Phosphatidyl Ethanolamine Binding Protein FLOWERING LOCUS T-like 12 (OsFTL12) Regulates the Rice Heading Date under Different Day-Length Conditions
Abstract
1. Introduction
2. Results
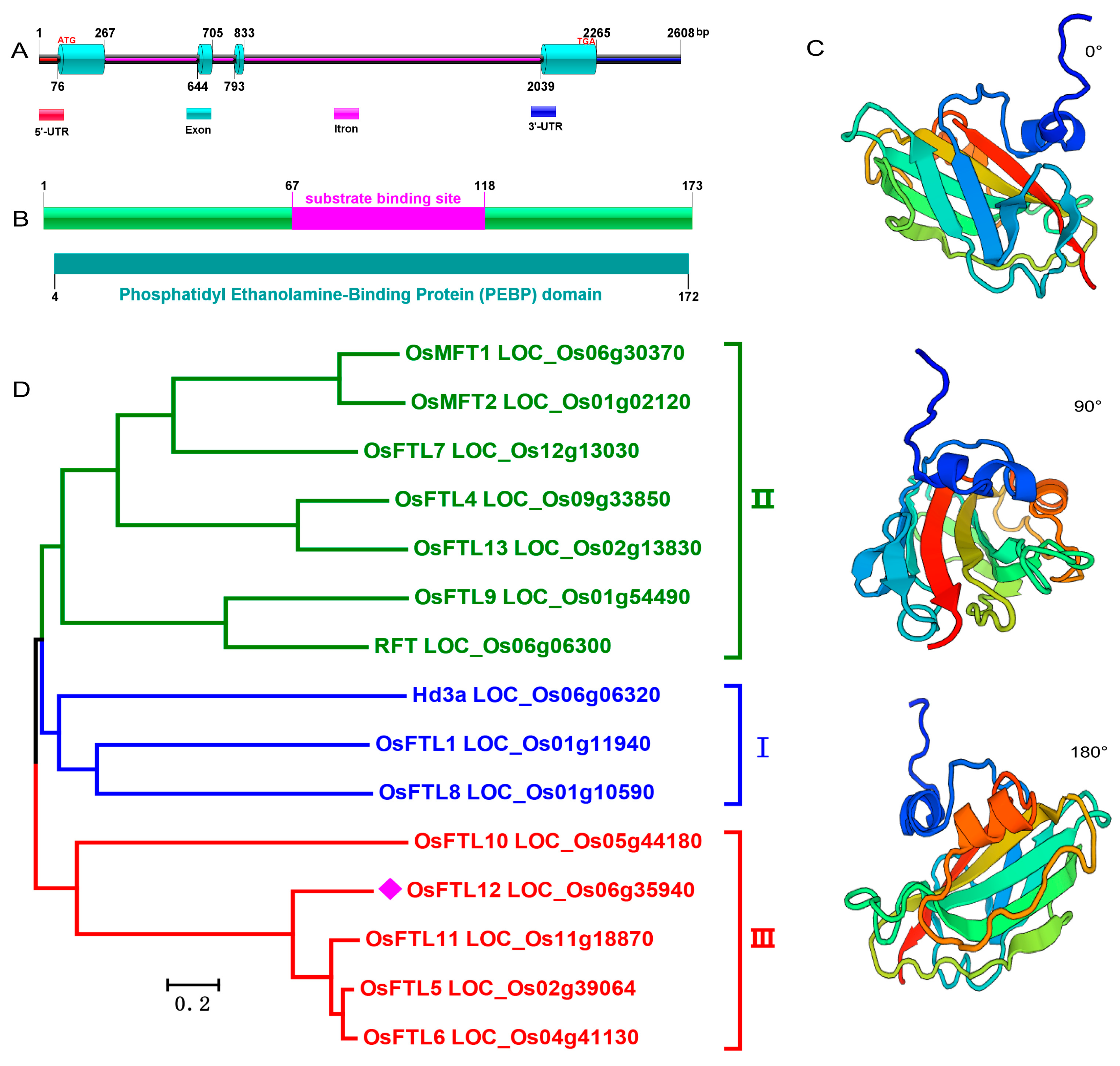
2.1. Isolation of OsFTL12 from Rice Genome
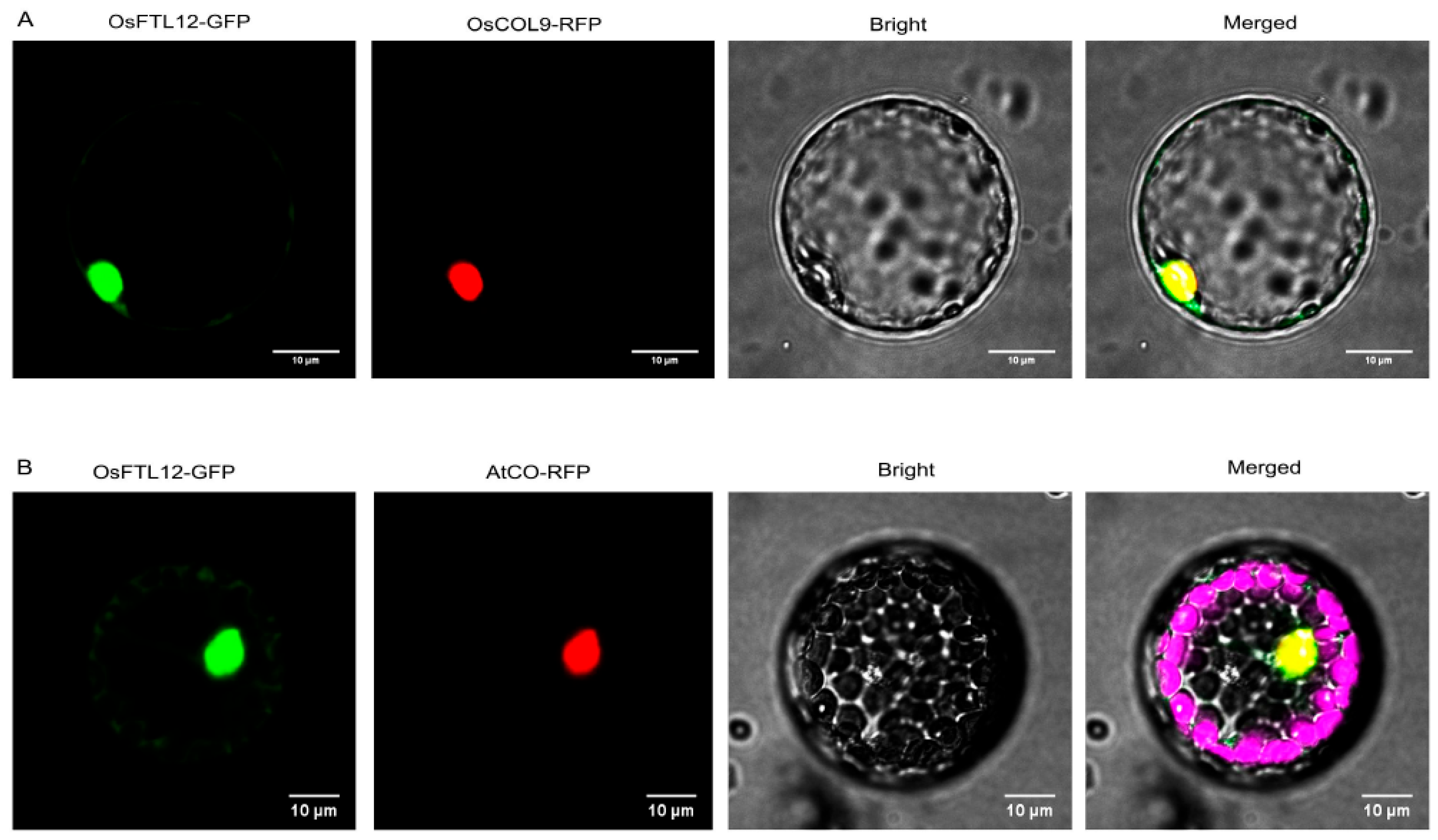
2.2. Subcellular Localization Identified That the OsFTL12 Protein Is Targeted to the Nucleus
2.3. OsFTL12 Promoter Activity Analysis
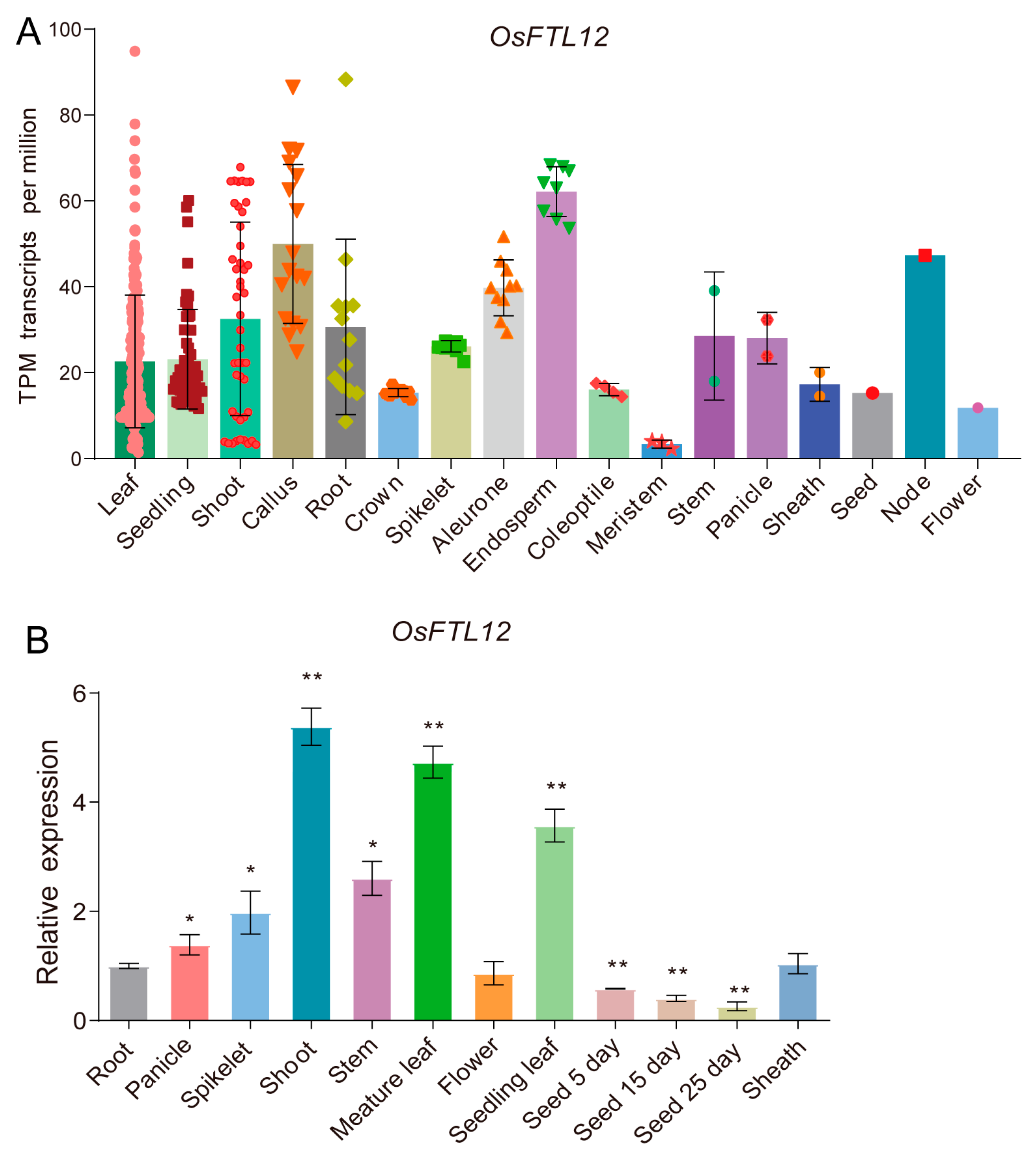
2.4. Identification of the Expression Pattern of OsFTL12
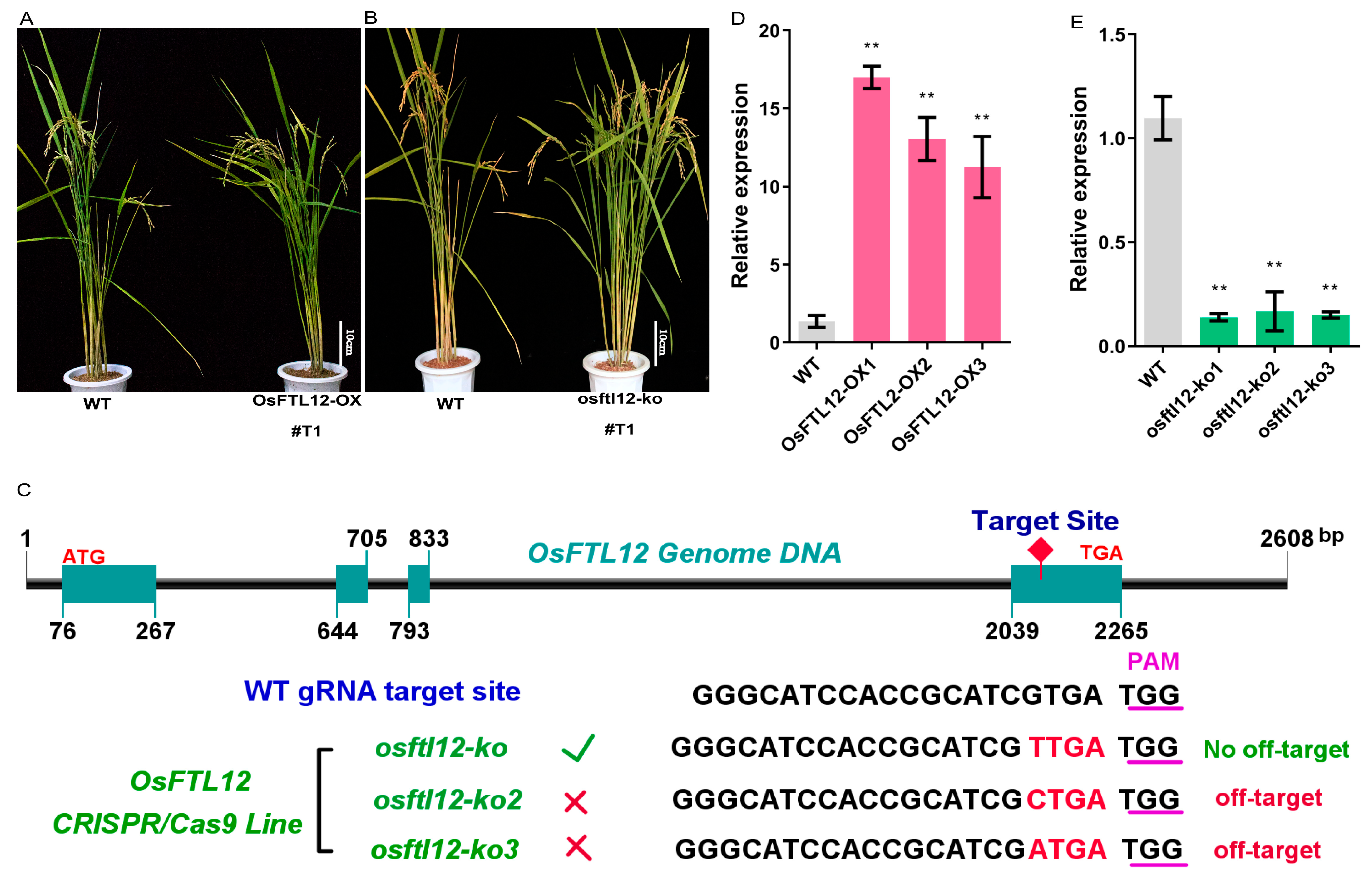
2.5. Construction of OsFTL12 Over-Expression and Knock-Down Transgenic Plants
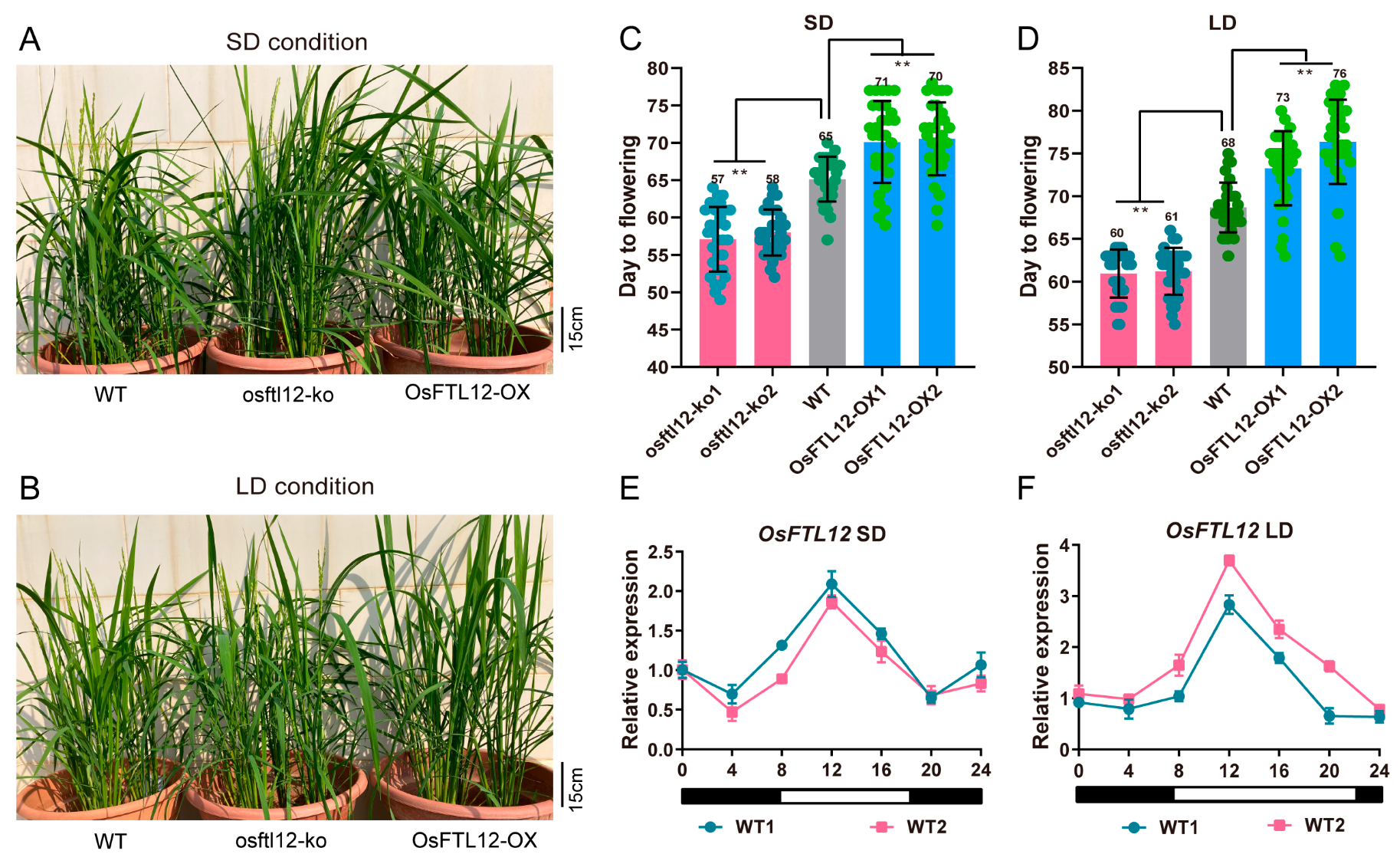
2.6. OsFTL12 Over-Expression Delays Heading Date in Rice
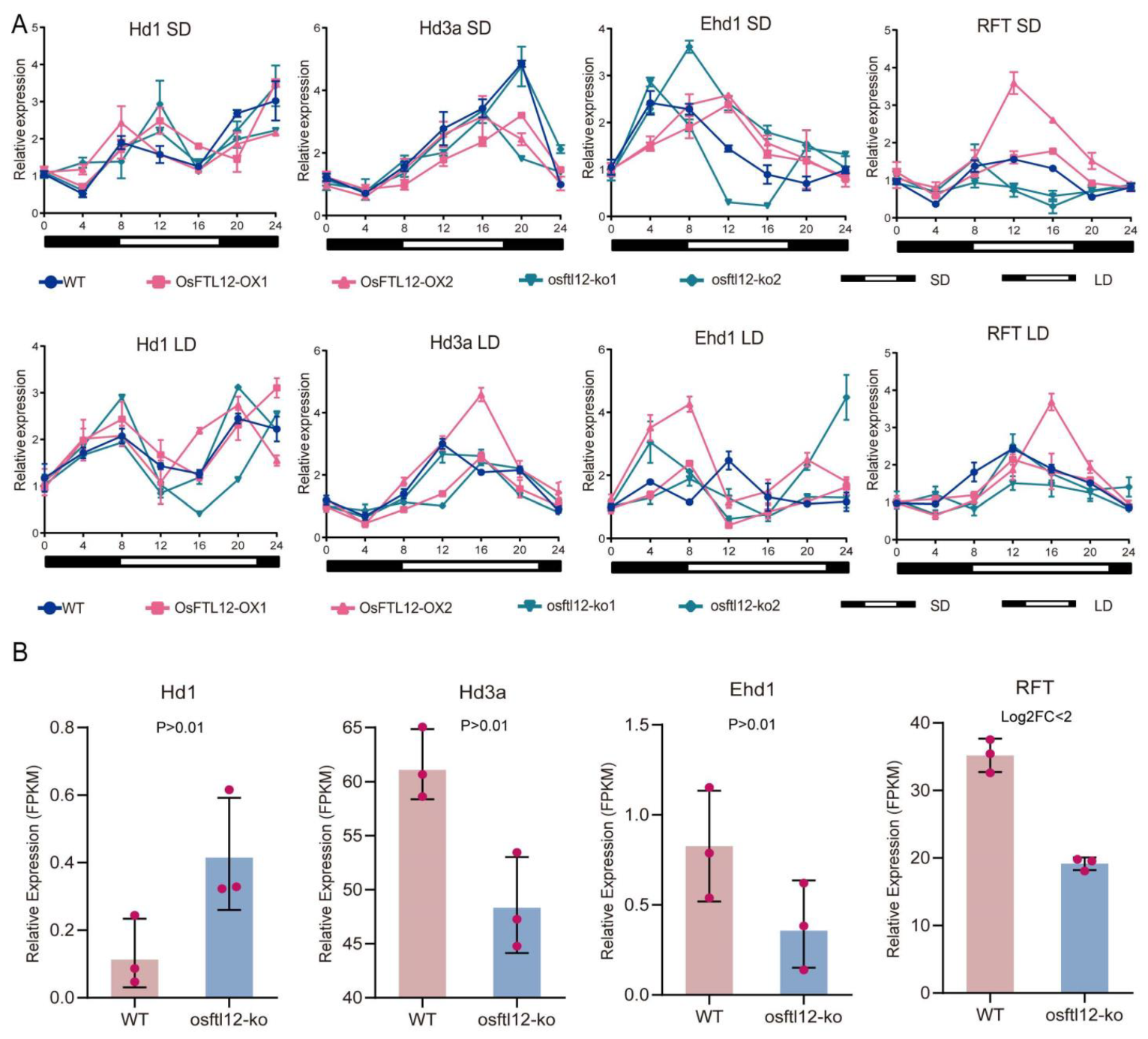
2.7. RNA-Seq Identified the Differentially Expressed Genes (DEGs) in osftl12-ko Plants
2.8. OsFTL12 Independently Regulate Rice Early Flowering
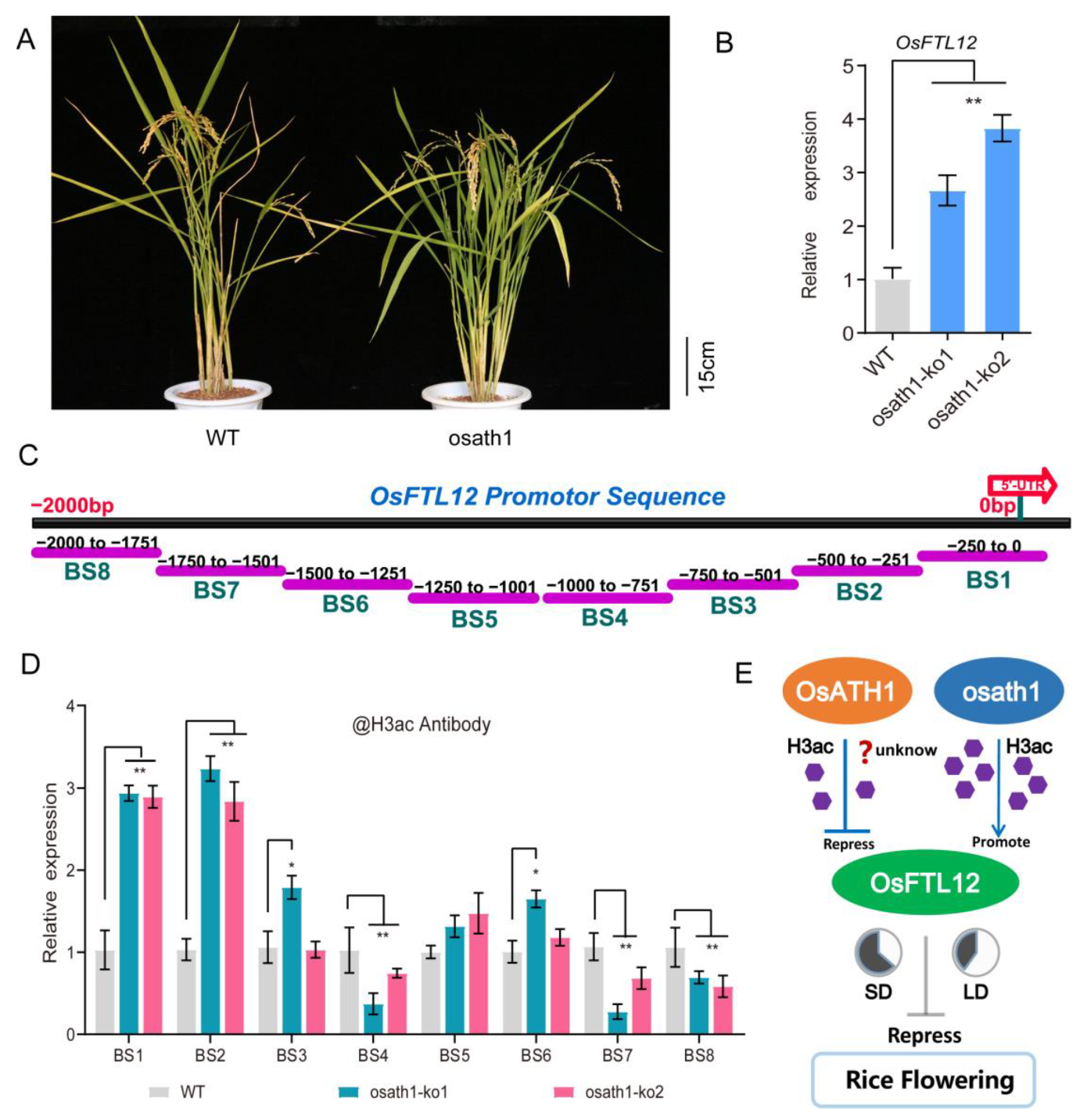
2.9. Knock-Out OsATH1 Enhances the H3 Acetylation Level in OsFTL12 Promoter Region
3. Discussion
4. Materials and Methods
4.1. Experiment Materials and Growth Conditions
4.2. Bioinformatics Analysis
4.3. Subcellular Localization Analysis
4.4. GUS Assay
4.5. Total RNA Extraction, Real-Time PCR Analysis of Gene Expression
4.6. Generation of the Transgenic Plants
4.7. RNA-Seq
