Exploring the Potential of Bioactive Peptides: From Natural Sources to Therapeutics
Abstract
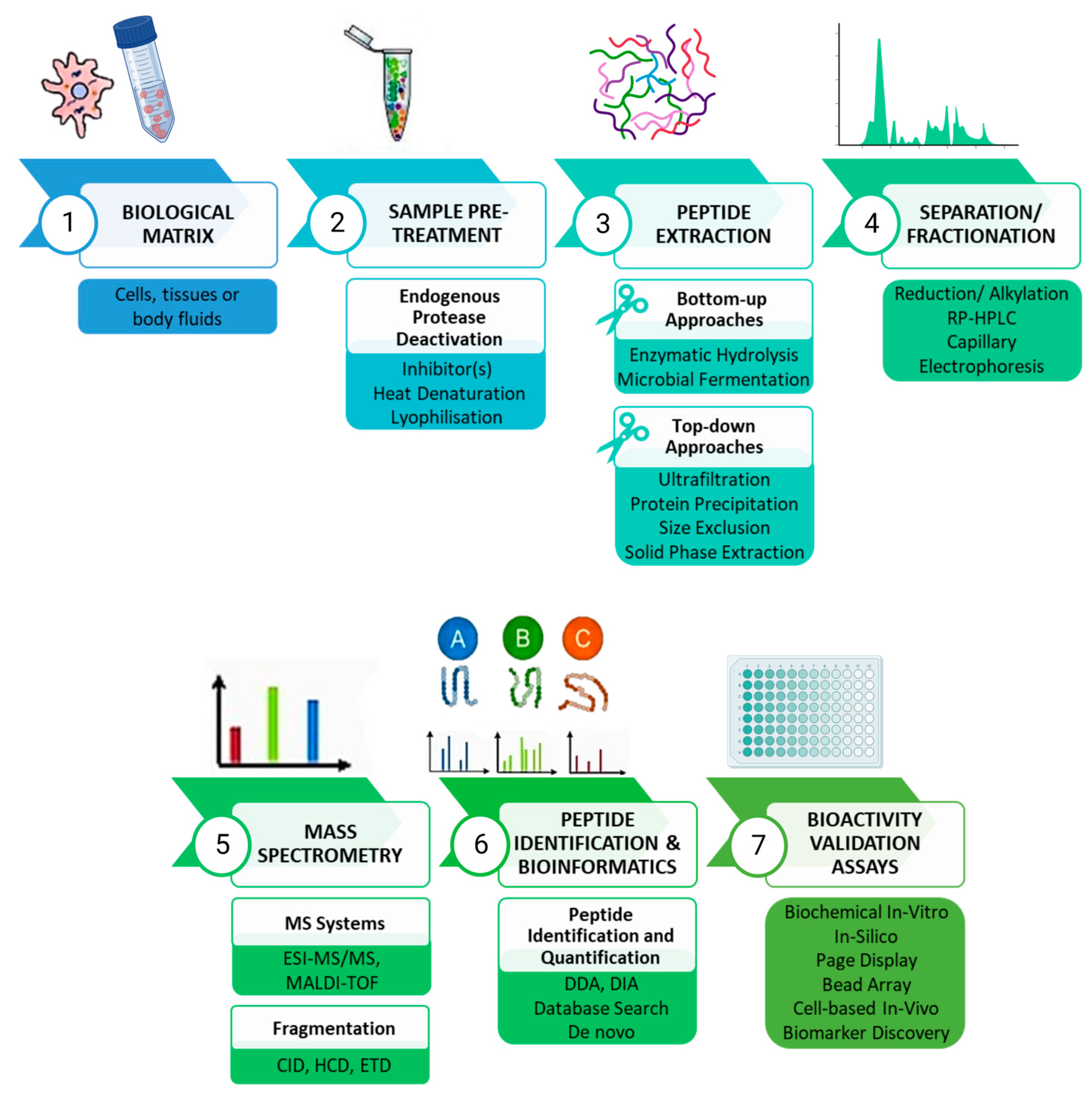
:1. Introduction
2. Peptide Discovery
2.1. Natural Sources
2.2. Peptidomics and Search for Novel Peptides
3. Peptide Extraction Approaches
3.1. Bottom-Up Approaches
3.1.1. Enzymatic Hydrolysis
3.1.2. Microbial Fermentation
3.2. Top-Down Approaches
3.2.1. Ultrafiltration
3.2.2. Protein Precipitation
3.2.3. Size Exclusion Chromatography (SEC)
3.2.4. Solid-Phase Extraction (SPE)
3.3. Peptides with Disulphide Linkages
4. Mass Spectrometric Analysis
4.1. Separation and Fractionation of Peptide Mixtures
4.1.1. Liquid Chromatography
4.1.2. Capillary Electrophoresis
4.2. Untargeted Peptide Analysis and Bioinformatics
4.2.1. Data Acquisition
4.2.2. Peptide Identification
5. Peptide Bioactivity
5.1. Biochemical In Vitro Assays
5.1.1. Ligand Binding Assays
5.1.2. Fluorescence Assays
5.2. In Silico Assays
5.3. Phage Display
5.4. Bead Array
5.5. Cell-Based In Vivo Assays
5.5.1. Cell Viability Assay
5.5.2. Reporter Gene Assays
5.5.3. Protein–Protein Interactions
5.5.4. Label-Free Detection
5.5.5. High-Throughput Electrophysiology Assays
5.6. AI-Driven Approaches for Peptide Discovery
6. Structure–Activity Relationship (SAR)
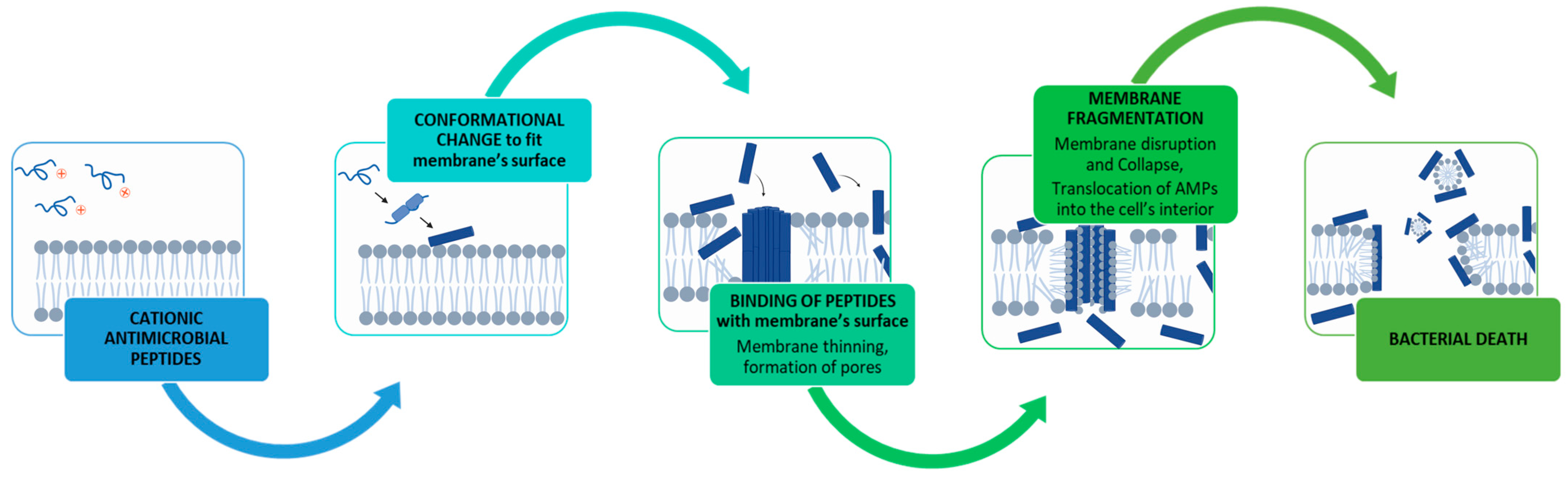
6.1. Antimicrobial Peptides (AMPs)
- α-helix and β-sheet structures are favourable for antimicrobial activity.
- The cationic nature of peptides favours antimicrobial activity.
- Aspartic acid (D)-rich anionic AMPs exist. However, their mode of action is not known.
6.2. ACE-Inhibitory Peptides (ACEs)
- Strong chelating groups to efficiently coordinate with the zinc II ion in the active site.
- Suitable functional groups that can form strong hydrogen bonds with appropriate sub-sites in the ACE active site.
- A hydrophobic residue with an aromatic side chain, such as phenylalanine.
- Proline-containing sequences for effective binding.
- Preferably, a short peptide chain with 2–4 residues (a tripeptide may be the best fit).
6.3. Antioxidative Peptides (AOPs)
- The properties of the residue next to the C-terminal position play a key role in determining the antioxidant activity of the sequence. For high activity, hydrophilic (Q, N, T, and S) and hydrogen bonding residues (E, D, H, K, D, P, Y, C, and R) are favoured in this position. Specifically, it has been demonstrated that hydrophobic residues are not suitable for this position.
- N-terminal hydrophobic residues (A, V, L) and C-terminal polar residues (W, E, Y, Q) are favourable for potent antioxidant peptides.
- The limitation of the model that only tri- and tetrapeptides are considered.
- The possible relationship of the antioxidant activities to the secondary structures of peptides.
