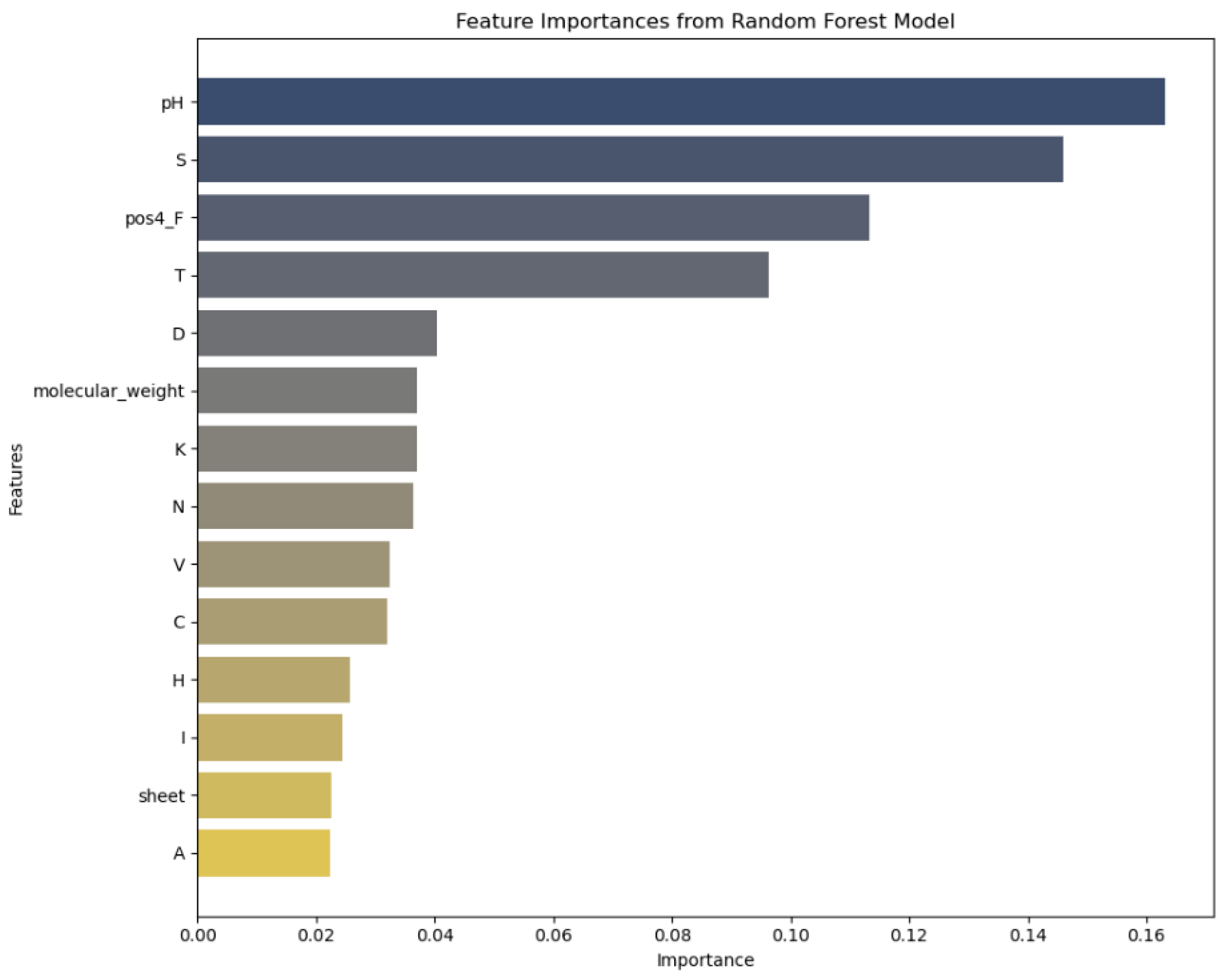
2. Results
2.1. Model Performance
2.2. Feature Importance
2.3. Determination of Structural Features of Laccases—The Catalytic Domain Architecture
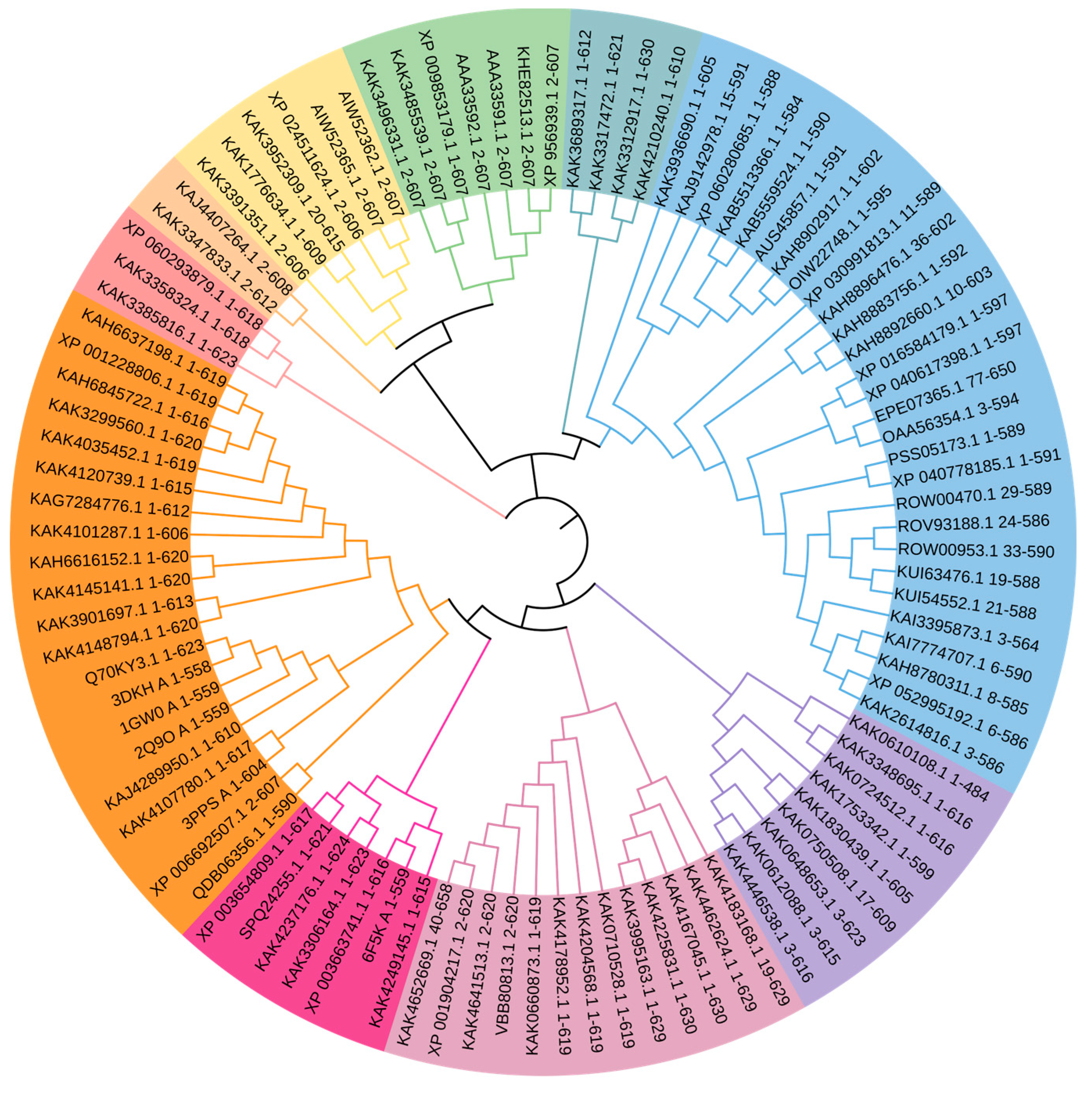
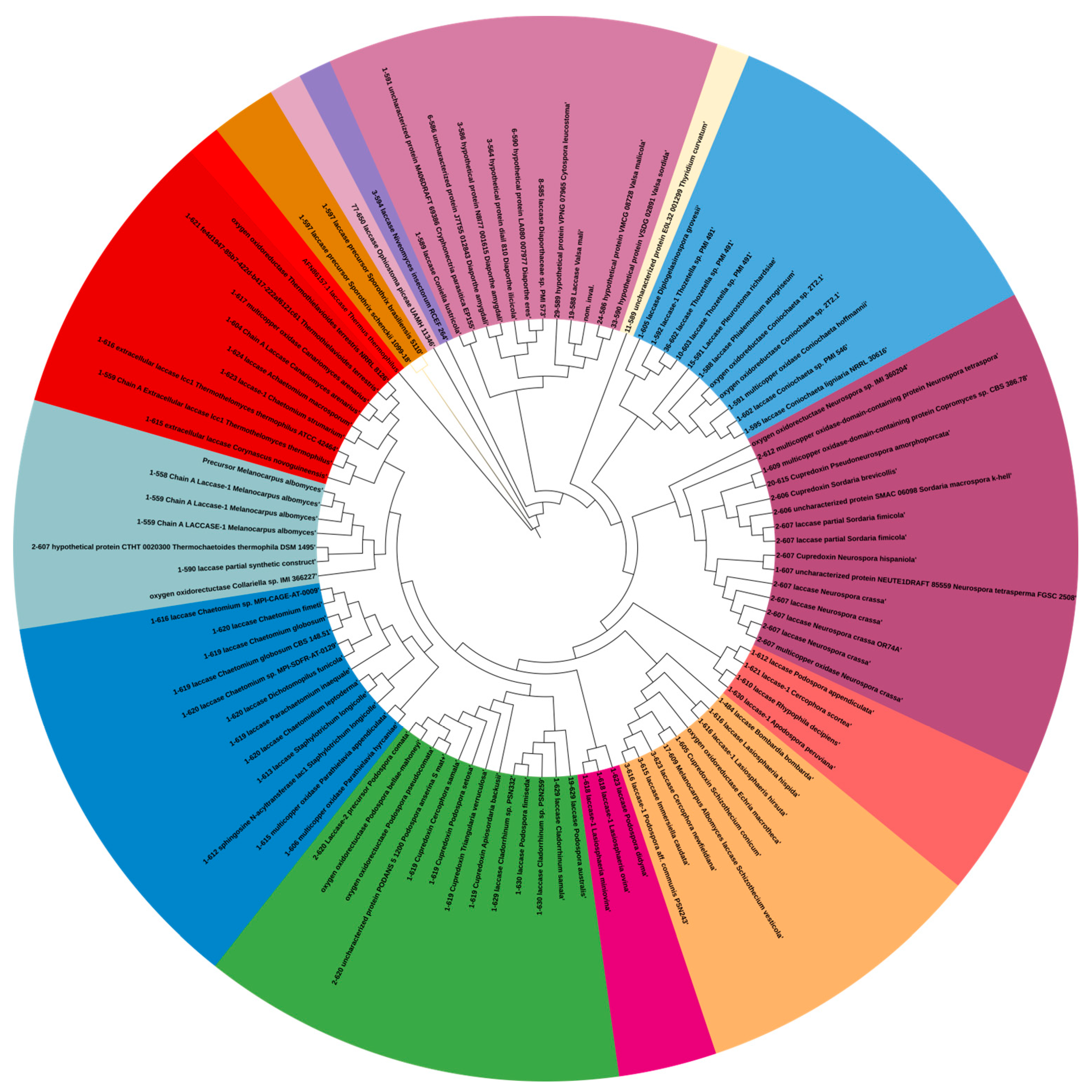
2.4. Evolutionary Aspects of Laccase Stability
2.5. Amino Acid Profiling
| Organism | Family | Domain | Homologous Superfamily | Conserved Site | Binding Site |
|---|---|---|---|---|---|
| Thermothelomces thermophilus | Multi-copper oxidase F Cu-oxidase_fam-IPR045087 multi-copper oxidase-PTHR11709 | Cu-oxidase_CCu-oxidase_2 D Cu-oxidase_C-IPR011706 Cu-oxidase_2-PF07731 | Cupredoxin H Cupredoxin-IPR008972 Cupredoxins-SSF49503 | S Cu_oxidase_CS-IPR033138 MULTICOPPER_OXIDASE1-PS00079 | S Cu_oxidase_Cu_BS-IPR002355 multicopper_oxidase2-PS00080 |
| Parachaetomium inaequale | Multi-copper oxidase F Cu-oxidase_fam-IPR045087 multi-copper oxidase-PTHR11709 | Cu-oxidase_2ndCu-oxidase D Cu-oxidase_2nd-IPR001117 Cu-oxidase-PF00394 | Cupredoxin H Cupredoxin-IPR008972 Cupredoxins-SSF49503 | S Cu_oxidase_CS-IPR033138 multicopper_oxidase1-PS00079 | S Cu_oxidase_Cu_BS-IPR002355 multicopper_oxidase2-PS00080 |