4.8. Chromatin Immunoprecipitation (ChIP) qPCR Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Davila-Velderrain, J.; Martinez-Garcia, J.C.; Alvarez-Buylla, E.R. Dynamic network modelling to understand flowering transition and floral patterning. J. Exp. Bot. 2016, 67, 2565–2572. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Ma, H. The flowering transition and florigen. Curr. Biol. 2001, 11, R815. [Google Scholar] [CrossRef] [PubMed]
- Zicola, J.; Liu, L.; Tanzler, P.; Turck, F. Targeted DNA methylation represses two enhancers of FLOWERING LOCUS T in Arabidopsis thaliana. Nat. Plants 2019, 5, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Payyavula, R.S.; Chen, L.; Zhang, J.; Zhang, C.; Turgeon, R. FLOWERING LOCUS T mRNA is synthesized in specialized companion cells in Arabidopsis and Maryland Mammoth tobacco leaf veins. Proc. Natl. Acad. Sci. USA 2018, 115, 2830–2835. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Chen, T.; Zeng, X.; He, D.; He, Y. Feedback Regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5’ FLC Promoter Region in Arabidopsis. Mol. Plant 2019, 12, 285–288. [Google Scholar] [CrossRef]
- Benlloch, R.; Kim, M.C.; Sayou, C.; Thevenon, E.; Parcy, F.; Nilsson, O.O. Integrating long-day flowering signals: A LEAFY binding site is essential for proper photoperiodic activation of APETALA1. Plant J. 2011, 67, 1094–1102. [Google Scholar] [CrossRef]
- Goretti, D.; Silvestre, M.; Collani, S.; Langenecker, T.; Mendez, C.; Madueno, F.; Schmid, M. TERMINAL FLOWER1 Functions as a Mobile Transcriptional Cofactor in the Shoot Apical Meristem. Plant Physiol. 2020, 182, 2081–2095. [Google Scholar] [CrossRef]