6.4. Anticancer Peptides (ACPs)
| Name | Primary Structure | Source | References |
|---|---|---|---|
| Naturally occurring peptides | |||
| Chlorotoxin | MCMPCFTTDHQMARKCDDCCGGKGRGKCYGPQCLCR | Scorpion venom (L. quinqestriatus) | [27,222] |
| BmKn2 | FIGAIARLLSKIF | Scorpion venom (M. martensii) | [27,228] |
| Pantinin-2 (P2) | IFGAIWKGISSLL | Scorpion venom (P. imperator) | [27,223] |
| Pantanin-3 (P3) | FLSTIWNGIKSLL | Scorpion venom (P. imperator) | [27,223] |
| RK1 | IDCSKVNLTAECSS | Scorpion venom (B. occitanus tunetanus) | [27,224] |
| TsAP1 | FLSLIPSLVGGSISAFK | Brazilian yellow scorpion (T. serrulatus) | [27,229] |
| TsAP2 | FLGMIPGLIGGLISAFK | Brazilian yellow scorpion (T. serrulatus) | [27,229] |
| Smp24 | IWSFLIKAATKLLPSLFGGGKKDS | Scorpion venom (S. Maurus palmatus) | [27,230] |
| Bf-CATH30 | KFFRKLKKSVKKRAKEFFKKPRVIGVSIPF | Snake venom (Bungarus fasciatus) | [225] |
| Cdt-CATH | KRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTIPF | Snake venom (Crotalus durissus terrificus) | [226] |
| ∆Pb-CATH4 | TRSRWRRFIRGAGRFARRYGWRIA | Snake venom (Python bivittatus) | [227] |
6.5. Antiparasitic Peptides (APPs)
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Preet, P. Peptides: A new therapeutic approach. Int. J. Curr. Pharm. Res. 2018, 10, 29–34. [Google Scholar] [CrossRef]
- Uhlig, T.; Kyprianou, T.; Martinelli, F.G.; Oppici, C.A.; Heiligers, D.; Hills, D.; Calvo, X.R.; Verhaert, P. The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Proteom. 2014, 4, 58–69. [Google Scholar] [CrossRef]
- Craik, D.J.; Fairlie, D.P.; Liras, S.; Price, D. The Future of Peptide-based Drugs. Chem. Biol. Drug Des. 2013, 81, 136–147. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Matrisian, L.M. Emerging roles of proteases in tumour suppression. Nat. Rev. Cancer 2007, 7, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Quintal-Bojórquez, N.; Segura-Campos, M.R. Bioactive Peptides as Therapeutic Adjuvants for Cancer. Nutr. Cancer 2021, 73, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Fields, K.; Falla, T.J.; Rodan, K.; Bush, L. Bioactive peptides: Signaling the future. J. Cosmet. Dermatol. 2009, 8, 8–13. [Google Scholar] [CrossRef]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef]
- Marqus, S.; Pirogova, E.; Piva, T.J. Evaluation of the use of therapeutic peptides for cancer treatment. J. Biomed. Sci. 2017, 24, 21. [Google Scholar] [CrossRef]
- Rossino, G.; Marchese, E.; Galli, G.; Verde, F.; Finizio, M.; Serra, M.; Linciano, P.; Collina, S. Peptides as Therapeutic Agents: Challenges and Opportunities in the Green Transition Era. Molecules 2023, 28, 7165. [Google Scholar] [CrossRef]
- Naeem, M.; Malik, M.I.; Umar, T.; Ashraf, S.; Ahmad, A. A Comprehensive Review About Bioactive Peptides: Sources to Future Perspective. Int. J. Pept. Res. Ther. 2022, 28, 155. [Google Scholar] [CrossRef]
- Daliri, E.B.-M.; Oh, D.H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Alzaydi, A.; Barbhuiya, R.I.; Routray, W.; Elsayed, A.; Singh, A. Bioactive peptides: Synthesis, applications, and associated challenges. Food Bioeng. 2023, 2, 273–290. [Google Scholar] [CrossRef]
- Bhandari, D.; Rafiq, S.; Gat, Y.; Gat, P.; Waghmare, R.; Kumar, V. A Review on Bioactive Peptides: Physiological Functions, Bioavailability and Safety. Int. J. Pept. Res. Ther. 2020, 26, 139–150. [Google Scholar] [CrossRef]
- Du, Z.; Li, Y. Review and perspective on bioactive peptides: A roadmap for research, development, and future opportunities. J. Agric. Food Res. 2022, 9, 100353. [Google Scholar] [CrossRef]
- Zaky, A.A.; Simal-Gandara, J.; Eun, J.B.; Shim, J.H.; Abd El-Aty, A.M. Bioactivities, Applications, Safety, and Health Benefits of Bioactive Peptides From Food and By-Products: A Review. Front. Nutr. 2021, 8, 815640. [Google Scholar] [CrossRef] [PubMed]
- Akbarian, M.; Khani, A.; Eghbalpour, S.; Uversky, V.N. Bioactive Peptides: Synthesis, Sources, Applications, and Proposed Mechanisms of Action. Int. J. Mol. Sci. 2022, 23, 1445. [Google Scholar] [CrossRef]
- Sable, R.; Parajuli, P.; Jois, S. Peptides, peptidomimetics, and polypeptides from marine sources: A wealth of natural sources for pharmaceutical applications. Mar. Drugs 2017, 15, 124. [Google Scholar] [CrossRef]
- Bond, A. Exenatide (Byetta) as a Novel Treatment Option for Type 2 Diabetes Mellitus. In Baylor University Medical Center Proceedings; Taylor & Francis: Oxfordshire, UK, 2006; Volume 19, pp. 218–284. [Google Scholar] [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 3, 16–18. [Google Scholar] [CrossRef]
- Agrawal, S.; Adholeya, A.; Deshmukh, S.K. The pharmacological potential of non-ribosomal peptides from marine sponge and tunicates. Front. Pharmacol. 2016, 7, 333. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Jahandideh, F.; Wu, J. Food-Derived Bioactive Peptides on Inflammation and Oxidative Stress. Biomed Res. Int. 2014, 2014, 608979. [Google Scholar] [CrossRef] [PubMed]
- Chatupheeraphat, C.; Roytrakul, S.; Phaonakrop, N.; Deesrisak, K.; Krobthong, S.; Anurathapan, U.; Tanyong, D. A Novel Peptide Derived from Ginger Induces Apoptosis through the Modulation of p53, BAX, and BCL2 Expression in Leukemic Cell Lines. Planta Med. 2021, 87, 560–569. [Google Scholar] [CrossRef] [PubMed]
- De Lumen, B.O. Lunasin: A cancer-preventive soy peptide. Nutr. Rev. 2005, 63, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Sompinit, K.; Lersiripong, S.; Reamtong, O.; Pattarayingsakul, W.; Patikarnmonthon, N.; Panbangred, W. In vitro study on novel bioactive peptides with antioxidant and antihypertensive properties from edible rhizomes. Food Sci. Technol. 2020, 134, 110227. [Google Scholar] [CrossRef]
- Kang, N.J.; Jin, H.-S.; Lee, S.-E.; Kim, H.J.; Koh, H.; Lee, D.-W. New approaches towards the discovery and evaluation of bioactive peptides from natural resources. Crit. Rev. Environ. Sci. Technol. 2020, 50, 72–103. [Google Scholar] [CrossRef]
- Xia, Z.; He, D.; Wu, Y.; Kwok, H.F.; Cao, Z. Scorpion venom peptides: Molecular diversity, structural characteristics, and therapeutic use from channelopathies to viral infections and cancers. Pharmacol. Res. 2023, 197, 106978. [Google Scholar] [CrossRef]
- Kawano, K.; Yoneya, T.; Miyata, T.; Yoshikawa, K.; Tokunaga, F.; Terada, Y.; Iwanaga, S. Antimicrobial peptide, tachyplesin I, isolated from hemocytes of the horseshoe crab (Tachypleus tridentatus). NMR determination of the beta-sheet structure. J. Biol. Chem. 1990, 265, 15365–15367. [Google Scholar] [CrossRef]
- Wang, X.; Song, Y.; Li, J.; Liu, H.; Xu, X.; Lai, R.; Zhang, K. A new family of antimicrobial peptides from skin secretions of Rana pleuraden. Peptides 2007, 28, 2069–2074. [Google Scholar] [CrossRef]
- Grassi, L.; Maisetta, G.; Maccari, G.; Esin, S.; Batoni, G. Analogs of the Frog-skin Antimicrobial Peptide Temporin 1Tb Exhibit a Wider Spectrum of Activity and a Stronger Antibiofilm Potential as Compared to the Parental Peptide. Front. Chem. 2017, 5, 24. [Google Scholar] [CrossRef]
- Malkoski, M.; Dashper, S.G.; O’Brien-Simpson, N.M.; Talbo, G.H.; Macris, M.; Cross, K.J.; Reynolds, E.C. Kappacin, a novel antibacterial peptide from bovine milk. Antimicrob. Agents Chemother. 2001, 45, 2309–2315. [Google Scholar] [CrossRef]
- Pellegrini, A.; Thomas, U.; Bramaz, N.; Klauser, S.; Hunziker, P.; von Fellenberg, R. Identification and isolation of a bactericidal domain in chicken egg white lysozyme. J. Appl. Microbiol. 1997, 82, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Zenezini Chiozzi, R.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Piovesana, S.; Samperi, R.; Laganà, A. Purification and identification of endogenous antioxidant and ACE-inhibitory peptides from donkey milk by multidimensional liquid chromatography and nanoHPLC-high resolution mass spectrometry. Anal. Bioanal. Chem. Res. 2016, 408, 5657–5666. [Google Scholar] [CrossRef] [PubMed]
- Mudgil, P.; Kamal, H.; Yuen, G.C.; Maqsood, S. Characterization and identification of novel antidiabetic and anti-obesity peptides from camel milk protein hydrolysates. Food Chem. 2018, 259, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Mudgil, P.; Baby, B.; Ngoh, Y.-Y.; Vijayan, R.; Gan, C.-Y.; Maqsood, S. Identification and molecular docking study of novel cholesterol esterase inhibitory peptides from camel milk proteins. Int. J. Dairy Sci. 2019, 102, 10748–10759. [Google Scholar] [CrossRef] [PubMed]
- Macedo, M.W.F.S.; Cunha, N.B.d.; Carneiro, J.A.; Costa, R.A.d.; Alencar, S.A.d.; Cardoso, M.H.; Franco, O.L.; Dias, S.C. Marine Organisms as a Rich Source of Biologically Active Peptides. Front. Mar. Sci. 2021, 8, 667764. [Google Scholar] [CrossRef]
- Chen, J.; Ryu, B.; Zhang, Y.; Liang, P.; Li, C.; Zhou, C.; Yang, P.; Hong, P.; Qian, Z.J. Comparison of an angiotensin-I-converting enzyme inhibitory peptide from tilapia (Oreochromis niloticus) with captopril: Inhibition kinetics, in vivo effect, simulated gastrointestinal digestion and a molecular docking study. J. Sci. Food Agric. 2020, 100, 315–324. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X. Separation, antitumor activities, and encapsulation of polypeptide from Chlorella pyrenoidosa. Biotechnol. Prog. 2013, 29, 681–687. [Google Scholar] [CrossRef]
- Sheih, I.C.; Fang, T.J.; Wu, T.-K.; Lin, P.-H. Anticancer and Antioxidant Activities of the Peptide Fraction from Algae Protein Waste. J. Agric. Food Chem. 2010, 58, 1202–1207. [Google Scholar] [CrossRef]
- Wu, Q.; Cai, Q.-F.; Yoshida, A.; Sun, L.-C.; Liu, Y.-X.; Liu, G.-M.; Su, W.-J.; Cao, M.-J. Purification and characterization of two novel angiotensin I-converting enzyme inhibitory peptides derived from R-phycoerythrin of red algae (Bangia fusco-purpurea). Eur. Food Res. Technol. 2017, 243, 779–789. [Google Scholar] [CrossRef]
- Je, J.-Y.; Qian, Z.-J.; Byun, H.-G.; Kim, S.-K. Purification and characterization of an antioxidant peptide obtained from tuna backbone protein by enzymatic hydrolysis. Process Biochem. 2007, 42, 840–846. [Google Scholar] [CrossRef]