| Parathielavia appendiculata | Multi-copper oxidase F Cu-oxidase_fam-IPR045087 multi-copper oxidase-PTHR11709 | Cu-oxidase_CCu-oxidase_2 D Cu-oxidase_C-IPR011706 Cu-oxidase_2-PF07731 | Cupredoxin H Cupredoxin-IPR008972 Cupredoxins-SSF49503 | S Cu_oxidase_CS-IPR033138 multicopper_oxidase1-PS00079 | S Cu_oxidase_Cu_BS-IPR002355 multicopper_oxidase2-PS00080 |
| Chaetomium globosum CBS 148.51 | Multi-copper oxidase F Cu-oxidase_fam-IPR045087 multi-copper oxidase-PTHR11709 | Cu-oxidase_2ndCu-oxidase D Cu-oxidase_2nd-IPR001117 Cu-oxidase-PF00394 | Cupredoxin H Cupredoxin-IPR008972 Cupredoxins-SSF49503 | S Cu_oxidase_CS-IPR033138 multicopper_oxidase1-PS00079 | S Cu_oxidase_Cu_BS-IPR002355 multicopper_oxidase2-PS00080 |
| Corynascus novoguineensis | Multi-copper oxidase F Cu-oxidase_fam-IPR045087 multi-copper oxidase-PTHR11709 | Cu-oxidase_CCu-oxidase_2 D Cu-oxidase_C-IPR011706 Cu-oxidase_2-PF07731 | Cupredoxin H Cupredoxin-IPR008972 Cupredoxins-SSF49503 | S Cu_oxidase_CS-IPR033138 multicopper_oxidase1-PS00079 | S Cu_oxidase_Cu_BS-IPR002355 multicopper_oxidase2-PS00080 |
