The QTL and Candidate Genes Regulating the Early Tillering Vigor Traits of Late-Season Rice in Double-Cropping Systems
Abstract
1. Introduction
2. Results
2.1. Phenotypic Variation in Rice Tillering Vigor
2.2. Genetic Linkage Map Construction
2.3. QTLs of the ETV Traits
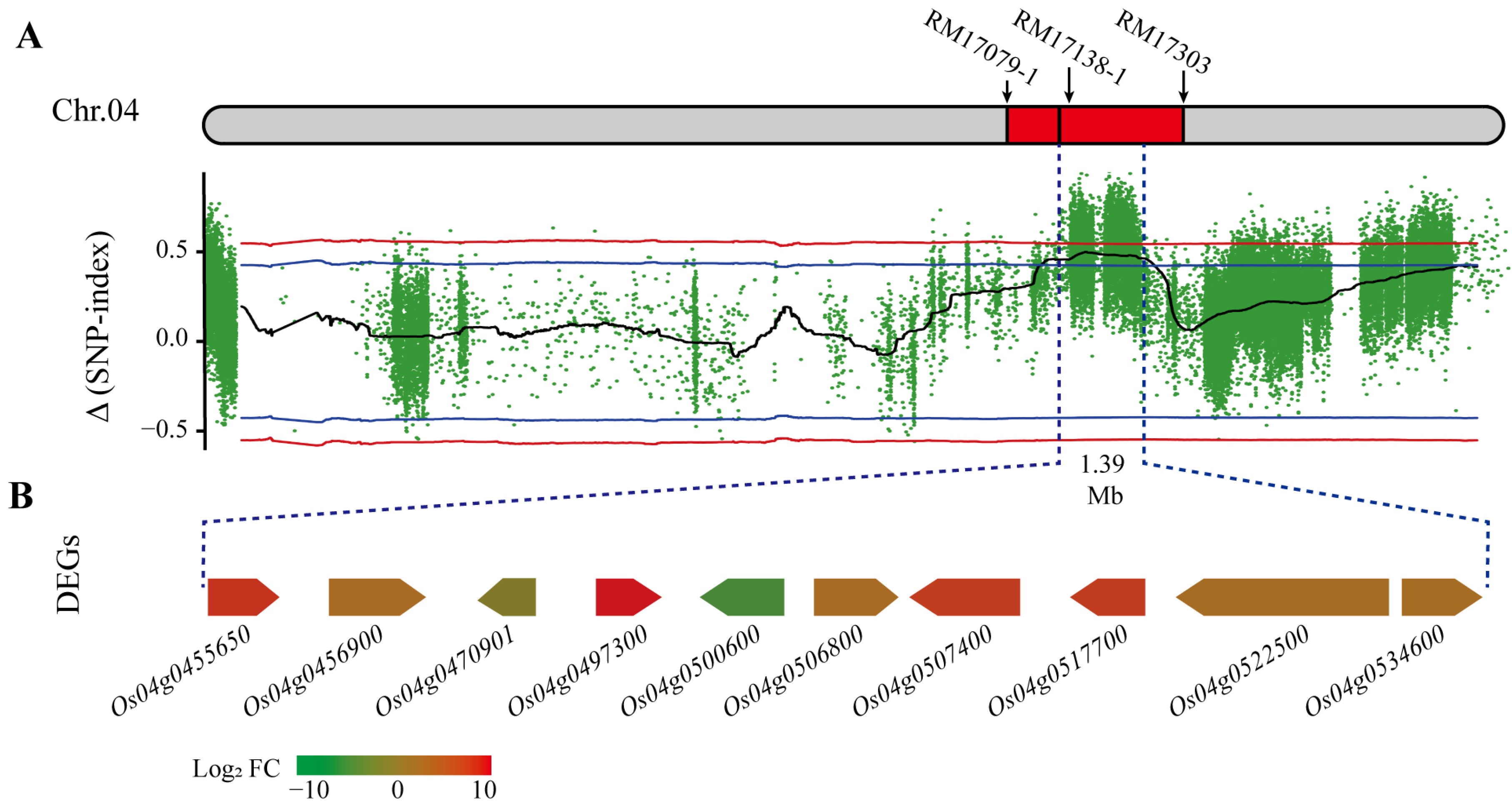
2.4. SNP/InDel Calling Via BSA-seq Method
2.5. The Overlapping Regions
2.6. The Candidate Genes Revealed by RNA-seq
2.7. Missense Base Mutation for the Candidate Genes
3. Discussion
3.1. The Candidate Genes Regulating Rice ETV Trait
3.2. Ethylene May Play an Important Role in ETV Traits of Rice
3.3. The ST-Like Protein Unigene OsSTLP3 Plays Important Roles in Plant ETV
3.4. Functional Unknown Genes
4. Materials and Methods
4.1. Plant Materials
4.2. Field Trial and Phenotypic Evaluation
4.3. Linkage Map Construction and QTL Analysis
4.4. BSA-seq Mapping Analysis
4.5. Analysis of the Overlapping Regions
4.6. Expression Pattern Analysis for the Genes in the Overlapping Regions
4.7. Analysis of Missense Base Mutation for the DEGs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shi, J.; An, G.; Weber, A.P.M.; Zhang, D. Prospects for rice in 2050. Plant Cell Environ. 2023, 46, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, F.; Zhang, G. Difference in Grain Yield and Quality among Tillers in Rice Genotypes Differing in Tillering Capacity. Rice Sci. 2007, 14, 135–140. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J. Branching in rice. Curr. Opin. Plant Biol. 2011, 14, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Shao, G.; Lu, Z.; Xiong, J.; Wang, B.; Jing, Y.; Meng, X.; Liu, G.; Ma, H.; Liang, Y.; Chen, F.; et al. Tiller Bud Formation Regulators MOC1 and MOC3 Cooperatively Promote Tiller Bud Outgrowth by Activating FON1 Expression in Rice. Mol. Plant. 2019, 12, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, L.; Lee, S.; Pasriga, R.; Tun, W.; Yang, J.; Yoon, H.; Jeong, H.J.; Jeon, J.; An, G. Chromatin Interacting Factor OsVIL2 Is Required for Outgrowth of Axillary Buds in Rice. Mol. Cells 2019, 42, 858–868. [Google Scholar] [PubMed]