- Wang, W.; De Mejia, E.G. A new frontier in soy bioactive peptides that may prevent age-related chronic diseases. Compr. Rev. Food Sci. Food Saf. 2005, 4, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Wen, H.; Zhai, L.; Yu, Y.; Li, Y.; Yu, W.; Cheng, A.; Wang, C.; Kou, X. Antioxidant activity and anti-proliferative effect of a bioactive peptide from chickpea (Cicer arietinum L.). Food Res. Int. 2015, 77, 75–81. [Google Scholar] [CrossRef]
- Adebiyi, A.P.; Adebiyi, A.O.; Ogawa, T.; Muramoto, K. Purification and characterisation of antioxidative peptides from unfractionated rice bran protein hydrolysates. Int. J. Food Sci. Technol. 2008, 43, 35–43. [Google Scholar] [CrossRef]
- Kudo, K.; Onodera, S.; Takeda, Y.; Benkeblia, N.; Shiomi, N. Antioxidative activities of some peptides isolated from hydrolyzed potato protein extract. J. Funct. Foods 2009, 1, 170–176. [Google Scholar] [CrossRef]
- Coda, R.; Rizzello, C.G.; Pinto, D.; Gobbetti, M. Selected lactic acid bacteria synthesize antioxidant peptides during sourdough fermentation of cereal flours. Appl. Environ. Microbiol. 2012, 78, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M. Origins, Technological Development, and Applications of Peptidomics. Methods Mol. Biol. 2018, 1719, 3–39. [Google Scholar] [CrossRef] [PubMed]
- Baggerman, G.; Verleyen, P.; Clynen, E.; Huybrechts, J.; De Loof, A.; Schoofs, L. Peptidomics. J. Chromatogr. B Biomed. Appl. 2004, 803, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhang, H.; Niu, H.; Wu, R.a. Peptidomic analyses: The progress in enrichment and identification of endogenous peptides. TrAC-Trends Anal. Chem. 2020, 125, 115835. [Google Scholar] [CrossRef]
- Foreman, R.E.; George, A.L.; Reimann, F.; Gribble, F.M.; Kay, R.G. Peptidomics: A Review of Clinical Applications and Methodologies. J. Proteome Res. 2021, 20, 3782–3797. [Google Scholar] [CrossRef]
- Andrews, S.J.; Rothnagel, J.A. Emerging evidence for functional peptides encoded by short open reading frames (vol 15, pg 193, 2014). Nat. Rev. Genet. 2014, 15, 286. [Google Scholar] [CrossRef]
- Payre, F.; Desplan, C. Small peptides control heart activity. Sci. Adv. 2016, 351, 226–227. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Jing, Y.; Xu, H. Mining for missed sORF-encoded peptides. Expert Rev. Proteom. 2019, 16, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Hoofnagle, A.N.; Wener, M.H. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J. Immunol. Methods 2009, 347, 3–11. [Google Scholar] [CrossRef]
- Lee, A.Y.H.; Chappell, D.L.; Bak, M.J.; Judo, M.; Liang, L.; Churakova, T.; Ayanoglu, G.; Castro-Perez, J.; Zhou, H.; Previs, S.; et al. Multiplexed quantification of proglucagon-derived peptides by immunoaffinity enrichment and tandem mass spectrometry after a meal tolerance test. Clin. Chem. 2016, 62, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Purcell, A.W.; Ramarathinam, S.H.; Ternette, N. Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 2019, 14, 1687–1707. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Simpson, R.J. A centrifugal ultrafiltration strategy for isolating the low-molecular weight (≤25 K) component of human plasma proteome. J. Proteom. 2010, 73, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Baker, H.; Hancock, W.S. Analysis of the low molecular weight serum peptidome using ultrafiltration and a hybrid ion trap-Fourier transform mass spectrometer. J. Chromatogr. A 2006, 1120, 173–184. [Google Scholar] [CrossRef]
- Finoulst, I.; Pinkse, M.; Van Dongen, W.; Verhaert, P. Sample Preparation Techniques for the Untargeted LC-MS-Based Discovery of Peptides in Complex Biological Matrices. J. Biomed. Biotechnol. 2011, 2011, 245291. [Google Scholar] [CrossRef]
- Aristoteli, L.P.; Molloy, M.P.; Baker, M.S. Evaluation of Endogenous Plasma Peptide Extraction Methods for Mass Spectrometric Biomarker Discovery. J. Proteome Res. 2007, 6, 571–581. [Google Scholar] [CrossRef]
- Sharma, M.; Gat, Y.; Arya, S.; Kumar, V.; Panghal, A.; Kumar, A. A review on microbial alkaline protease: An essential tool for various industrial approaches. Ind. Biotechnol. 2019, 15, 69–78. [Google Scholar] [CrossRef]
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- Nimalaratne, C.; Bandara, N.; Wu, J. Purification and characterization of antioxidant peptides from enzymatically hydrolyzed chicken egg white. Food Chem. 2015, 188, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, C.; Chen, Y.; Zheng, B. Purification and characterization of antioxidant peptides of Pseudosciaena crocea protein hydrolysates. Molecules 2017, 22, 57. [Google Scholar] [CrossRef]
- Salampessy, J.; Reddy, N.; Kailasapathy, K.; Phillips, M. Functional and potential therapeutic ACE-inhibitory peptides derived from bromelain hydrolysis of trevally proteins. J. Funct. Foods 2015, 14, 716–725. [Google Scholar] [CrossRef]
- Kadam, S.U.; Tiwari, B.K.; Álvarez, C.; O’Donnell, C.P. Ultrasound applications for the extraction, identification and delivery of food proteins and bioactive peptides. Trends Food Sci. Technol. 2015, 46, 60–67. [Google Scholar] [CrossRef]
- Hajfathalian, M.; Ghelichi, S.; García-Moreno, P.J.; Moltke Sørensen, A.-D.; Jacobsen, C. Peptides: Production, bioactivity, functionality, and applications. Crit. Rev. Food Sci. Nutr. 2018, 58, 3097–3129. [Google Scholar] [CrossRef] [PubMed]
- Kristinsson, H. Aquatic food protein hydrolysates. In Maximising the Value of Marine by-Products; Elsevier: Amsterdam, The Netherlands, 2007; pp. 229–248. [Google Scholar]
- Giri, A.; Nasu, M.; Ohshima, T. Bioactive properties of Japanese fermented fish paste, fish miso, using koji inoculated with Aspergillus oryzae. Int. J. Nutr. Food Sci 2012, 1, 13–22. [Google Scholar] [CrossRef]
- Korhonen, H.J. Technological and health aspects of bioactive components of milk. Int. Dairy J. 2006, 16, 1227–1228. [Google Scholar] [CrossRef]
- Sanjukta, S.; Rai, A.K. Production of bioactive peptides during soybean fermentation and their potential health benefits. Trends Food Sci. Technol. 2016, 50, 1–10. [Google Scholar] [CrossRef]
- Sieber, R.; Bütikofer, U.; Egger, C.; Portmann, R.; Walther, B.; Wechsler, D. ACE-inhibitory activity and ACE-inhibiting peptides in different cheese varieties. Dairy Sci. Technol. 2010, 90, 47–73. [Google Scholar] [CrossRef]
- El-Fattah, A.A.; Sakr, S.; El-Dieb, S.; Elkashef, H. Angiotensin-converting enzyme inhibition and antioxidant activity of commercial dairy starter cultures. Food Sci. Biotechnol. 2016, 25, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Sanjukta, S.; Rai, A.K.; Muhammed, A.; Jeyaram, K.; Talukdar, N.C. Enhancement of antioxidant properties of two soybean varieties of Sikkim Himalayan region by proteolytic Bacillus subtilis fermentation. J. Funct. Foods 2015, 14, 650–658. [Google Scholar] [CrossRef]
- Li, N.; Zhou, Y.; Wang, J.; Niu, L.; Zhang, Q.; Sun, L.; Ding, X.; Guo, X.; Xie, Z.; Zhu, N. Sequential precipitation and delipidation enables efficient enrichment of low-molecular weight proteins and peptides from human plasma. J. Proteome Res. 2020, 19, 3340–3351. [Google Scholar] [CrossRef] [PubMed]
- Zougman, A.; Pilch, B.; Podtelejnikov, A.; Kiehntopf, M.; Schnabel, C.; Kumar, C.; Mann, M. Integrated Analysis of the Cerebrospinal Fluid Peptidome and Proteome. J. Proteome Res. 2008, 7, 386–399. [Google Scholar] [CrossRef] [PubMed]
- Kay, R.; Barton, C.; Ratcliffe, L.; Matharoo-Ball, B.; Brown, P.; Roberts, J.; Teale, P.; Creaser, C. Enrichment of low molecular weight serum proteins using acetonitrile precipitation for mass spectrometry based proteomic analysis. Rapid Commun. Mass Spectrom. 2008, 22, 3255–3260. [Google Scholar] [CrossRef] [PubMed]
- Polson, C.; Sarkar, P.; Incledon, B.; Raguvaran, V.; Grant, R. Optimization of protein precipitation based upon effectiveness of protein removal and ionization effect in liquid chromatography–tandem mass spectrometry. J. Chromatogr. 2003, 785, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Van Midwoud, P.M.; Rieux, L.; Bischoff, R.; Verpoorte, E.; Niederländer, H.A.G. Improvement of Recovery and Repeatability in Liquid Chromatography−Mass Spectrometry Analysis of Peptides. J. Proteome Res. 2007, 6, 781–791. [Google Scholar] [CrossRef]
- Tian, R.; Ren, L.; Ma, H.; Li, X.; Hu, L.; Ye, M.; Wu, R.a.; Tian, Z.; Liu, Z.; Zou, H. Selective enrichment of endogenous peptides by chemically modified porous nanoparticles for peptidome analysis. J. Chromatogr. A 2009, 1216, 1270–1278. [Google Scholar] [CrossRef]
- Hennion, M.-C. Solid-phase extraction: Method development, sorbents, and coupling with liquid chromatography. J. Chromatogr. A 1999, 856, 3–54. [Google Scholar] [CrossRef]
- Wijte, D.; McDonnell, L.A.; Balog, C.I.A.; Bossers, K.; Deelder, A.M.; Swaab, D.F.; Verhaagen, J.; Mayboroda, O.A. A novel peptidomics approach to detect markers of Alzheimer’s disease in cerebrospinal fluid. Methods 2012, 56, 500–507. [Google Scholar] [CrossRef]
- Chen, Z.; Caulfield, M.P.; McPhaul, M.J.; Reitz, R.E.; Taylor, S.W.; Clarke, N.J. Quantitative insulin analysis using liquid chromatography-tandem mass spectrometry in a high-throughput clinical laboratory. Clin. Chem. 2013, 59, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Nadler, W.M.; Waidelich, D.; Kerner, A.; Hanke, S.; Berg, R.; Trumpp, A.; Rösli, C. MALDI versus ESI: The Impact of the Ion Source on Peptide Identification. J. Proteome Res. 2017, 16, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Pilo, A.L.; Bu, J.; McLuckey, S.A. Transformation of [M+2H]2+ Peptide Cations to [M − H]+, [M+H+O]+, and M+• Cations via Ion/Ion Reactions: Reagent Anions Derived from Persulfate. J. Am. Soc. Mass Spectrom. 2015, 26, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Nwosu, C.C.; Strum, J.S.; An, H.J.; Lebrilla, C.B. Enhanced detection and identification of glycopeptides in negative ion mode mass spectrometry. Anal. Chem. 2010, 82, 9654–9662. [Google Scholar] [CrossRef] [PubMed]