| Chaetomidium leptoderma | Multi-copper oxidase F Cu-oxidase_fam-IPR045087 multi-copper oxidase-PTHR11709 | Cu-oxidase-like…Cu-oxidase_3 D Cu-oxidase-like_N-IPR011707 Cu-oxidase_3-PF07732 | Cupredoxin H Cupredoxin-IPR008972 Cupredoxins-SSF49503 | S Cu_oxidase_CS-IPR033138 multicopper_oxidase1-PS00079 | S Cu_oxidase_Cu_BS-IPR002355 multicopper_oxidase2-PS00080 |
| Cladorrhinum samala | Multi-copper oxidase F Cu-oxidase_fam-IPR045087 multi-copper oxidase-PTHR11709 | Cu-oxidase_2ndCu-oxidase D Cu-oxidase_2nd-IPR001117 Cu-oxidase-PF00394 | Cupredoxin H Cupredoxin-IPR008972 Cupredoxins-SSF49503 | S Cu_oxidase_CS-IPR033138 multicopper_oxidase1-PS00079 | S Cu_oxidase_Cu_BS-IPR002355 multicopper_oxidase2-PS00080 |
| Staphylotrichum longicolle | Multi-copper oxidase F Cu-oxidase_fam-IPR045087 multi-copper oxidase-PTHR11709 | Cu-oxidase-like_NCu-oxidase_3 D Cu-oxidase-like_N-IPR011707 Cu-oxidase_3-PF07732 | Cupredoxin H Cupredoxin-IPR008972 Cupredoxins-SSF49503 | S Cu_oxidase_CS-IPR033138 multicopper_oxidase1-PS00079 | S Cu_oxidase_Cu_BS-IPR002355 multicopper_oxidase2-PS00080 |