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.Y. Axillary Meristem Formation in Rice Requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184. [Google Scholar] [CrossRef]
- Zha, M.; Zhao, Y.; Wang, Y.; Chen, B.; Tan, Z. Strigolactones and Cytokinin Interaction in Buds in the Control of Rice Tillering. Front. Plant Sci. 2022, 13, 837136. [Google Scholar] [CrossRef]
- Yan, Y.; Ding, C.; Zhang, G.; Hu, J.; Zhu, L.; Zeng, D.; Qian, Q.; Ren, D. Genetic and environmental control of rice tillering. Crop J. 2023, 11, 1287–1302. [Google Scholar] [CrossRef]
- Hu, Y.; Xue, J.; Li, L.; Cong, S.; Yu, E.; Xu, K.; Zhang, H. Influence of dynamic high temperature during grain filling on starch fine structure and functional properties of semi-waxy japonica rice. J. Cereal Sci. 2021, 101, 103319. [Google Scholar] [CrossRef]
- Tong, H.; Duan, H.; Wang, S.; Su, J.; Sun, Y.; Liu, Y.; Tang, L.; Liu, X.; Chen, W. Moderate drought alleviates the damage to grain quality at high temperatures by improving the starch synthesis of inferior grains in japonica rice. J. Integr. Agric. 2022, 21, 3094–3101. [Google Scholar] [CrossRef]
- Saud, S.; Wang, D.; Fahad, S.; Alharby, H.F.; Bamagoos, A.A.; Mjrashi, A.; Alabdallah, N.M.; AlZahrani, S.S.; AbdElgawad, H.; Adnan, M.; et al. Comprehensive Impacts of Climate Change on Rice Production and Adaptive Strategies in China. Front. Microbiol. 2022, 13, 926059. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Wang, C.; Linderholm, H.W.; Fu, Y.; Cai, W.; Xu, J.; Zhuang, L.; Wu, M.; Shi, Y.; Wang, G.; et al. The negative impact of increasing temperatures on rice yields in southern China. Sci. Total Environ. 2022, 820, 153262. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Tan, X.; Huang, S.; Pan, X.; Zeng, Y.; Zhang, J.; Cheng, S.; Zeng, Y. Grain yield and quality performances of different late-season rice cultivars in response to experimental warming in subtropical China. Front. Plant Sci. 2023, 14, 1136564. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Liu, S.; Zheng, X.; Chu, G.; Xu, C.; Zhang, X.; Wang, D.; Chen, S. Solar radiation-use characteristics of indica/japonica hybrid rice (Oryza sativa L.) in the late season in southeast China. Crop J. 2021, 9, 427–439. [Google Scholar] [CrossRef]