- Jin, S.; Jung, H.S.; Chung, K.S.; Lee, J.H.; Ahn, J.H. FLOWERING LOCUS T has higher protein mobility than TWIN SISTER OF FT. J. Exp. Bot. 2015, 66, 6109–6117. [Google Scholar] [CrossRef]
- Harig, L.; Beinecke, F.A.; Oltmanns, J.; Muth, J.; Muller, O.; Ruping, B.; Twyman, R.M.; Fischer, R.; Prufer, D.; Noll, G.A. Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J. 2012, 72, 908–921. [Google Scholar] [CrossRef]
- Du, A.; Tian, W.; Wei, M.; Yan, W.; He, H.; Zhou, D.; Huang, X.; Li, S.; Ouyang, X. The DTH8-Hd1 Module Mediates Day-Length-Dependent Regulation of Rice Flowering. Mol. Plant 2017, 10, 948–961. [Google Scholar] [CrossRef]
- Pasriga, R.; Yoon, J.; Cho, L.H. Overexpression of RICE FLOWERING LOCUS T 1 (RFT1) Induces Extremely Early Flowering in Rice. Mol. Cells 2019, 42, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Komiya, R.; Yokoi, S.; Shimamoto, K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 2009, 136, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, H.; Ren, D.; Tang, H.; Qiu, R.; Feng, J.; Long, Y.; Niu, B.; Chen, D.; Zhong, T.; et al. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). N. Phytol. 2015, 208, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Damerval, C.C. Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 2005, 61, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Hanzawa, Y. FLOWERING LOCUS T/TERMINAL FLOWER 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Kim, S.Y.; Ahn, J.H. TWIN SISTER OF FT (TSF) Interacts with FRUCTOKINASE6 and Inhibits Its Kinase Activity in Arabidopsis. Front. Plant Sci. 2017, 8, 1807. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef]