- Penanes, P.A.; Gorshkov, V.; Ivanov, M.V.; Gorshkov, M.V.; Kjeldsen, F. Potential of Negative-Ion-Mode Proteomics: An MS1-Only Approach. J. Proteome Res. 2023, 22, 2734–2742. [Google Scholar] [CrossRef] [PubMed]
- Walch, A.; Rauser, S.; Deininger, S.-O.; Höfler, H. MALDI imaging mass spectrometry for direct tissue analysis: A new frontier for molecular histology. Histochem. Cell Biol. 2008, 130, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhao, R.; Berger, S.J.; Anderson, G.A.; Rodriguez, N.; Smith, R.D. High-Efficiency Nanoscale Liquid Chromatography Coupled On-Line with Mass Spectrometry Using Nanoelectrospray Ionization for Proteomics. Anal. Chem. 2002, 74, 4235–4249. [Google Scholar] [CrossRef]
- Kay, R.G.; Gregory, B.; Grace, P.B.; Pleasance, S. The application of ultra-performance liquid chromatography/tandem mass spectrometry to the detection and quantitation of apolipoproteins in human serum. Rapid Commun. Mass Spectrom. 2007, 21, 2585–2593. [Google Scholar] [CrossRef]
- Latosinska, A.; Siwy, J.; Mischak, H.; Frantzi, M. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: The past, the present, and the future. Electrophoresis 2019, 40, 2294–2308. [Google Scholar] [CrossRef]
- Minerva, L.; Boonen, K.; Menschaert, G.; Landuyt, B.; Baggerman, G.; Arckens, L. Linking Mass Spectrometric Imaging and Traditional Peptidomics: A Validation in the Obese Mouse Model. Anal. Chem. 2011, 83, 7682–7691. [Google Scholar] [CrossRef]
- Klein, J.; Papadopoulos, T.; Mischak, H.; Mullen, W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis 2014, 35, 1060–1064. [Google Scholar] [CrossRef] [PubMed]
- Stolz, A.; Jooß, K.; Höcker, O.; Römer, J.; Schlecht, J.; Neusüß, C. Recent advances in capillary electrophoresis-mass spectrometry: Instrumentation, methodology and applications. Electrophoresis 2019, 40, 79–112. [Google Scholar] [CrossRef] [PubMed]
- Piovesana, S.; Capriotti, A.L.; Cerrato, A.; Crescenzi, C.; La Barbera, G.; Laganá, A.; Montone, C.M.; Cavaliere, C. Graphitized Carbon Black Enrichment and UHPLC-MS/MS Allow to Meet the Challenge of Small Chain Peptidomics in Urine. Anal. Chem. 2019, 91, 11474–11481. [Google Scholar] [CrossRef] [PubMed]
- Piovesana, S.; Cerrato, A.; Antonelli, M.; Benedetti, B.; Capriotti, A.L.; Cavaliere, C.; Montone, C.M.; Laganà, A. A clean-up strategy for identification of circulating endogenous short peptides in human plasma by zwitterionic hydrophilic liquid chromatography and untargeted peptidomics identification. J. Chromatogr. A 2020, 1613, 460699. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zheng, J.; Zheng, F.; Cai, Z.; Yu, Q. Advancing serum peptidomic profiling by data-independent acquisition for clear-cell renal cell carcinoma detection and biomarker discovery. J. Proteom. 2020, 215, 103671. [Google Scholar] [CrossRef]
- Dallas, D.C.; Guerrero, A.; Parker, E.A.; Robinson, R.C.; Gan, J.; German, J.B.; Barile, D.; Lebrilla, C.B. Current peptidomics: Applications, purification, identification, quantification, and functional analysis. Proteomics 2015, 15, 1026–1038. [Google Scholar] [CrossRef]
- Nardo, A.E.; Añón, M.C.; Parisi, G. Large-scale mapping of bioactive peptides in structural and sequence space. PLoS ONE 2018, 13, e0191063. [Google Scholar] [CrossRef]
- Ogrinc Potočnik, N.; Fisher, G.L.; Prop, A.; Heeren, R.M.A. Sequencing and Identification of Endogenous Neuropeptides with Matrix-Enhanced Secondary Ion Mass Spectrometry Tandem Mass Spectrometry. Anal. Chem. 2017, 89, 8223–8227. [Google Scholar] [CrossRef]
- Knickelbine, J.J.; Konop, C.J.; Viola, I.R.; Rogers, C.B.; Messinger, L.A.; Vestling, M.M.; Stretton, A.O.W. Different Bioactive Neuropeptides are Expressed in Two Sub-Classes of GABAergic RME Nerve Ring Motorneurons in Ascaris suum. ACS Chem. Neurosci. 2018, 9, 2025–2040. [Google Scholar] [CrossRef]
- Abreu, T.F.; Sumitomo, B.N.; Nishiyama, M.Y.; Oliveira, U.C.; Souza, G.H.M.F.; Kitano, E.S.; Zelanis, A.; Serrano, S.M.T.; Junqueira-de-Azevedo, I.; Silva, P.I.; et al. Peptidomics of Acanthoscurria gomesiana spider venom reveals new toxins with potential antimicrobial activity. J. Proteom. 2017, 151, 232–242. [Google Scholar] [CrossRef]
- Blay, V.; Tolani, B.; Ho, S.P.; Arkin, M.R. High-Throughput Screening: Today’s biochemical and cell-based approaches. Drug Discov. Today 2020, 25, 1807–1821. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, Y.; Shen, H.; You, J. Antioxidant activities and functional properties of grass carp (Ctenopharyngodon idellus) protein hydrolysates. J. Sci. Food Agric. 2012, 92, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Hirai, S.; Yamanaka, M.; Kawachi, H.; Matsui, T.; Yano, H. Activin A inhibits differentiation of 3T3-L1 preadipocyte. Mol. Cell. Endocrinol. 2005, 232, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Cronk, D.; Shearer, J. High-throughput screening. In Drug Discovery and Development, 2nd ed.; Churchill Livingstone: London, UK, 2013; pp. 95–117. [Google Scholar]
- Harrison, C.; Traynor, J.R. The [35S]GTPγS binding assay: Approaches and applications in pharmacology. Life Sci. 2003, 74, 489–508. [Google Scholar] [CrossRef] [PubMed]
- Strange, P.G. Use of the GTPγS ([35S]GTPγS and Eu-GTPγS) binding assay for analysis of ligand potency and efficacy at G protein-coupled receptors. Br. J. Pharmacol. 2010, 161, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Cooper, T.; McMurchie, E.J.; Leifert, W.R. [35S]GTPγS Binding in G Protein-Coupled Receptor Assays. In G Protein-Coupled Receptors in Drug Discovery; Leifert, W.R., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 143–151. [Google Scholar]
- Inglese, J.; Johnson, R.L.; Simeonov, A.; Xia, M.; Zheng, W.; Austin, C.P.; Auld, D.S. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007, 3, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Quinn, C.M.; Gagnon, A.I.; Talanian, R. Homogeneous time-resolved fluorescence and its applications for kinase assays in drug discovery. Anal. Biochem. 2006, 356, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Karvinen, J.; Hurskainen, P.; Gopalakrishnan, S.; Burns, D.; Warrior, U.; Hemmilä, I. Homogeneous Time-Resolved Fluorescence Quenching Assay (LANCE) for Caspase-3. SLAS Discov. 2002, 7, 223–231. [Google Scholar] [CrossRef]
- Achard, S.; Jean, A.; Lorphelin, D.; Amoravain, M.; Claret, E.J. Homogeneous assays allow direct “in well” cytokine level quantification. Assay Drug Dev. Technol. 2003, 1, 181–185. [Google Scholar] [CrossRef]
- He, R.; Aluko, R.E.; Ju, X.-R. Evaluating molecular mechanism of hypotensive peptides interactions with renin and angiotensin converting enzyme. PLoS ONE 2014, 9, e91051. [Google Scholar] [CrossRef]
- Velarde-Salcedo, A.J.; Barrera-Pacheco, A.; Lara-González, S.; Montero-Morán, G.M.; Díaz-Gois, A.; González de Mejia, E.; Barba de la Rosa, A.P. In vitro inhibition of dipeptidyl peptidase IV by peptides derived from the hydrolysis of amaranth (Amaranthus hypochondriacus L.) proteins. Food Chem. 2013, 136, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Azkargorta, M.; Soria, J.; Ojeda, C.; Guzmán, F.; Acera, A.; Iloro, I.; Suárez, T.; Elortza, F. Human Basal Tear Peptidome Characterization by CID, HCD, and ETD Followed by in Silico and in Vitro Analyses for Antimicrobial Peptide Identification. J. Proteome Res. 2015, 14, 2649–2658. [Google Scholar] [CrossRef] [PubMed]
- Pande, J.; Szewczyk, M.M.; Grover, A.K. Phage display: Concept, innovations, applications and future. Biotechnol. Adv. 2010, 28, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Puentes, P.R.; Henao, M.C.; Torres, C.E.; Gómez, S.C.; Gómez, L.A.; Burgos, J.C.; Arbeláez, P.; Osma, J.F.; Muñoz-Camargo, C.; Reyes, L.H.; et al. Design, screening, and testing of non-rational peptide libraries with antimicrobial activity: In silico and experimental approaches. Antibiotics 2020, 9, 854. [Google Scholar] [CrossRef] [PubMed]
- Zani, M.-L.; Moreau, T. Phage display as a powerful tool to engineer protease inhibitors. Biochimie 2010, 92, 1689–1704. [Google Scholar] [CrossRef] [PubMed]
- Bahudhanapati, H.; Zhang, Y.; Sidhu, S.S.; Brew, K. Phage display of tissue inhibitor of metalloproteinases-2 (TIMP-2): Identification of selective inhibitors of collagenase-1 (metalloproteinase 1 (MMP-1)). J. Biol. Chem. 2011, 286, 31761–31770. [Google Scholar] [CrossRef]
- Lin, S.-H.; Chang, D.-K.; Chou, M.-J.; Huang, K.-J.; Shiuan, D. Peptide inhibitors of human HMG-CoA reductase as potential hypocholesterolemia agents. Biochem. Biophys. Res. Commun. 2015, 456, 104–109. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Hsiao, N.-W.; Tseng, T.-S.; Chen, W.-C.; Lin, H.-H.; Leu, S.-J.; Yang, E.-W.; Tsai, K.-C. Phage display—Mediated discovery of novel tyrosinase-targeting tetrapeptide inhibitors reveals the significance of N-terminal preference of cysteine residues and their functional sulfur atom. Mol. Pharmacol. 2015, 87, 218–230. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Z.; Rothschild, K.J.; Lim, M.J. Proteome-wide drug screening using mass spectrometric imaging of bead-arrays. Sci. Rep. 2016, 6, 26125. [Google Scholar] [CrossRef]
- Lim, M.J.; Liu, Z.; Braunschweiger, K.I.; Awad, A.; Rothschild, K.J. Correlated matrix-assisted laser desorption/ionization mass spectrometry and fluorescent imaging of photocleavable peptide-coded random bead-arrays. Rapid Commun. Mass Spectrom. 2014, 28, 49–62. [Google Scholar] [CrossRef]
- Aslantürk, Ö.S. In Vitro Cytotoxicity and Cell Viability Assays: Principles, Advantages, and Disadvantages; Intech: Sydney, Australia, 2018. [Google Scholar] [CrossRef]
- Invitrogen, Molecular Probes, Inc. Propidium Iodide Nucleic Acid Stain. 2006. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/mp01304.pdf (accessed on 30 November 2023).