- Wang, J.; Wu, B.; Lu, K.; Wei, Q.; Qian, J.; Chen, Y.; Fang, Z. The Amino Acid Permease 5 (OsAAP5) Regulates Tiller Number and Grain Yield in Rice. Plant Physiol. 2019, 180, 1031–1045. [Google Scholar] [CrossRef]
- Wen, X.; Sun, L.; Chen, Y.; Xue, P.; Yang, Q.; Wang, B.; Yu, N.; Cao, Y.; Zhang, Y.; Gong, K.; et al. Rice dwarf and low tillering 10 (OsDLT10) regulates tiller number by monitoring auxin homeostasis. Plant Sci. 2020, 297, 110502. [Google Scholar] [CrossRef]
- Xia, T.; Chen, H.; Dong, S.; Ma, Z.; Ren, H.; Zhu, X.; Fang, X.; Chen, F. OsWUS promotes tiller bud growth by establishing weak apical dominance in rice. Plant J. 2020, 104, 1635–1647. [Google Scholar] [CrossRef]
- Feng, F.; Guo, X.; Zhu, X.; Hu, Y.; Chen, Y.; Sun, H.; Li, J.; Zhao, C.; Sun, H.; Zhao, Q. OsPIN2 is involved in OsSPL14/17-inhibited tiller bud outgrowth in response to phosphate deficiency in rice. Environ. Exp. Bot. 2023, 209, 105297. [Google Scholar] [CrossRef]
- Hang, J.; Wu, B.; Qiu, D.; Yang, G.; Fang, Z.; Zhang, M. OsNPF3.1, a nitrate, abscisic acid and gibberellin transporter gene, is essential for rice tillering and nitrogen utilization efficiency. J. Integr. Agric. 2023, in press. [Google Scholar] [CrossRef]
- Lin, Q.; Wang, D.; Dong, H.; Gu, S.; Cheng, Z.; Gong, J.; Qin, R.; Jiang, L.; Li, G.; Wang, J.L.; et al. Rice APC/CTE controls tillering by mediating the degradation of MONOCULM 1. Nat. Commun. 2012, 3, 752. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, Y.; Yu, Y.; Duan, J.; Liao, Z.; Xiong, G.; Meng, X.; Liu, G.; Qian, Q.; Li, J. Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat. Commun. 2012, 3, 750. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Yu, H.; Duan, J.; Yuan, K.; Yu, C.; Meng, X.; Kou, L.; Chen, M.; Jing, Y.; Liu, G.; et al. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 2019, 10, 2738. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Han, T.; Song, Q.; Ye, W.; Song, X.; Chu, J.; Li, J.; Chen, Z.J. The Rice Circadian Clock Regulates Tiller Growth and Panicle Development Through Strigolactone Signaling and Sugar Sensing. Plant Cell 2020, 32, 3124–3138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sun, X.; Ma, X.; Xu, B.; Zhao, Y.; Ma, Z.; Li, G.; Khan, N.U.; Pan, Y.; Liang, Y.; et al. GNP6, a novel allele of MOC1, regulates panicle and tiller development in rice. Crop J. 2021, 9, 57–67. [Google Scholar] [CrossRef]
- Li, G.; Tang, J.; Zheng, J.; Chu, C. Exploration of rice yield potential: Decoding agronomic and physiological traits. Crop J. 2021, 9, 577–589. [Google Scholar] [CrossRef]
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y.; et al. MONOCULM 3, an Ortholog of WUSCHEL in Rice, Is Required for Tiller Bud Formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef]
- Zhang, X.; Liang, S.; Luo, B.; Zhou, Z.; Bao, J.; Fang, R.; Wang, F.; Song, X.; Liao, Z.; Chen, G.; et al. Transcriptomic and Metabolomic Investigation on Leaf Necrosis Induced by ZmWus2 Transient Overexpression in Nicotiana benthamiana. Int. J. Mol. Sci. 2023, 24, 11190. [Google Scholar] [CrossRef]
- Huang, G.; Kilic, A.; Karady, M.; Zhang, J.; Mehra, P.; Song, X.; Sturrock, C.J.; Zhu, W.; Qin, H.; Hartman, S.; et al. Ethylene inhibits rice root elongation in compacted soil via ABA- and auxin-mediated mechanisms. Proc. Natl. Acad. Sci. USA 2022, 119, e2093895177. [Google Scholar] [CrossRef]
- Qin, H.; Pandey, B.K.; Li, Y.; Huang, G.; Wang, J.; Quan, R.; Zhou, J.; Zhou, Y.; Miao, Y.; Zhang, D.; et al. Orchestration of ethylene and gibberellin signals determines primary root elongation in rice. Plant Cell 2022, 34, 1273–1288. [Google Scholar] [CrossRef]
- Yin, C.C.; Huang, Y.H.; Zhang, X.; Zhou, Y.; Chen, S.Y.; Zhang, J.S. Ethylene-mediated regulation of coleoptile elongation in rice seedlings. Plant Cell Environ. 2023, 46, 1060–1074. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Sun, F.; Wang, Q.; Chen, M.; Huang, Y.; Feng, Y.Q.; Luo, X.; Yang, J. Rice Ethylene-Response AP2/ERF Factor OsEATB Restricts Internode Elongation by Down-Regulating a Gibberellin Biosynthetic Gene. Plant Physiol. 2011, 157, 216–228. [Google Scholar] [CrossRef]