3. Discussion
4. Materials and Methods
4.1. Data Collection
4.2. Feature Extraction and Preprocessing
4.2.1. Amino Acid Composition
4.2.2. Molecular Weight
4.2.3. Aromaticity
4.2.4. Isoelectric Point (pI)
4.2.5. Secondary Structural Content
4.2.6. Extraction Technique
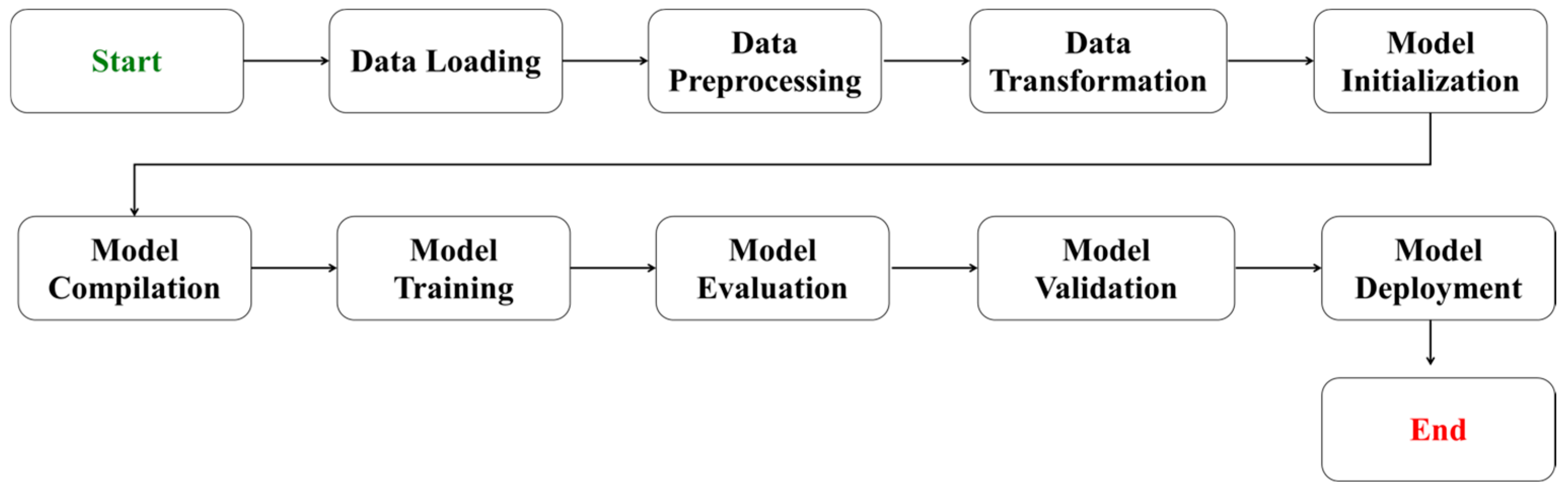
4.3. Machine Learning Models
4.3.1. RF Regressor
- The dataset was first split into training, test, and validation sets to ensure a robust evaluation of the model’s performance.
- Feature columns, excluding identifiers and the target variable (‘temp_melt’), were extracted as inputs for the model.
- The RF model was initialized with 100 estimators, leveraging the ensemble learning method to reduce overfitting and improve prediction accuracy.
- The model was trained on the training set, with its performance evaluated on both the test and validation sets to gauge its generalization ability.
- RF was selected for its effectiveness in handling high-dimensional data, like amino acid sequences in our study, and its capacity to model complex relationships between features and the target variable without extensive parameter tuning.
- The model’s inherent feature importance metric offered insights into which genomic features were most predictive of thermostability, aiding in the biological interpretation of the results.
4.3.2. Convolutional Neural Network (CNN)
- Input data were reshaped to fit CNN requirements, with each sequence represented as a two-dimensional (2D) array (features by 1) to mimic a single-channel image.
- Datasets were normalized to ensure efficient training of the neural network.
- Our CNN model comprised an input Conv1D layer with 64 filters and a kernel size of 3, followed by a MaxPooling1D layer to reduce dimensionality.
- A flattened layer was then applied to convert the pooled feature maps into a single vector per sample, which was followed by two Dense layers, with the final one outputting the predicted temperature melt value.
- The model was compiled using the Adam optimizer and MSE loss function, trained over 100 epochs with a batch size determined by default settings.