- Mulki, M.A.; Bi, X.; von Korff, M. FLOWERING LOCUS T3 Controls Spikelet Initiation But Not Floral Development. Plant Physiol. 2018, 178, 1170–1186. [Google Scholar] [CrossRef]
- Song, J.; Zhang, S.; Wang, X.; Sun, S.; Liu, Z.; Wang, K.; Wan, H.; Zhou, G.; Li, R.; Yu, H.; et al. Variations in Both FTL1 and SP5G, Two Tomato FT Paralogs, Control Day-Neutral Flowering. Mol. Plant 2020, 13, 939–942. [Google Scholar] [CrossRef]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Song, S.; Wang, G.; Wu, H.; Fan, X.; Liang, L.; Zhao, H.; Li, S.; Hu, Y.; Liu, H.; Ayaad, M.; et al. OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J. 2020, 103, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, S.; Li, M.; Gu, F.; Yang, G.; Guo, T.; Chen, Z.; Wang, J. The Class III peroxidase gene OsPrx30, transcriptionally modulated by the AT-hook protein OsATH1, mediates rice bacterial blight-induced ROS accumulation. J. Integr. Plant Biol. 2021, 63, 393–408. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Lin, Y.C.; Watanabe, S.; Liu, Y.C.; Katsuyama, K.; Kanehara, K.; Inaba, K. High-Resolution Crystal Structure of Arabidopsis FLOWERING LOCUS T Illuminates Its Phospholipid-Binding Site in Flowering. iScience 2019, 21, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gu, F.; Dong, S.; Liu, W.; Wang, H.; Chen, Z.; Wang, J. CONSTANS-like 9 (COL9) delays the flowering time in Oryza sativa by repressing the Ehd1 pathway. Biochem. Biophys. Res. Commun. 