- Meijer, K.; Vonk, R.J.; Priebe, M.G.; Roelofsen, H. Cell-based screening assay for anti-inflammatory activity of bioactive compounds. Food Chem. 2015, 166, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Michelini, E.; Cevenini, L.; Mezzanotte, L.; Coppa, A.; Roda, A. Cell-based assays: Fuelling drug discovery. Anal. Bioanal. Chem. Res. 2010, 398, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A comprehensive review on current advances in peptide drug development and design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [PubMed]
- Romei, M.G.; Boxer, S.G. Split Green Fluorescent Proteins: Scope, Limitations, and Outlook. Annu. Rev. Biophys. 2019, 48, 19–44. [Google Scholar] [CrossRef] [PubMed]
- Tebo, A.G.; Gautier, A. A split fluorescent reporter with rapid and reversible complementation. Nat. Commun. 2019, 10, 2822–2828. [Google Scholar] [CrossRef] [PubMed]
- Lievens, S.; Caligiuri, M.; Kley, N.; Tavernier, J. The use of mammalian two-hybrid technologies for high-throughput drug screening. Methods 2012, 58, 335–342. [Google Scholar] [CrossRef]
- Boute, N.; Jockers, R.; Issad, T. The use of resonance energy transfer in high-throughput screening: BRET versus FRET. Trends Pharmacol Sci. 2002, 23, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Frutos, A.G.; Verklereen, R. Label-free cell-based assays for GPCR screening. Comb. Chem. High Throughput Screen. 2008, 11, 357–369. [Google Scholar] [CrossRef]
- McGuinness, R. Impedance-based cellular assay technologies: Recent advances, future promise. Curr. Opin. Pharmacol. 2007, 7, 535–540. [Google Scholar] [CrossRef]
- Chernov-Rogan, T.; Li, T.; Lu, G.; Verschoof, H.; Khakh, K.; Jones, S.W.; Beresini, M.H.; Liu, C.; Ortwine, D.F.; McKerrall, S.J.; et al. Mechanism-specific assay design facilitates the discovery of Nav1.7-selective inhibitors. Proc. Natl. Acad. Sci. USA 2018, 115, E792–E801. [Google Scholar] [CrossRef]
- Xu, H.; Li, T.; Rohou, A.; Arthur, C.P.; Tzakoniati, F.; Wong, E.; Estevez, A.; Kugel, C.; Franke, Y.; Chen, J.; et al. Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin. Cell 2019, 176, 1238–1239. [Google Scholar] [CrossRef] [PubMed]
- Perpetuo, L.; Klein, J.; Ferreira, R.; Guedes, S.; Amado, F.; Leite-Moreira, A.; Silva, A.M.S.; Thongboonkerd, V.; Vitorino, R. How can artificial intelligence be used for peptidomics? Expert Rev. Proteom. 2021, 18, 527–556. [Google Scholar] [CrossRef] [PubMed]
- Terziyska, M.; Desseva, I.; Terziyski, Z. Food-Derived Bioactive Peptides And Artificial Intelligence Techniques For Their Prediction: A Brief Review. Int. J. Sci. Technol. Res. 2021, 10, 141–147. [Google Scholar]
- Li, G.; Iyer, B.; Prasath, V.S.; Ni, Y.; Salomonis, N. DeepImmuno: Deep learning-empowered prediction and generation of immunogenic peptides for T-cell immunity. Brief. Bioinform. 2021, 22, bbab160. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Jing, R.; Liu, F.; Luo, J.; Li, Y. DeepACP: A novel computational approach for accurate identification of anticancer peptides by deep learning algorithm. Mol. Ther. Nucleic Acids 2020, 22, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cheong, H.H.; Siu, S.W. xDeep-AcPEP: Deep learning method for anticancer peptide activity prediction based on convolutional neural network and multitask learning. J. Chem. Inf. Model. 2021, 61, 3789–3803. [Google Scholar] [CrossRef] [PubMed]
- Repecka, D.; Jauniskis, V.; Karpus, L.; Rembeza, E.; Rokaitis, I.; Zrimec, J.; Poviloniene, S.; Laurynenas, A.; Viknander, S.; Abuajwa, W. Expanding functional protein sequence spaces using generative adversarial networks. Nat. Mach. Intell. 2021, 3, 324–333. [Google Scholar] [CrossRef]
- Szymczak, P.; Mozejko, M.; Grzegorzek, T.; Jurczak, R.; Bauer, M.; Neubauer, D.; Sikora, K.; Michalski, M.; Sroka, J.; Setny, P.; et al. Discovering highly potent antimicrobial peptides with deep generative model HydrAMP. Nat. Commun. 2023, 14, 1453. [Google Scholar] [CrossRef]
- Dean, S.N.; Walper, S.A. Variational autoencoder for generation of antimicrobial peptides. ACS Omega 2020, 5, 20746–20754. [Google Scholar] [CrossRef]
- Tucs, A.; Tran, D.P.; Yumoto, A.; Ito, Y.; Uzawa, T.; Tsuda, K. Generating ampicillin-level antimicrobial peptides with activity-aware generative adversarial networks. ACS Omega 2020, 5, 22847–22851. [Google Scholar] [CrossRef]
- Dean, S.N.; Alvarez, J.A.E.; Zabetakis, D.; Walper, S.A.; Malanoski, A.P. PepVAE: Variational autoencoder framework for antimicrobial peptide generation and activity prediction. Front. Microbiol. 2021, 12, 725727. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Saravanan, K.M.; Wei, Y.; Jiao, Y.; Yang, Y.; Pan, Y.; Wu, X.; Zhang, J.Z.H. Deep Learning-Based Bioactive Therapeutic Peptide Generation and Screening. J. Chem. Inf. Model. 2023, 63, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Khosravian, M.; Kazemi Faramarzi, F.; Mohammad Beigi, M.; Behbahani, M.; Mohabatkar, H. Predicting antibacterial peptides by the concept of Chou’s pseudo-amino acid composition and machine learning methods. Protein Pept. Lett. 2013, 20, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, X.; Zou, Q.; Dong, Q.; Chen, Q. Protein remote homology detection by combining Chou’s pseudo amino acid composition and profile-based protein representation. Mol. Inform. 2013, 32, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Mohabatkar, H.; Mohammad Beigi, M.; Abdolahi, K.; Mohsenzadeh, S. Prediction of allergenic proteins by means of the concept of Chou’s pseudo amino acid composition and a machine learning approach. Med. Chem. 2013, 9, 133–137. [Google Scholar] [CrossRef] [PubMed]
- London, N.; Raveh, B.; Cohen, E.; Fathi, G.; Schueler-Furman, O. Rosetta FlexPepDock web server—High resolution modeling of peptide–protein interactions. Nucleic Acids Res. 2011, 39, W249–W253. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Hourai, Y.; Weng, Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE 2011, 6, e24657. [Google Scholar] [CrossRef]
- Ding, C.; Hao, M.; Ma, S.; Zhang, Y.; Yang, J.; Ding, Q.; Sun, S.; Zhang, J.; Zhang, Y.; Liu, W. Identification of peptides with antioxidant, anti-lipoxygenase, anti-xanthine oxidase and anti-tyrosinase activities from velvet antler blood. LWT 2022, 168, 113889. [Google Scholar] [CrossRef]
- Möller, N.P.; Scholz-Ahrens, K.E.; Roos, N.; Schrezenmeir, J. Bioactive peptides and proteins from foods: Indication for health effects. Eur. J. Nutr. 2008, 47, 171–182. [Google Scholar] [CrossRef]
- Singh, B.P.; Vij, S.; Hati, S. Functional significance of bioactive peptides derived from soybean. Peptides 2014, 54, 171–179. [Google Scholar] [CrossRef]
- Benkerroum, N. Antimicrobial peptides generated from milk proteins: A survey and prospects for application in the food industry. A review. Int. J. Dairy Technol. 2010, 63, 320–338. [Google Scholar] [CrossRef]
- Kamran, F.; Phillips, M.; Reddy, N. Functional properties of Australian blue lupin (Lupinus angustifolius) protein and biological activities of protein hydrolysates. Legum. Sci. 2021, 3, e65. [Google Scholar] [CrossRef]
- Sila, A.; Hedhili, K.; Przybylski, R.; Ellouz-Chaabouni, S.; Dhulster, P.; Bougatef, A.; Nedjar-Arroume, N. Antibacterial activity of new peptides from barbel protein hydrolysates and mode of action via a membrane damage mechanism against Listeria monocytogenes. J. Funct. Foods 2014, 11, 322–329. [Google Scholar] [CrossRef]
- Théolier, J.; Hammami, R.; Labelle, P.; Fliss, I.; Jean, J. Isolation and identification of antimicrobial peptides derived by peptic cleavage of whey protein isolate. J. Funct. Foods 2013, 5, 706–714. [Google Scholar] [CrossRef]
- Balti, R.; Bougatef, A.; Sila, A.; Guillochon, D.; Dhulster, P.; Nedjar-Arroume, N. Nine novel angiotensin I-converting enzyme (ACE) inhibitory peptides from cuttlefish (Sepia officinalis) muscle protein hydrolysates and antihypertensive effect of the potent active peptide in spontaneously hypertensive rats. Food Chem. 2015, 170, 519–525. [Google Scholar] [CrossRef]
- Majumder, K.; Chakrabarti, S.; Morton, J.S.; Panahi, S.; Kaufman, S.; Davidge, S.T.; Wu, J. Egg-derived ACE-inhibitory peptides IQW and LKP reduce blood pressure in spontaneously hypertensive rats. J. Funct. Foods 2015, 13, 50–60. [Google Scholar] [CrossRef]
- Medina-Godoy, S.; Rodríguez-Yáñez, S.K.; Bobadilla, N.A.; Pérez-Villalva, R.; Valdez-Ortiz, R.; Hong, E.; Luna-Suárez, S.; Paredes-López, O.; Valdez-Ortiz, A. Antihypertensive activity of AMC3, an engineered 11S amaranth globulin expressed in Escherichia coli, in spontaneously hypertensive rats. J. Funct. Foods 2013, 5, 1441–1449. [Google Scholar] [CrossRef]
- Li, Y.-W.; Li, B.; He, J.; Qian, P. Quantitative structure–activity relationship study of antioxidative peptide by using different sets of amino acids descriptors. J. Mol. Struct. 2011, 998, 53–61. [Google Scholar] [CrossRef]