- Qi, H.; Liang, K.; Ke, Y.; Wang, J.; Yang, P.; Yu, F.; Qiu, F. Advances of Apetala2/Ethylene Response Factors in Regulating Development and Stress Response in Maize. Int. J. Mol. Sci. 2023, 24, 5416. [Google Scholar] [CrossRef] [PubMed]
- Takashima, S.; Abe, T.; Yoshida, S.; Kawahigashi, H.; Saito, T.; Tsuji, S.; Tsujimoto, M. Analysis of Sialyltransferase-Like Proteins from Oryza sativa. J Biochem. 2006, 139, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Dumont, M.; Lehner, A.; Bouton, S.; Kiefer-Meyer, M.C.; Voxeur, A.; Pelloux, J.; Lerouge, P.; Mollet, J.C. The cell wall pectic polymer rhamnogalacturonan-II is required for proper pollen tube elongation: Implications of a putative sialyltransferase-like protein. Ann. Bot. 2014, 114, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]

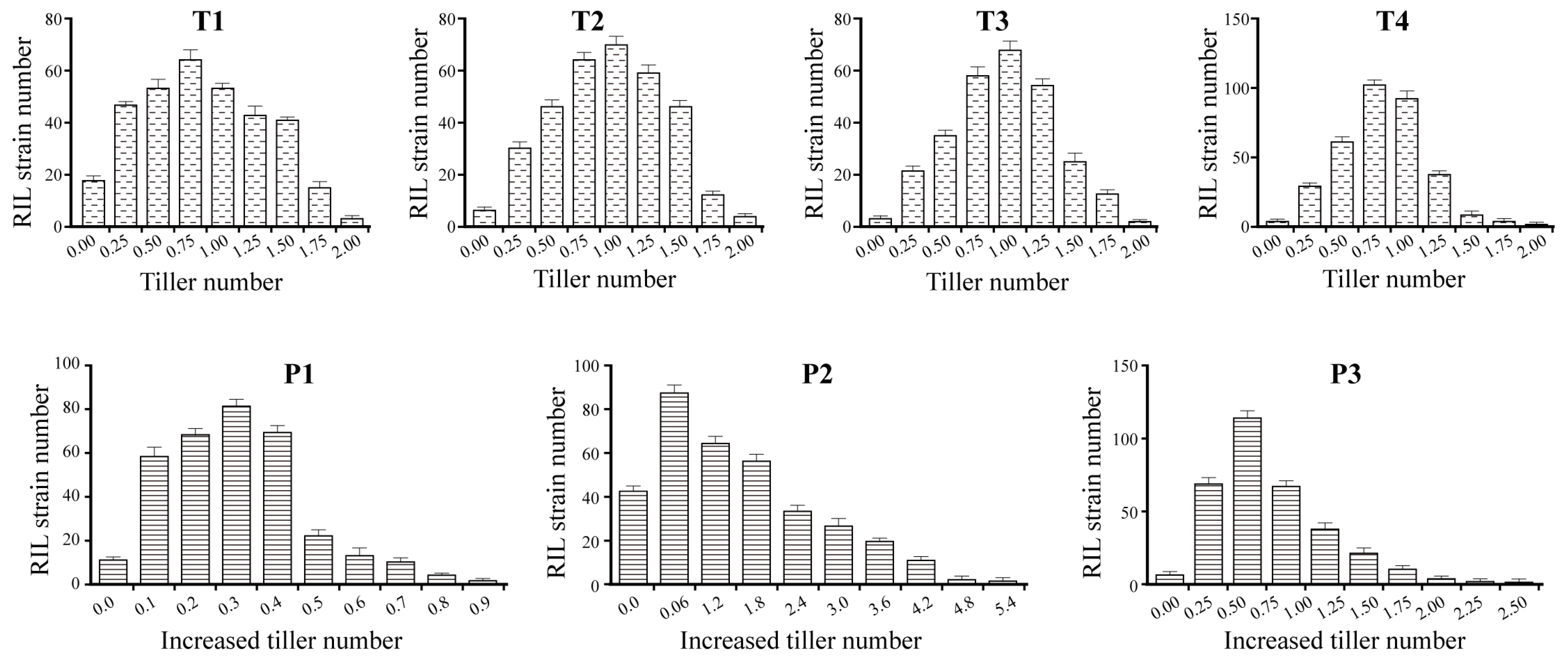

| Traits | Time | Parents | BIL Populations 6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T461 | WYG | t-Test | Mean | SD | Min | Max | Kurtosis | Skewness | |||
| TN 1 | T1 3 | 0.47 | 0.12 | *** 5 | 0.85 | 0.48 | 0 | 1.93 | −0.88 | 0.17 | |
| T2 | 0.61 | 0.15 | *** | 1.15 | 0.52 | 0.04 | 2.53 | −0.68 | 0.02 | ||
| T3 | 0.75 | 0.21 | *** | 1.30 | 0.56 | 0.08 | 2.80 | −0.55 | 0.01 | ||
| T4 | 1.89 | 0.87 | *** | 1.95 | 0.78 | 0.10 | 5.52 | 1.43 | 0.52 | ||
| ITN 2 | P1 4 | 0.14 | 0.03 | *** | 0.30 | 0.17 | 0 | 0.86 | 0.41 | 0.68 | |
| P2 | 0.14 | 0.06 | *** | 0.15 | 0.12 | 0 | 0.54 | −0.08 | 0.77 | ||
| P3 | 1.14 | 0.66 | *** | 0.66 | 0.42 | 0 | 2.90 | 4.66 | 1.64 | ||
| QTL | Chr. 2 | Time | Marker Interval | CI 3 (cM) | Physical Location (Mb) | LOD 4 | PVE 5 (%) | Add 6 |
|---|---|---|---|---|---|---|---|---|
| qETV11 | 1 | T4 | RM11797-RM11873 | 216.2–219.5 | 34.52–36.43 | 3.60 | 4.23 | 0.17 7 |
| 1 | P3 | RM11797-RM11873 | 214.5–219.5 | 34.52–36.43 | 6.18 | 7.96 | 0.11 | |
| qETV3-1 | 3 | P1 | RM15861-RM15795 | 120.5–134.5 | 29.99–28.81 | 4.11 | 4.50 | −0.04 |
| qETV3-2 | 3 | P2 | RM15795-RM15711 | 127.5–136.5 | 28.81–27.60 | 3.52 | 3.23 | −0.03 |
| qETV4-1 | 4 | P2 | RM17079_1-RM17138 | 2.5–19.5 | 22.63–24.02 | 4.26 | 6.38 | 0.04 |
| qETV4-2 | 4 | T3 | RM17138_1-RM17303 | 24.5–37.5 | 24.02–27.27 | 7.57 | 12.98 | 0.18 |
