Exosomal Osteoclast-Derived miRNA in Rheumatoid Arthritis: From Their Pathogenesis in Bone Erosion to New Therapeutic Approaches
Abstract
1. Introduction
1.1. Introduction to Rheumatoid Arthritis
1.2. The Osteoclasts
1.3. Exosomal miRNAs
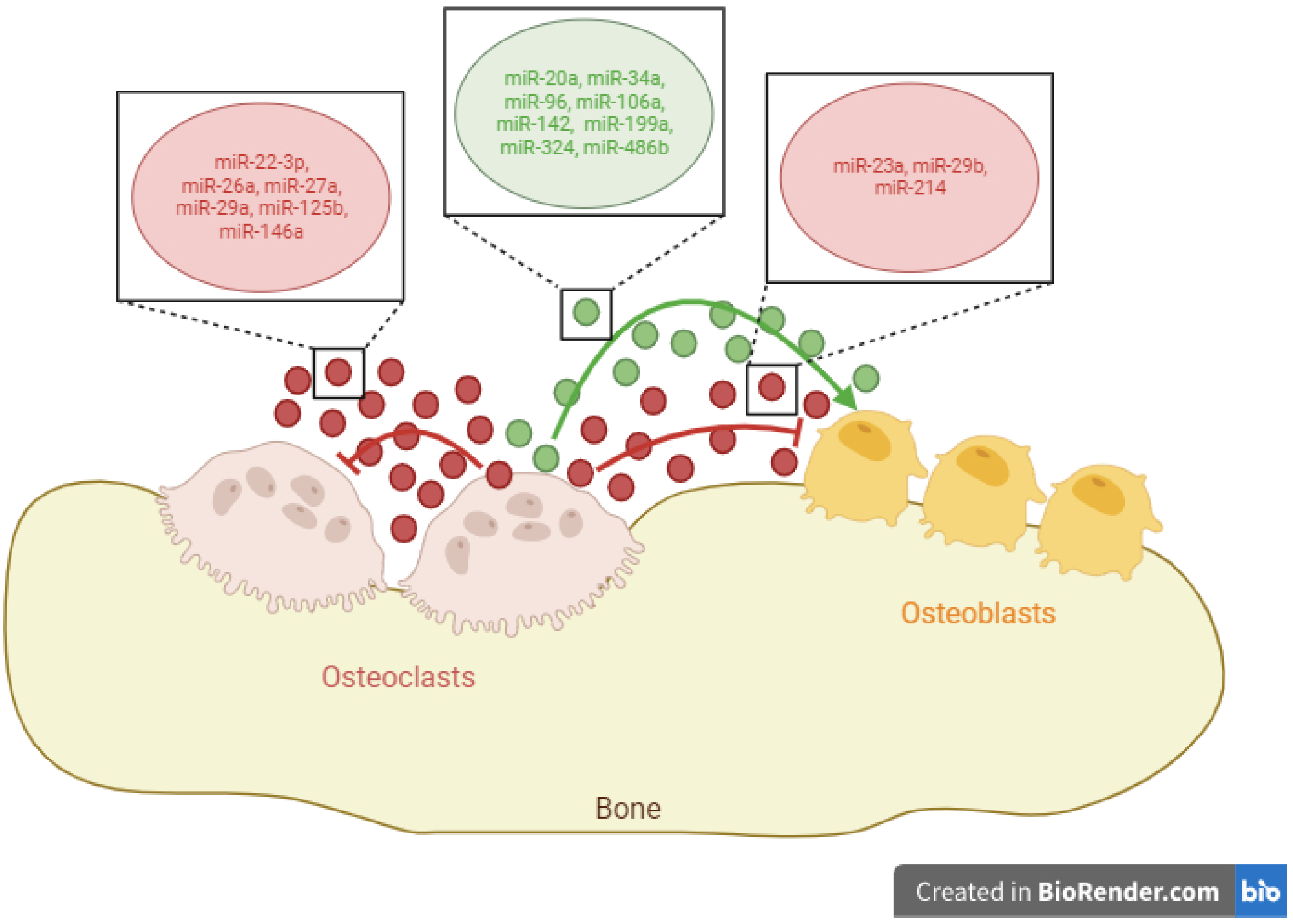
2. miRNAs Released by Osteoclasts
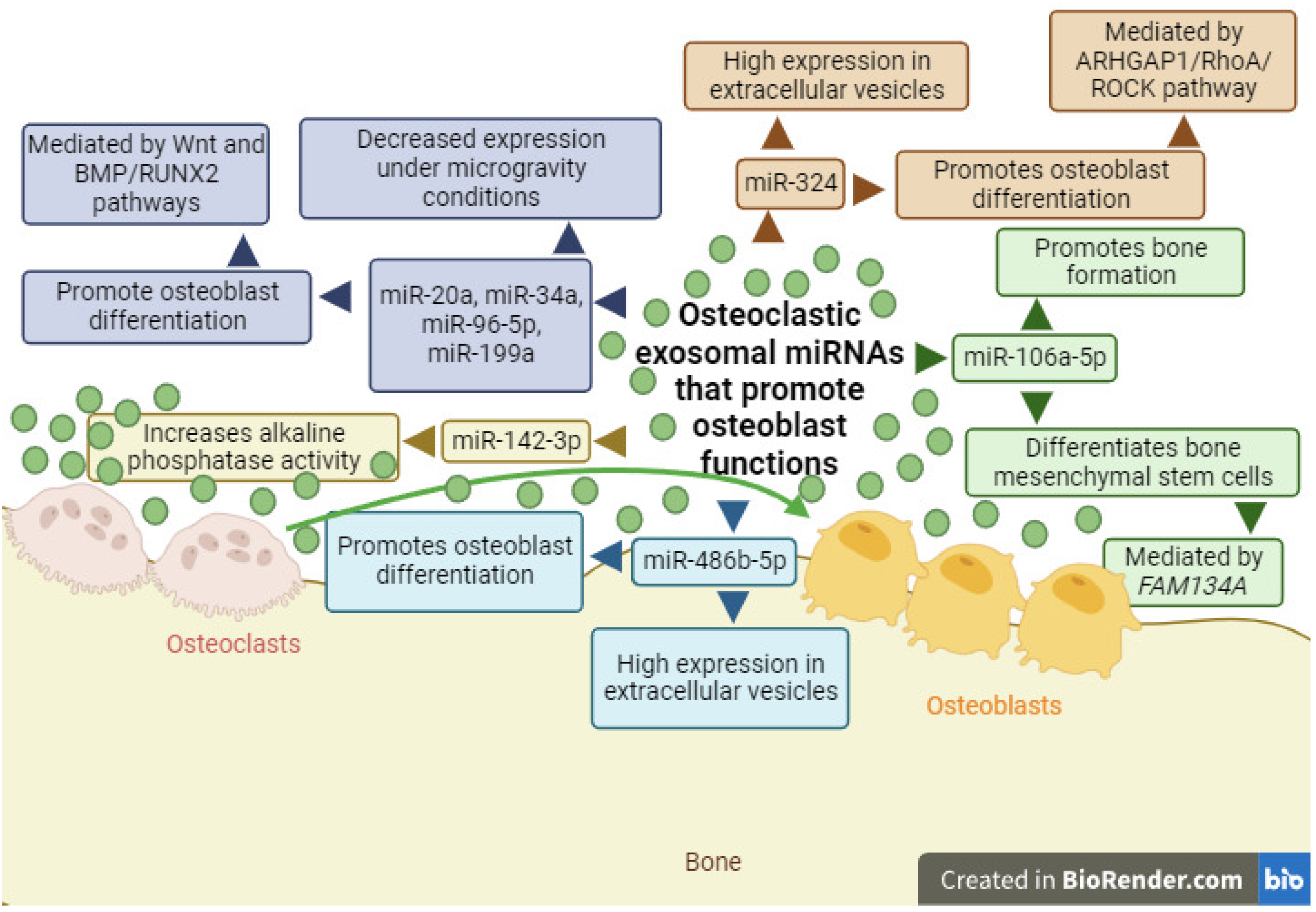