- The CNN was chosen for its ability to capture local dependencies and patterns within sequential data, a common characteristic of genomic sequences affecting protein function and stability.
- This approach is particularly suited for datasets like ours, where the spatial arrangement of features (e.g., amino acid sequences) plays a crucial role in determining the target variable (thermostability).
4.3.3. Model Evaluation
4.4. Model Training and Validation
4.4.1. Data Preparation
4.4.2. Model Training
4.4.3. Validation and Evaluation
4.4.4. Model Assessment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, C.-D.; Tiwari, A.; Anisha, G.S.; Chen, C.-W.; Singh, A.; Haldar, D.; Patel, A.K.; Singhania, R.R. Laccase: A Potential Biocatalyst for Pollutant Degradation. Environ. Pollut. 2023, 319, 120999. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Chandra, R. Ligninolytic Enzymes and Its Mechanisms for Degradation of Lignocellulosic Waste in Environment. Heliyon 2020, 6, e03170. [Google Scholar] [CrossRef] [PubMed]
- Maghraby, Y.R.; El-Shabasy, R.M.; Ibrahim, A.H.; Azzazy, H.M.E.-S. Enzyme Immobilization Technologies and Industrial Applications. ACS Omega 2023, 8, 5184–5196. [Google Scholar] [CrossRef]
- Li, C.; Zhang, R.; Wang, J.; Wilson, L.M.; Yan, Y. Protein Engineering for Improving and Diversifying Natural Products Biosynthesis. Trends Biotechnol. 2020, 38, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, Q.; Zhang, W.; Mu, W. Overview of Strategies for Developing High Thermostability Industrial Enzymes: Discovery, Mechanism, Modification and Challenges. Crit. Rev. Food Sci. Nutr. 2021, 63, 2057–2073. [Google Scholar] [CrossRef]
- López-López, O.; Cerdán, M.-E.; González-Siso, M.-I. Thermus Thermophilus as a Source of Thermostable Lipolytic Enzymes. Microorganisms 2015, 3, 792–808. [Google Scholar] [CrossRef]
- Kumwenda, B.; Litthauer, D.; Bishop, Ö.T.; Reva, O. Analysis of Protein Thermostability Enhancing Factors in Industrially Important Thermus Bacteria Species. Evol. Bioinform. Online 2013, 9, 327–342. [Google Scholar] [CrossRef]
- Mehra, R.; Muschiol, J.; Meyer, A.S.; Kepp, K.P. A Structural-Chemical Explanation of Fungal Laccase Activity. Sci. Rep. 2018, 8, 17285. [Google Scholar] [CrossRef]
- Maestre-Reyna, M.; Liu, W.-C.; Jeng, W.-Y.; Lee, C.-C.; Hsu, C.-A.; Wen, T.-N.; Wang, A.H.-J.; Shyur, L.-F. Structural and Functional Roles of Glycosylation in Fungal Laccase from Lentinus Sp. PLoS ONE 2015, 10, e0120601. [Google Scholar] [CrossRef]
- Cázares-García, S.V.; Vázquez-Garcidueñas, M.S.; Vázquez-Marrufo, G. Structural and Phylogenetic Analysis of Laccases from Trichoderma: A Bioinformatic Approach. PLoS ONE 2013, 8, e55295. [Google Scholar] [CrossRef]
- Kumari, A.; Kishor, N.; Guptasarma, P. Characterization of a Mildly Alkalophilic and Thermostable Recombinant Thermus Thermophilus Laccase with Applications in Decolourization of Dyes. Biotechnol. Lett. 2018, 40, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Chen, C.-W.; Haldar, D.; Patel, A.K.; Dong, C.-D.; Singhania, R.R. Laccase in Biorefinery of Lignocellulosic Biomass. Appl. Sci. 2023, 13, 4673. [Google Scholar] [CrossRef]