- Bougatef, A.; Nedjar-Arroume, N.; Manni, L.; Ravallec, R.; Barkia, A.; Guillochon, D.; Nasri, M. Purification and identification of novel antioxidant peptides from enzymatic hydrolysates of sardinelle (Sardinella aurita) by-products proteins. Food Chem. 2010, 118, 559–565. [Google Scholar] [CrossRef]
- Li, Y.-W.; Li, B. Characterization of structure–antioxidant activity relationship of peptides in free radical systems using QSAR models: Key sequence positions and their amino acid properties. J. Theor. Biol. 2013, 318, 29–43. [Google Scholar] [CrossRef]
- Bonomi, F.; Brandt, R.; Favalli, S.; Ferranti, P.; Fierro, O.; Frøkiær, H.; Ragg, E.; Iametti, S. Structural determinants of the immunomodulatory properties of the C-terminal region of bovine β-casein. Int. Dairy J. 2011, 21, 770–776. [Google Scholar] [CrossRef]
- Haney, E.F.; Hancock, R.E.W. Peptide design for antimicrobial and immunomodulatory applications. Biopolymers 2013, 100, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, M.; Jenssen, H.; Kindrachuk, J.; Scott, W.R.P.; Elliott, M.; Hilpert, K.; Cheng, J.T.J.; Hancock, R.E.W.; Straus, S.K. Structural Studies of a Peptide with Immune Modulating and Direct Antimicrobial Activity. Chem. Biol. 2010, 17, 970–980. [Google Scholar] [CrossRef] [PubMed]
- Aimutis, W.R. Bioactive Properties of Milk Proteins with Particular Focus on Anticariogenesis. J. Nutr. 2004, 134, 989S–995S. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.E.; Kim, H.H.; Kim, J.Y.; Kang, Y.I.; Woo, H.J.; Lee, H.J. Anticancer activity of hydrophobic peptides from soy proteins. BioFactors 2000, 12, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.M. Bioorganic chemistry: Peptides and proteins. J. Chem. Educ. 1998, 76, 905. [Google Scholar]
- Barbé, F.; Le Feunteun, S.; Rémond, D.; Ménard, O.; Jardin, J.; Henry, G.; Laroche, B.; Dupont, D. Tracking the in vivo release of bioactive peptides in the gut during digestion: Mass spectrometry peptidomic characterization of effluents collected in the gut of dairy matrix fed mini-pigs. Food Res. Int. 2014, 63, 147–156. [Google Scholar] [CrossRef]
- Chary, K.V.R. NMR in Biological Systems: From Molecules to Human; Springer: Berlin, Germany, 2008; Volume 6. [Google Scholar]
- Mar Contreras, M.d.; Lopez-Exposito, I.; Hernandez-Ledesma, B.; Ramos, M.; Recio, I. Application of mass spectrometry to the characterization and quantification of food-derived bioactive peptides. J. AOAC Int. 2008, 91, 981–993. [Google Scholar] [CrossRef]
- Kamran, F.; Salampessy, J.; Reddy, N. Application of NMR spectroscopy for structural characterization of bioactive peptides derived from food protein. In Application of NMR Spectroscopy Food Science; Bentham Science Publishers: Sharjah, United Arab Emirates, 2016; Volume 5, pp. 3–76. [Google Scholar]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- López-Expósito, I.; Gómez-Ruiz, J.Á.; Amigo, L.; Recio, I. Identification of antibacterial peptides from ovine α s2-casein. Int. Dairy J. 2006, 16, 1072–1080. [Google Scholar] [CrossRef]
- Losito, I.; Carbonara, T.; De Bari, M.D.; Gobbetti, M.; Palmisano, F.; Rizzello, C.G.; Zambonin, P.G. Identification of peptides in antimicrobial fractions of cheese extracts by electrospray ionization ion trap mass spectrometry coupled to a two-dimensional liquid chromatographic separation. Rapid Commun. Mass Spectrom. 2006, 20, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, A.; Dettling, C.; Thomas, U.; Hunziker, P. Isolation and characterization of four bactericidal domains in the bovine β-lactoglobulin. Biochim Biophys Acta Gen Subj. 2001, 1526, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Pihlanto, A. Bioactive peptides. In Encyclopedia of Dairy Sciences; Fuquay, J.W., Fox, P.F., McSweeney, P.L.H., Eds.; Academic Press Inc.: Cambridge, MA, USA, 2006; Volume 3. [Google Scholar]
- Zucht, H.-D.; Raida, M.; Adermann, K.; Mägert, H.-J.; Forssmann, W.-G. Casocidin-I: A casein-α s2 derived peptide exhibits antibacterial activity. FEBS Lett. 1995, 372, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A.; Ackermann, M.; McCray, P.B.; Tack, B.F. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 2003, 22, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Vizioli, J.; Salzet, M. Antimicrobial peptides from animals: Focus on invertebrates. Trends Pharmacol. 2002, 23, 494–496. [Google Scholar] [CrossRef]
- Powers, J.-P.S.; Hancock, R.E.W. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef]
- Mine, Y.; Shahidi, F. Nutraceutical Proteins and Peptides in Health and Disease; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- McCann, K.B.; Shiell, B.J.; Michalski, W.P.; Lee, A.; Wan, J.; Roginski, H.; Coventry, M.J. Isolation and characterisation of a novel antibacterial peptide from bovine α S1-casein. Int. Dairy J. 2006, 16, 316–323. [Google Scholar] [CrossRef]
- Hancock, R.E.W. Cationic peptides: Effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 2001, 1, 156–164. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Meisel, H.; Walsh, D.; Murray, B.; FitzGerald, R. ACE inhibitory peptides. In Nutraceutical Proteins and Peptides in Health and Disease; Mine, Y., Shahidi, F., Eds.; CRC Press; Taylor and Francis Group: New York, NY, USA, 2006; pp. 269–315. [Google Scholar]
- Bhuyan, B.J.; Mugesh, G. Antioxidant activity of peptide-based angiotensin converting enzyme inhibitors. Org. Biomol. Chem. 2012, 10, 2237–2247. [Google Scholar] [CrossRef]
- Sturrock, E.D.; Acharya, K.R.; Natesh, R.; Schwager, S.L.U. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature 2003, 421, 551–554. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural Requirements of Angiotensin I-Converting Enzyme Inhibitory Peptides: Quantitative Structure-Activity Relationship Modeling of Peptides Containing 4-10 Amino Acid Residues. QSAR Comb. Sci. 2006, 25, 873–880. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural Requirements of Angiotensin I-Converting Enzyme Inhibitory Peptides: Quantitative Structure−Activity Relationship Study of Di- and Tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Yamamoto, N.; Sakai, K.; Okubo, A.; Yamazaki, S.; Takano, T. Purification and Characterization of Angiotensin I-Converting Enzyme Inhibitors from Sour Milk. J. Dairy Sci. 1995, 78, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Majumder, K.; Wu, J. A new approach for identification of novel antihypertensive peptides from egg proteins by QSAR and bioinformatics. Food Res. Int. 2010, 43, 1371–1378. [Google Scholar] [CrossRef]
- Chen, J.; Liu, S.; Ye, R.; Cai, G.; Ji, B.; Wu, Y. Angiotensin-I converting enzyme (ACE) inhibitory tripeptides from rice protein hydrolysate: Purification and characterization. J. Funct. Foods 2013, 5, 1684–1692. [Google Scholar] [CrossRef]
- Saito, Y.; Wanezaki, K.; Kawato, A.; Imayasu, S. Structure and Activity of Angiotensin I Converting Enzyme Inhibitory Peptides from Sake and Sake Lees. Biosci. Biotechnol. Biochem. 1994, 58, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Suzuki, K.; Funatsu, G. Isolation from α-Zein of Thermolysin Peptides with Angiotensin I-Converting Enzyme Inhibitory Activity. Biosci. Biotechnol. Biochem. 1996, 60, 661–663. [Google Scholar] [CrossRef]
- Matsufuji, H.; Matsui, T.; Seki, E.; Osajima, K.; Nakashima, M.; Osajima, Y. Angiotensin I-converting enzyme inhibitory peptides in an alkaline protease hydrolyzate derived from sardine muscle. Biosci. Biotechnol. Biochem. 1994, 58, 2244–2245. [Google Scholar] [CrossRef]
- Cheung, H.S.; Wang, F.L.; Ondetti, M.A.; Sabo, E.F.; Cushman, D.W. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH-terminal dipeptide sequence. J. Biol. Chem. 1980, 255, 401–407. [Google Scholar] [CrossRef]
- Tomatsu, M.; Shimakage, A.; Shinbo, M.; Yamada, S.; Takahashi, S. Novel angiotensin I-converting enzyme inhibitory peptides derived from soya milk. Food Chem. 2013, 136, 612–616. [Google Scholar] [CrossRef]
- Salampessy, J.; Reddy, N.; Phillips, M.; Kailasapathy, K. Isolation and characterization of nutraceutically potential ACE-Inhibitory peptides from leatherjacket (Meuchenia sp.) protein hydrolysates. Food Sci. Technol. 2017, 80, 430–436. [Google Scholar] [CrossRef]
- Kodama, R.T.; Cajado-Carvalho, D.; Kuniyoshi, A.K.; Kitano, E.S.; Tashima, A.K.; Barna, B.F.; Takakura, A.C.; Serrano, S.M.T.; Dias-Da-Silva, W.; Tambourgi, D.V.; et al. New proline-rich oligopeptides from the venom of African adders: Insights into the hypotensive effect of the venoms. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1180–1187. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.H.; Bartelt, D.C.; Greene, L.J. Isolation of bradykinin-potentiating peptides from Bothrops jararaca venom. Biochemistry 1970, 9, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.A.F.; Möllring, T.; Lebrun, F.L.A.S.; Raida, M.; Znottka, R.; Habermehl, G.G. Structure and effects of A kinin potentiating fraction F (AppF) isolated from Agkistrodon piscivorus piscivorus venom. Toxicon 1995, 33, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Faria Ferreira, L.A.; Auer, H.; Haslinger, E.; Fedele, C.; Habermehl, G.G. Spatial structures of the bradykinin potentiating peptide F from Agkistrodon piscivorus piscivoris venom. Toxicon 1999, 37, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Politi, V.; De Luca, G.; Di Stazio, G.; Schinina, E.; Bossa, F. A new peptide from Crotalus atrox snake venom. Peptides 1985, 6, 343–346. [Google Scholar] [CrossRef]