2016, 479, 173–178. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Ran, X.; Zhao, F.; Wang, Y.; Liu, J.; Zhuang, Y.; Ye, L.; Qi, M.; Cheng, J.; Zhang, Y. Plant Regulomics: A data-driven interface for retrieving upstream regulators from plant multi-omics data. Plant J. 2020, 101, 237–248. [Google Scholar] [CrossRef]
- Xia, L.; Zou, D.; Sang, J.; Xu, X.; Yin, H.; Li, M.; Wu, S.; Hu, S.; Hao, L.; Zhang, Z. Rice Expression Database (RED): An integrated RNA-Seq-derived gene expression database for rice. J. Genet. Genom. 2017, 44, 235–241. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Xie, X.; Ma, X.; Zhu, Q.; Zeng, D.; Li, G.; Liu, Y.G. CRISPR-GE: A Convenient Software Toolkit for CRISPR-Based Genome Editing. Mol. Plant 2017, 10, 1246–1249. [Google Scholar] [CrossRef] [PubMed]
- Osugi, A.; Itoh, H.; Ikeda-Kawakatsu, K.; Takano, M.; Izawa, T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 2011, 157, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Kim, Y.S.; Jung, J.H.; Seo, P.J.; Park, C.M. The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J. Biol. Chem. 2012, 287, 15307–15316. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, L.; Yawen, L.; Hongtao, L. Flowering responses to light and temperature. Sci. China Life Sci. 2016, 59, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Gaudinier, A.; Blackman, B.K. Evolutionary processes from the perspective of flowering time diversity. N. Phytol. 2020, 225, 1883–1898. [Google Scholar] [CrossRef] [PubMed]