- Clément, R.; Wang, X.; Biaso, F.; Ilbert, M.; Mazurenko, I.; Lojou, E. Mutations in the Coordination Spheres of T1 Cu Affect Cu2+-Activation of the Laccase from Thermus thermophilus. Biochimie 2021, 182, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Fernandez, B.J.; Risso, V.A.; Rueda, A.; Sanchez-Ruiz, J.M.; Alcalde, M. Ancestral Resurrection and Directed Evolution of Fungal Mesozoic Laccases. Appl. Environ. Microbiol. 2020, 86, e00778-20. [Google Scholar] [CrossRef]
- Kolyadenko, I.; Tishchenko, S.; Gabdulkhakov, A. Structural Insight into the Amino Acid Environment of the Two-Domain Laccase’s Trinuclear Copper Cluster. Int. J. Mol. Sci. 2023, 24, 11909. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkołazka, A.; Paszczyński, A. Lignin Degradation: Microorganisms, Enzymes Involved, Genomes Analysis and Evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef]
- Miyazaki, K. A Hyperthermophilic Laccase from Thermus Thermophilus HB27. Extremophiles 2005, 9, 415–425. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; Herrera de los Santos, M.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.N.; et al. Laccases: Structure, Function, and Potential Application in Water Bioremediation. Microb. Cell Factories 2019, 18, 200. [Google Scholar] [CrossRef]
- Makam, P.; Yamijala, S.S.R.K.C.; Bhadram, V.S.; Shimon, L.J.W.; Wong, B.M.; Gazit, E. Single Amino Acid Bionanozyme for Environmental Remediation. Nat. Commun. 2022, 13, 1505. [Google Scholar] [CrossRef]
- Long, S.; Zhang, X.; Rao, Z.; Chen, K.; Xu, M.; Yang, T.; Yang, S. Amino Acid Residues Adjacent to the Catalytic Cavity of Tetramer L-Asparaginase II Contribute Significantly to Its Catalytic Efficiency and Thermostability. Enzym. Microb. Technol. 2016, 82, 15–22. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Lakizadeh, A.; Agha-Golzadeh, P.; Ebrahimie, E.; Ebrahimi, M. Prediction of Thermostability from Amino Acid Attributes by Combination of Clustering with Attribute Weighting: A New Vista in Engineering Enzymes. PLoS ONE 2011, 6, e23146. [Google Scholar] [CrossRef] [PubMed]
- About-InterPro. Available online: https://www.ebi.ac.uk/interpro/about/interproscan/ (accessed on 14 April 2024).
- Gromiha, M.M.; Oobatake, M.; Sarai, A. Important Amino Acid Properties for Enhanced Thermostability from Mesophilic to Thermophilic Proteins. Biophys. Chem. 1999, 82, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Modarres, H.P.; Mofrad, M.R.; Sanati-Nezhad, A. Protein Thermostability Engineering. RSC Adv. 2016, 6, 115252–115270. [Google Scholar] [CrossRef]
- Gollan, M.; Black, G.; Munoz-Munoz, J. A Computational Approach to Optimising Laccase-Mediated Polyethylene Oxidation through Carbohydrate-Binding Module Fusion. BMC Biotechnol. 2023, 23, 18. [Google Scholar] [CrossRef]
- GenBank Overview. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 14 April 2024).
- UniProt. Available online: https://www.uniprot.org/ (accessed on 14 April 2024).
- Mycocosm. Available online: https://mycocosm.jgi.doe.gov/mycocosm/home (accessed on 14 April 2024).