- Puchalska, P.; Marina, M.L.; García, M.C. Isolation and identification of antioxidant peptides from commercial soybean-based infant formulas. Food Chem. 2014, 148, 147–154. [Google Scholar] [CrossRef]
- Puchalska, P.; Concepción García, M.; Luisa Marina, M. Identification of native angiotensin-I converting enzyme inhibitory peptides in commercial soybean based infant formulas using HPLC-Q-ToF-MS. Food Chem. 2014, 157, 62–69. [Google Scholar] [CrossRef]
- Chen, H.-M.; Muramoto, K.; Yamauchi, F. Structural Analysis of Antioxidative Peptides from Soybean.beta.-Conglycinin. J. Agric. Food Chem. 1995, 43, 574–578. [Google Scholar] [CrossRef]
- Je, J.-Y.; Park, P.-J.; Kim, S.-K. Antioxidant activity of a peptide isolated from Alaska pollack (Theragra chalcogramma) frame protein hydrolysate. Food Res. Int. 2005, 38, 45–50. [Google Scholar] [CrossRef]
- Pihlanto, A. Antioxidative peptides derived from milk proteins. Int. Dairy J. 2006, 16, 1306–1314. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Je, J.-Y.; Kim, S.-K. Purification and characterization of antioxidant peptide from hoki (Johnius belengerii) frame protein by gastrointestinal digestion. J. Nutr. Biochem. 2007, 18, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhao, M.; Shi, J.; Wang, J.; Jiang, Y.; Cui, C.; Kakuda, Y.; Xue, S.J. Purification and identification of antioxidant peptides from grass carp muscle hydrolysates by consecutive chromatography and electrospray ionization-mass spectrometry. Food Chem. 2008, 108, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Rival, S.G.; Boeriu, C.G.; Wichers, H.J. Caseins and Casein Hydrolysates. 2. Antioxidative Properties and Relevance to Lipoxygenase Inhibition. J. Agric. Food Chem. 2001, 49, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ledesma, B.; Dávalos, A.; Bartolomé, B.; Amigo, L. Preparation of Antioxidant Enzymatic Hydrolysates from α-Lactalbumin and β-Lactoglobulin. Identification of Active Peptides by HPLC-MS/MS. J. Agric. Food Chem. 2005, 53, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Kamran, F.; Phillips, M.; Harman, D.G.; Reddy, N. Antioxidant activities of lupin (Lupinus angustifolius) protein hydrolysates and their potential for nutraceutical and functional foods. Food Chem. Adv. 2023, 2, 100297. [Google Scholar] [CrossRef]
- Guo, C.; Hu, Y.; Li, J.; Liu, Y.; Li, S.; Yan, K.; Wang, X.; Liu, J.; Wang, H. Identification of multiple peptides with antioxidant and antimicrobial activities from skin and its secretions of Hylarana taipehensis, Amolops lifanensis, and Amolops granulosus. Biochimie 2014, 105, 192–201. [Google Scholar] [CrossRef]
- Lu, Z.; Zhai, L.; Wang, H.; Che, Q.; Wang, D.; Feng, F.; Zhao, Z.; Yu, H. Novel families of antimicrobial peptides with multiple functions from skin of Xizang plateau frog, Nanorana parkeri. Biochimie 2010, 92, 475–481. [Google Scholar] [CrossRef]
- Pérez-Peinado, C.; Defaus, S.; Andreu, D. Hitchhiking with Nature: Snake Venom Peptides to Fight Cancer and Superbugs. Toxins 2020, 12, 255. [Google Scholar] [CrossRef]
- Deshane, J.; Garner, C.C.; Sontheimer, H. Chlorotoxin inhibits glioma cell invasion via matrix metalloproteinase-2. J. Biol. Chem. 2003, 278, 4135–4144. [Google Scholar] [CrossRef] [PubMed]
- Crusca, E., Jr.; Basso, L.G.M.; Altei, W.F.; Marchetto, R. Biophysical characterization and antitumor activity of synthetic Pantinin peptides from scorpion’s venom. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Khamessi, O.; Ben Mabrouk, H.; ElFessi-Magouri, R.; Kharrat, R. RK1, the first very short peptide from Buthus occitanus tunetanus inhibits tumor cell migration, proliferation and angiogenesis. Biochem. Biophys. Res. Commun. 2018, 499, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ke, M.; Tian, Y.; Wang, J.; Li, B.; Wang, Y.; Dou, J.; Zhou, C. BF-30 selectively inhibits melanoma cell proliferation via cytoplasmic membrane permeabilization and DNA-binding in vitro and in B16F10-bearing mice. Eur. J. Pharmacol. 2013, 707, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Falcao, C.B.; Pérez-Peinado, C.; de la Torre, B.G.; Mayol, X.; Zamora-Carreras, H.; Jiménez, M.A.; Rádis-Baptista, G.; Andreu, D. Structural Dissection of Crotalicidin, a Rattlesnake Venom Cathelicidin, Retrieves a Fragment with Antimicrobial and Antitumor Activity. J. Med. Chem. 2015, 58, 8553–8563. [Google Scholar] [CrossRef]
- Kim, D.; Soundrarajan, N.; Lee, J.; Cho, H.-s.; Choi, M.; Cha, S.-Y.; Ahn, B.; Jeon, H.; Le, M.T.; Song, H. Genomewide analysis of the antimicrobial peptides in Python bivittatus and characterization of cathelicidins with potent antimicrobial activity and low cytotoxicity. Antimicrob. Agents Chemother. 2017, 61, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Satitmanwiwat, S.; Changsangfa, C.; Khanuengthong, A.; Promthep, K.; Roytrakul, S.; Arpornsuwan, T.; Saikhun, K.; Sritanaudomchai, H. The scorpion venom peptide BmKn2 induces apoptosis in cancerous but not in normal human oral cells. Biomed. Pharmacother. 2016, 84, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ma, C.; Du, Q.; Wei, R.; Wang, L.; Zhou, M.; Chen, T.; Shaw, C. Two peptides, TsAP-1 and TsAP-2, from the venom of the Brazilian yellow scorpion, Tityus serrulatus: Evaluation of their antimicrobial and anticancer activities. Biochimie 2013, 95, 1784–1794. [Google Scholar] [CrossRef]
- Nguyen, T.; Guo, R.; Chai, J.; Wu, J.; Liu, J.; Chen, X.; Abdel-Rahman, M.A.; Xia, H.; Xu, X. Smp24, a scorpion-venom peptide, exhibits potent antitumor effects against hepatoma HepG2 cells via multi-mechanisms in vivo and in vitro. Toxins 2022, 14, 717. [Google Scholar] [CrossRef]
- Lacerda, A.F.; Pelegrini, P.B.; de Oliveira, D.M.; Vasconcelos, É.A.; Grossi-de-Sá, M.F. Anti-parasitic peptides from arthropods and their application in drug therapy. Fron. Microbiol. 2016, 7, 91. [Google Scholar] [CrossRef]
- Fieck, A.; Hurwitz, I.; Kang, A.S.; Durvasula, R. Trypanosoma cruzi: Synergistic cytotoxicity of multiple amphipathic anti-microbial peptides to T. cruzi and potential bacterial hosts. Exp. Parasitol. 2010, 125, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Rangel, M.; Cabrera, M.P.d.S.; Kazuma, K.; Ando, K.; Wang, X.; Kato, M.; Nihei, K.-i.; Hirata, I.Y.; Cross, T.; Garcia, A.N.; et al. Chemical and biological characterization of four new linear cationic α-helical peptides from the venoms of two solitary eumenine wasps. Toxicon 2011, 57, 1081–1092. [Google Scholar] [CrossRef]
- Marr, A.K.; McGwire, B.S.; McMaster, W.R. Modes of action of Leishmanicidal antimicrobial peptides. Future Microbiol. 2012, 7, 1047–1059. [Google Scholar] [CrossRef]
- Gao, B.; Xu, J.; Rodriguez Mdel, C.; Lanz-Mendoza, H.; Hernández-Rivas, R.; Du, W.; Zhu, S. Characterization of two linear cationic antimalarial peptides in the scorpion Mesobuthus eupeus. Biochimie 2010, 92, 350–359. [Google Scholar] [CrossRef]
- Vale, N.; Aguiar, L.; Gomes, P. Antimicrobial peptides: A new class of antimalarial drugs? Front. Pharmacol. 2014, 5, 275. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, H.; Chattopadhyay, A. Melittin: A membrane-active peptide with diverse functions. Biosci. Rep. 2007, 27, 189–223. [Google Scholar] [CrossRef]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T.; et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect. Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef]
- Sengupta, D.; Leontiadou, H.; Mark, A.E.; Marrink, S.J. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim. Biophys. Acta 2008, 1778, 2308–2317. [Google Scholar] [CrossRef] [PubMed]
- Mor, A. Multifunctional host defense peptides: Antiparasitic activities. FEBS J. 2009, 276, 6474–6482. [Google Scholar] [CrossRef]
- Boman, H.G.; Wade, D.; Boman, I.A.; Wåhlin, B.; Merrifield, R.B. Antibacterial and antimalarial properties of peptides that are cecropin-melittin hybrids. FEBS Lett. 1989, 259, 103–106. [Google Scholar] [CrossRef]
- Gwadz, R.W.; Kaslow, D.; Lee, J.Y.; Maloy, W.L.; Zasloff, M.; Miller, L.H. Effects of magainins and cecropins on the sporogonic development of malaria parasites in mosquitoes. Infect. Immun. 1989, 57, 2628–2633. [Google Scholar] [CrossRef] [PubMed]

| Name | Primary Structure | Source | Activity | Reference |