- Andres, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic flowering: Time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef]
- O’Maoileidigh, D.S.; Graciet, E.; Wellmer, F. Gene networks controlling Arabidopsis thaliana flower development. N. Phytol. 2014, 201, 16–30. [Google Scholar] [CrossRef]
- Noh, H.S.; Hah, Y.S.; Zada, S.; Ha, J.H.; Sim, G.; Hwang, J.S.; Lai, T.H.; Nguyen, H.Q.; Park, J.Y.; Kim, H.J.; et al. PEBP1, a RAF kinase inhibitory protein, negatively regulates starvation-induced autophagy by direct interaction with LC3. Autophagy 2016, 12, 2183–2196. [Google Scholar] [CrossRef]
- Hogekamp, C.; Arndt, D.; Pereira, P.A.; Becker, J.D.; Hohnjec, N.; Kuster, H. Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol. 2011, 157, 2023–2043. [Google Scholar] [CrossRef]
- Zhu, C.; Peng, Q.; Fu, D.; Zhuang, D.; Yu, Y.; Duan, M.; Xie, W.; Cai, Y.; Ouyang, Y.; Lian, X.; et al. The E3 Ubiquitin Ligase HAF1 Modulates Circadian Accumulation of EARLY FLOWERING3 to Control Heading Date in Rice under Long-Day Conditions. Plant Cell 2018, 30, 2352–2367. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Jeong, D.H.; Lee, D.Y.; Yi, J.; Ryu, C.H.; Kim, S.L.; Jeong, H.J.; Choi, S.C.; Jin, P.; Yang, J.; et al. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 2010, 63, 18–30. [Google Scholar] [CrossRef]