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Hormoz, S. Amino Acid Composition of Proteins Reduces Deleterious Impact of Mutations. Sci. Rep. 2013, 3, 2919. [Google Scholar] [CrossRef]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between Stability of a Protein and Its Dipeptide Composition: A Novel Approach for Predicting in Vivo Stability of a Protein from Its Primary Sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Dudek, W.M.; Ostrowski, S.; Dobrowolski, J.C. On Aromaticity of the Aromatic α-Amino Acids and Tuning of the NICS Indices to Find the Aromaticity Order. J. Phys. Chem. A 2022, 126, 3433–3444. [Google Scholar] [CrossRef]
- Behrens, C.; Hartmann, K.; Sunderhaus, S.; Braun, H.-P.; Eubel, H. Approximate Calculation and Experimental Derivation of Native Isoelectric Points of Membrane Protein Complexes of Arabidopsis Chloroplasts and Mitochondria. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2013, 1828, 1036–1046. [Google Scholar] [CrossRef]
- Khrustalev, V.V.; Khrustaleva, T.A.; Poboinev, V.V. Amino Acid Content of Beta Strands and Alpha Helices Depends on Their Flanking Secondary Structure Elements. Biosystems 2018, 168, 45–54. [Google Scholar] [CrossRef]
- Biau, G. Analysis of a Random Forests Model. J. Mach. Learn. Res. 2010, 13, 1063–1095. [Google Scholar]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a Convolutional Neural Network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar]


| UniProtKB ID /PDB/GenBank Accession No. | Source of Laccase | Total Amino Acid Residues | Theoretical pI | % of Acidic Amino Acid Residues | % of Sulfur Amino Acids | C + F + T % | D + E | % of Basic Amino Acid Residues | R + K | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aspartic Acid (D) | Glutamic Acid (E) | Cysteine (C) | Phenylalanine (F) | Threonine (T) | Arginine (R) | Lysine (K) | |||||||
| XP_003663741 | Thermothelomyces thermophilus ATCC 42464 | 616 | 5.28 | 7.3 | 4.1 | 1.1 | 3.4 | 7.0 | 11.5 | 70 | 4.7 | 2.8 | 46 |
| 6F5K | Thermothelomyces thermophilus | 559 | 5.08 | 7.9 | 3.2 | 1.3 | 3.6 | 7.0 | 11.9 | 62 | 4.5 | 2.3 | 38 |
| KAK4035452 | Parachaetomium inaequale | 619 | 6.33 | 6.0 | 2.6 | 1.1 | 4.5 | 7.4 | 13.0 | 53 | 5.3 | 2.1 | 46 |
| KAK4120739 | Parathielavia appendiculata | 615 | 6.64 | 6.0 | 2.3 | 1.1 | 4.4 | 7.5 | 13.0 | 51 | 5.2 | 2.4 | 47 |
| XP_001228806 | Chaetomium globosum CBS 148.51 | 619 | 6.11 | 5.8 | 3.1 | 1.3 | 4.5 | 8.2 | 14.0 | 55 | 5.3 | 1.9 | 45 |
| KAK4249145 | Corynascus novoguineensis | 615 | 5.14 | 7.6 | 4.4 | 1.1 | 3.6 | 6.7 | 11.4 | 74 | 4.7 | 2.6 | 45 |
| KAK4148794 | Chaetomidium leptoderma | 620 | 6.76 | 5.5 | 2.4 | 1.3 | 4.7 | 8.4 | 14.4 | 49 | 4.8 | 2.6 | 46 |
| KAK4462624 | Cladorrhinum samala | 629 | 6.92 | 6.0 | 2.2 | 1.1 | 3.7 | 7.6 | 12.4 | 52 | 4.8 | 3.2 | 50 |
| KAK3901697 | Staphylotrichum tortipilum | 613 | 6.46 | 5.5 | 2.4 | 1.3 | 4.9 | 8.5 | 14.7 | 51 | 4.4 | 3.1 | 46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwari, A.; Krisnawati, D.I.; Widodo; Cheng, T.-M.; Kuo, T.-R. Precision Thermostability Predictions: Leveraging Machine Learning for Examining Laccases and Their Associated Genes. Int. J. Mol. Sci. 2024, 25, 13035. https://doi.org/10.3390/ijms252313035
Tiwari A, Krisnawati DI, Widodo, Cheng T-M, Kuo T-R. Precision Thermostability Predictions: Leveraging Machine Learning for Examining Laccases and Their Associated Genes. International Journal of Molecular Sciences. 2024; 25(23):13035. https://doi.org/10.3390/ijms252313035
Chicago/Turabian StyleTiwari, Ashutosh, Dyah Ika Krisnawati, Widodo, Tsai-Mu Cheng, and Tsung-Rong Kuo. 2024. "Precision Thermostability Predictions: Leveraging Machine Learning for Examining Laccases and Their Associated Genes" International Journal of Molecular Sciences 25, no. 23: 13035. https://doi.org/10.3390/ijms252313035
APA StyleTiwari, A., Krisnawati, D. I., Widodo, Cheng, T.-M., & Kuo, T.-R. (2024). Precision Thermostability Predictions: Leveraging Machine Learning for Examining Laccases and Their Associated Genes. International Journal of Molecular Sciences, 25(23), 13035. https://doi.org/10.3390/ijms252313035



_Kim.png)