|---|---|---|---|---|
| Animal Sources | ||||
| VGINVKCKHSGQCLKPCKDAGMRFGKCINGKCDCTPK | Scorpion venom | Anti-bacterial | [27] | |
| Tachyplesin I | KWCFRVCYRGICYRRCR | Horseshoe crab (Tachypleus tridentatus) | Anti-bacterial | [28] |
| SIITMTKEAKLPQLWKQIAC-RLYNTC | Yunnan frog, Rana pleuraden | Antioxidant | [29] | |
| Temporin 1Tb (TB) | ALWKTMLKKLGTMALHAGKAALGAAADTISQGTQ | Frog skin | Antimicrobial | [30] |
| Kappacin | AVESTVATLEDƩPEVIESPPE | Bovine milk | Anti-bacterial | [31] |
| IVSDGNGMNAWVAWR | Chicken egg | Anti-bacterial | [32] | |
| GQGAKDMWR | Donkey milk | Antioxidant | [33] | |
| EWFTFLKEAGQGAKDMWR | Donkey milk | Antioxidant | [33] | |
| Hydrolysates of camel milk protein | KDLWDDFKGL | Camel milk | Anti-diabetic | [34] |
| MPSKPPLL | Camel milk | Anti-diabetic | [34] | |
| KFQWGY | Camel milk | Inhibition of cholesterol esterase | [35] | |
| SQDWSFY | Camel milk | Inhibition of cholesterol esterase | [35] | |
| YWYPPQ | Camel milk | Inhibition of cholesterol esterase | [35] | |
| Marine Sources | ||||
| AERQ | Coral (Sarcophyton glaucum) | Anticancer | [36] | |
| RDTQ | Coral (Sarcophyton glaucum) | Anticancer | [36] | |
| AGAPGG | Coral (Sarcophyton glaucum) | Anticancer | [36] | |
| LSGYGP | Tilapia (O. niloticus) skin | ACE inhibitory | [37] | |
| CPAP | Chlorella pyrenoidosa | Anticancer | [38] | |
| VECYGPNRPQF | Chlorella vulgaris | Antioxidant | [39] | |
| ALLAGDPSVLEDR | Bangia fusco purpurea | Antihypertensive | [40] | |
| VVGGTGPVDEWGIAGAR | Bangia fusco purpurea | Antihypertensive | [40] | |
| VKAGFAWTANQQLS | Tuna backbone | Antioxidant | [41] | |
| Plant Sources | ||||
| FFL | Soy | ACE inhibitory | [42] | |
| RQSHFANAQP | Chickpea (Cicer anetinum) | Anticancer | [43] | |
| AIRQGDVF | Crude rice bran | Antioxidant | [44] | |
| FGER | Potato | Antioxidant | [45] | |
| DAQEFKR | Kamut (Triticum turanicum Jakubz.) | Antioxidant | [46] | |
| DNIPIVIR | Wheat (Triticum aestivum L.) | Antioxidant | [46] | |
| GNQEKVLELVQR | Spelt (Triticum spelta L.) | Antioxidant | [46] | |
| Extraction Method | Principle | Pros | Cons | References | |
|---|---|---|---|---|---|
| Bottom-Up Approaches | Enzymatic hydrolysis | Proteolytic cleavage by peptidases at specific sites to yield peptide fragments | (a) Enzyme-specific cleavage sites | (a) Loss of critical structural information of intact peptides and/or truncated proteins | [50] |
| (b) Fast process | |||||
| Microbial fermentation | Proteolytic cleavage by microbial peptidases at specific sites to yield peptide fragments | (a) Enzyme-specific cleavage sites | (a) Loss of critical structural information of intact peptides and/or truncated proteins | [26] | |
| (b) Presence of biological impurities and immunogenicity concerns | |||||
| (c) Differential proteolytic activity expressed by each culture—unpredictability of peptides obtained | |||||
| (d) Long reaction times and difficult to scale up operations | |||||
| Top-Down Approaches | Ultrafiltration | Molecular weight cut-off: Membrane-based separation allows molecules only under the specified limit to pass through | (a) Simple and cost-effective | (a) Non-specific binding by peptides and clogging membrane pores affect recovery | [50,57,58,59] |
| (b) Fast process | (b) Peptides with molecular weights closer to the cut-off may be lost | ||||
| (c) Possible filtrate contamination | |||||
| Organic protein precipitation | Solvent addition causes the precipitation of larger proteins, leaving peptide fragments in the supernatant | (a) Simple, efficient, and cost-effective | (a) The precipitating solvent chosen must be optimized for every biological matrix to ensure maximum peptide extraction | [50,59] | |
| (b) Entrapment of smaller peptides within large protein aggregates—inconsistencies in extraction yield | |||||
| Acidic protein precipitation | Solvent addition causes the precipitation of larger proteins, leaving peptide fragments in the supernatant | (a) Simple, efficient, and cost-effective | (a) Affected by peptide solubility at acidic pH | [50] | |
| (b) Rapid protein precipitation process, avoiding protease-related degradation | |||||
| Size exclusion chromatography (SEC) | Chromatographic columns with specific pore sizes exclude and cause early elution of high-molecular-weight biomolecules, while smaller peptides enter the pores and elute out later | (a) Simple and reproducible | (a) High elution volume leading to sample dilution and increased cost—low throughput | [49,50,59] | |
| (b) Higher sample load requirement | |||||
| (c) Low resolution—needs to be coupled with other separation techniques | |||||
| Solid-phase extraction (SPE) | Peptides of interest interacting with the stationary phase SPE sorbent are retained, while other interfering molecules are washed off with the solvent(s) | (a) Compatible with MS and other high-throughput techniques | (a) Newer versions such as mixed-mode or restricted access matrix sorbents are exceptionally selective and unsuitable for global peptidomics | [50,59,60] | |
| (b) High resolution | (b) Possibility of slight protein co-elution | ||||
| Combination of solvent precipitation and solid-phase extraction (SPE) | Organic solvent precipitation removes large molecules from the sample, while the subsequent SPE removes the leftover small hydrophobic interfering biomolecules | (a) Highly effective in removing all interfering molecules | (a) Lengthy process with increased extraction steps | [50] |
| Name | Primary Structure * | Source | References |
|---|---|---|---|
| Food-protein-derived peptides | |||
| TBS1 | IPP | Sour milk | [195] |
| VPP | Sour milk | [195] | |
| AR | Trevally (Pseudocaranx sp.) | [65] | |
| LKP | Egg white | [162,196] | |
| VNP | Rice | [197] | |
| VWP | Rice | [197] | |
| VY | Sake | [198] | |
| FY | α-zein | [199] | |
| IY | Sardine | [200] | |
| AF | Rabbit | [201] | |
| FFYY | Processed soya milk | [202] | |
| LPI5 | EPLYV | Leatherjacket (Meuchenia sp.) | [203] |
| LPI6 | DPHI | Leatherjacket (Meuchenia sp.) | [203] |
| LBI5 | AER | Leatherjacket (Meuchenia sp.) | [203] |
| Naturally occurring peptides | |||
| Bn-PRO-10a | pENWPRPKIPP | Bitis gabonica rhinoceros venom | [204] |
| Bj-PRO-10b | pENWPRPQIPP | Bothrops jararaca venom | [192,201,205] |
| Peptide F | pELWPRPHIPP | Agkistrodon piscivorus piscivoris venom | [206,207] |
| POL 236 | pELWPRPQIPP | Crotalus atrox snake venom | [208] |
| Br-PRO-10a | pENWPHPQVPP | Bitis gabonica rhinoceros venom | [204] |
| Bg-PRO-11a | pEWQRPGPEIPP | Bothrops jararaca venom | [204] |
| Bn-PRO-10c | pENWPRPKVPP | Bothrops jararaca venom | [204] |
| Name | Primary Structure | Source | Reference |
|---|---|---|---|
| Food-protein-derived peptides | |||
| LPHSGY | Alaska pollack (Theragra chalcogramma) | [212] | |
| GSTVPERTHPACPDFN | Hoki (Johnius belengerii) | [214] | |
| PSKYEPFV | Grass carp | [215] | |
| LHY | Sardinelle (Sardinella aurita) | [165] | |
| VKEAMAPK | Bovine β-casein | [216] | |
| AVPYPQR | Bovine β-casein | [216] | |
| YVEEL | Whey proteins | [217] | |
| TEINEGALLLPH | Lupin seed (Lupinus angustifolius) | [218] | |
| EAGTIETWNPN | Lupin seed (Lupinus angustifolius) | [218] | |
| Naturally occurring peptides | |||
| Brevinin-1TP2 | FLPGLIKAAVGIGSTIFCKISKKC | East Asian frog (Hylarana taipehensis) | [219] |
| Temporin-TP1 | FLPVLGKVIKLVGGLL | East Asian frog (Hylarana taipehensis) | [219] |
| Parkerin | GWANTLKNVAGGLCKITGAA | Xizang plateau frog (Nanorana parkeri) | [220] |
| Name | Primary Structure | Insect Source | References |
|---|---|---|---|
| Naturally occurring peptides | |||
| Cecropin A | KWKLFKKIEKVGQNIRDGIIKAGPAVAVVGQATQIAK | H. cecropia | [231,241] |
| Cecropin B | KWKVFKKIEKMGRNIRNGIVKAGPAIAVLGEAKAL | H. cecropia | [231,242] |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ | Apis melífera venom | [231,232] |
| Meucine-24 | GRGREFMSNLKEKLSGVKEKMKNS | A. melífera venom | [231,235] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Purohit, K.; Reddy, N.; Sunna, A. Exploring the Potential of Bioactive Peptides: From Natural Sources to Therapeutics. Int. J. Mol. Sci. 2024, 25, 1391. https://doi.org/10.3390/ijms25031391
Purohit K, Reddy N, Sunna A. Exploring the Potential of Bioactive Peptides: From Natural Sources to Therapeutics. International Journal of Molecular Sciences. 2024; 25(3):1391. https://doi.org/10.3390/ijms25031391
Chicago/Turabian StylePurohit, Kruttika, Narsimha Reddy, and Anwar Sunna. 2024. "Exploring the Potential of Bioactive Peptides: From Natural Sources to Therapeutics" International Journal of Molecular Sciences 25, no. 3: 1391. https://doi.org/10.3390/ijms25031391
APA StylePurohit, K., Reddy, N., & Sunna, A. (2024). Exploring the Potential of Bioactive Peptides: From Natural Sources to Therapeutics. International Journal of Molecular Sciences, 25(3), 1391. https://doi.org/10.3390/ijms25031391