- Gao, H.; Jin, M.; Zheng, X.M.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, E.; Brooks, A.M.; Concia, L.; Vera, D.; Wear, E.E.; LeBlanc, C.; Ramu, U.; Vaughn, M.W.; Bass, H.; Martienssen, R.; et al. Arabidopsis DNA replication initiates in intergenic, AT-rich open chromatin. Plant Physiol. 2020, 183, 206–220. [Google Scholar] [CrossRef] [PubMed]
- Street, I.H.; Shah, P.K.; Smith, A.M.; Avery, N.; Neff, M.M. The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 2008, 54, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Xu, X.F.; Zhu, J.; Gu, J.N.; Blackmore, S.; Yang, Z.N. The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat. Commun. 2014, 5, 3855. [Google Scholar] [CrossRef]
- Shi, J.; Dong, A.; Shen, W.H. Epigenetic regulation of rice flowering and reproduction. Front. Plant Sci. 2014, 5, 803. [Google Scholar] [CrossRef]
- Vittoria, B.; Fabio, F. Molecular Control of Flowering in Response to Day Length in Rice. J. Integr. Plant Biol. 2013, 55, 410–418. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.-H. Songnian Hu. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 15, 3359–3361. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, S.; Gu, F.; Liu, W.; Yang, G.; Huang, M.; Xiao, W.; Liu, Y.; Guo, T.; Wang, H.; et al. NBS-LRR Protein Pik-H4 Interacts with OsBIHD1 to Balance Rice Blast Resistance and Growth by Coordinating Ethylene-Brassinosteroid Pathway. Front. Plant Sci. 2017, 8, 127. [Google Scholar] [CrossRef][Green Version]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Tsuda, K.; Kurata, N.; Ohyanagi, H.; Hake, S. Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 2014, 26, 3488–3500. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Guo, J.; Sun, D.; Guo, Z.; Zheng, Z.; Wang, P.; Hong, Y.; Liu, H. Phosphatidyl Ethanolamine Binding Protein FLOWERING LOCUS T-like 12 (OsFTL12) Regulates the Rice Heading Date under Different Day-Length Conditions. Int. J. Mol. Sci. 2024, 25, 1449. https://doi.org/10.3390/ijms25031449
Huang Y, Guo J, Sun D, Guo Z, Zheng Z, Wang P, Hong Y, Liu H. Phosphatidyl Ethanolamine Binding Protein FLOWERING LOCUS T-like 12 (OsFTL12) Regulates the Rice Heading Date under Different Day-Length Conditions. International Journal of Molecular Sciences. 2024; 25(3):1449. https://doi.org/10.3390/ijms25031449
Chicago/Turabian StyleHuang, Yongxiang, Jianfu Guo, Dayuan Sun, Zhenhua Guo, Zihao Zheng, Ping Wang, Yanbin Hong, and Hao Liu. 2024. "Phosphatidyl Ethanolamine Binding Protein FLOWERING LOCUS T-like 12 (OsFTL12) Regulates the Rice Heading Date under Different Day-Length Conditions" International Journal of Molecular Sciences 25, no. 3: 1449. https://doi.org/10.3390/ijms25031449
APA StyleHuang, Y., Guo, J., Sun, D., Guo, Z., Zheng, Z., Wang, P., Hong, Y., & Liu, H. (2024). Phosphatidyl Ethanolamine Binding Protein FLOWERING LOCUS T-like 12 (OsFTL12) Regulates the Rice Heading Date under Different Day-Length Conditions. International Journal of Molecular Sciences, 25(3), 1449. https://doi.org/10.3390/ijms25031449