| qETV4-3 | 4 | T2 | RM17303-RM17445 | 38.5–52.5 | 27.27–30.36 | 4.67 | 6.02 | 0.16 |
| 4 | T4 | RM17303-RM17445_1 | 38.5–53.5 | 27.27–30.36 | 3.76 | 6.15 | 0.20 | |
| 4 | P1 | RM17303_1-RM17445 | 42.5–58.5 | 27.27–30.36 | 4.60 | 6.52 | 0.05 | |
| qETV9 | 9 | P3 | RM24631-RM24331_1 | 125.5–137.5 | 19.79–21.41 | 4.94 | 9.42 | 0.13 |
| QTLs | Chr. | Start (bp) | End (bp) | Interval (Mb) | Peak |
|---|---|---|---|---|---|
| bETV1 | 1 | 20,650,001 | 23,510,000 | 2.86 | 0.46 |
| bETV2 | 2 | 30,030,001 | 32,740,000 | 2.71 | 0.47 |
| bETV4-1 | 4 | 21,700,001 | 26,760,000 | 5.06 | 0.50 |
| bETV4-2 | 4 | 33,300,001 | 35,340,000 | 2.04 | 0.43 |
| bETV5 | 5 | 15,510,001 | 17,590,000 | 2.08 | 0.43 |
| bETV7 | 7 | 25,310,001 | 29,697,621 | 4.39 | 0.45 |
| bETV9 | 9 | 11,840,001 | 14,360,000 | 2.52 | −0.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Zhang, T.-T.; You, L.-L.; Wang, Z.-Y.; Du, S.-Q.; Song, H.-Y.; Wang, Z.-H.; Huang, Y.-J.; Liao, J.-L. The QTL and Candidate Genes Regulating the Early Tillering Vigor Traits of Late-Season Rice in Double-Cropping Systems. Int. J. Mol. Sci. 2024, 25, 1497. https://doi.org/10.3390/ijms25031497
Wu W, Zhang T-T, You L-L, Wang Z-Y, Du S-Q, Song H-Y, Wang Z-H, Huang Y-J, Liao J-L. The QTL and Candidate Genes Regulating the Early Tillering Vigor Traits of Late-Season Rice in Double-Cropping Systems. International Journal of Molecular Sciences. 2024; 25(3):1497. https://doi.org/10.3390/ijms25031497
Chicago/Turabian StyleWu, Wei, Tian-Tian Zhang, Li-Li You, Zi-Yi Wang, Si-Qi Du, Hai-Yan Song, Zao-Hai Wang, Ying-Jin Huang, and Jiang-Lin Liao. 2024. "The QTL and Candidate Genes Regulating the Early Tillering Vigor Traits of Late-Season Rice in Double-Cropping Systems" International Journal of Molecular Sciences 25, no. 3: 1497. https://doi.org/10.3390/ijms25031497
APA StyleWu, W., Zhang, T.-T., You, L.-L., Wang, Z.-Y., Du, S.-Q., Song, H.-Y., Wang, Z.-H., Huang, Y.-J., & Liao, J.-L. (2024). The QTL and Candidate Genes Regulating the Early Tillering Vigor Traits of Late-Season Rice in Double-Cropping Systems. International Journal of Molecular Sciences, 25(3), 1497. https://doi.org/10.3390/ijms25031497