2.1. Exosomal miRNAs with the Ability to Promote Osteoblast Differentiation
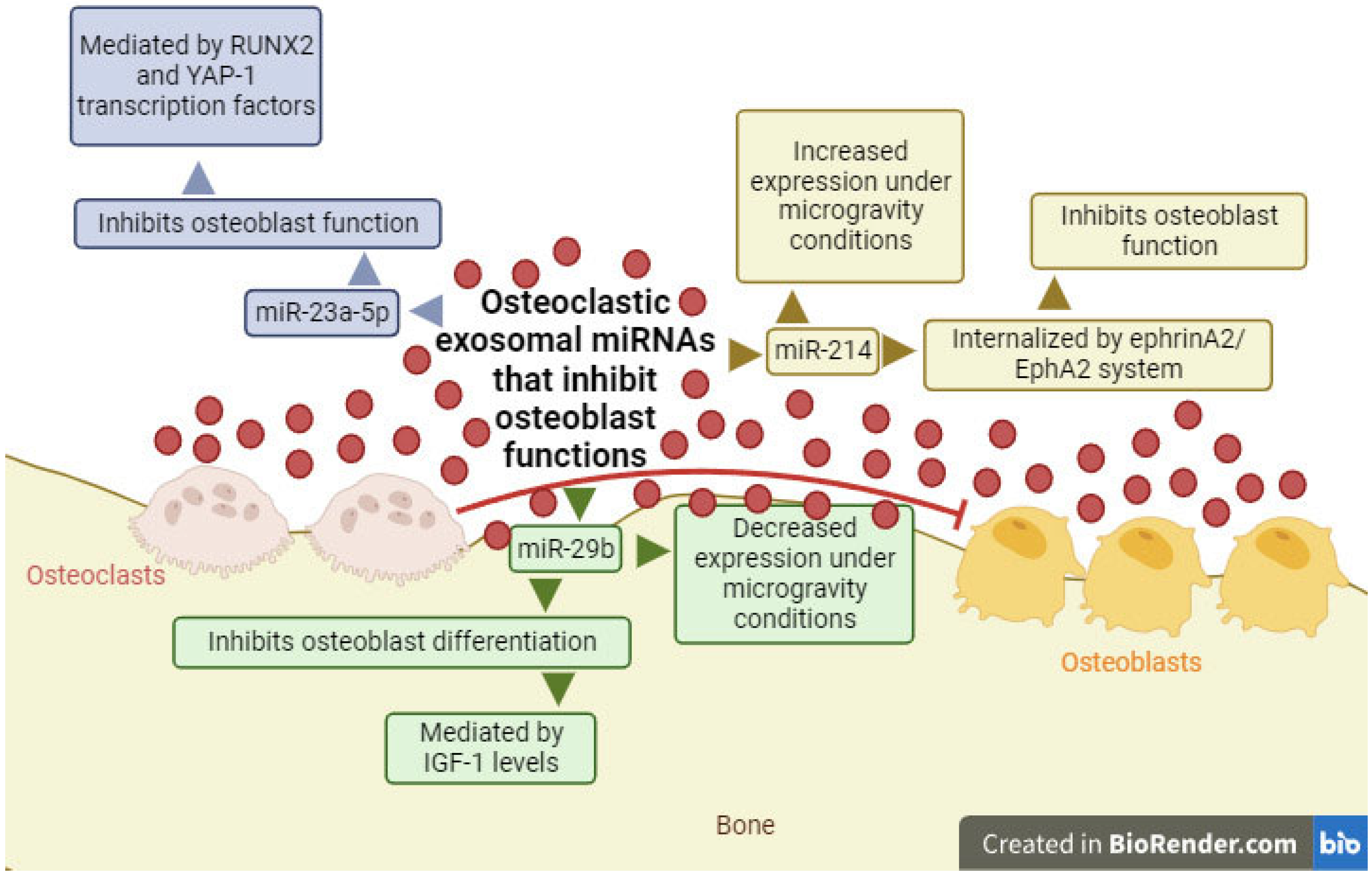
2.2. Exosomal miRNAs with the Ability to Inhibit Osteoblast Differentiation
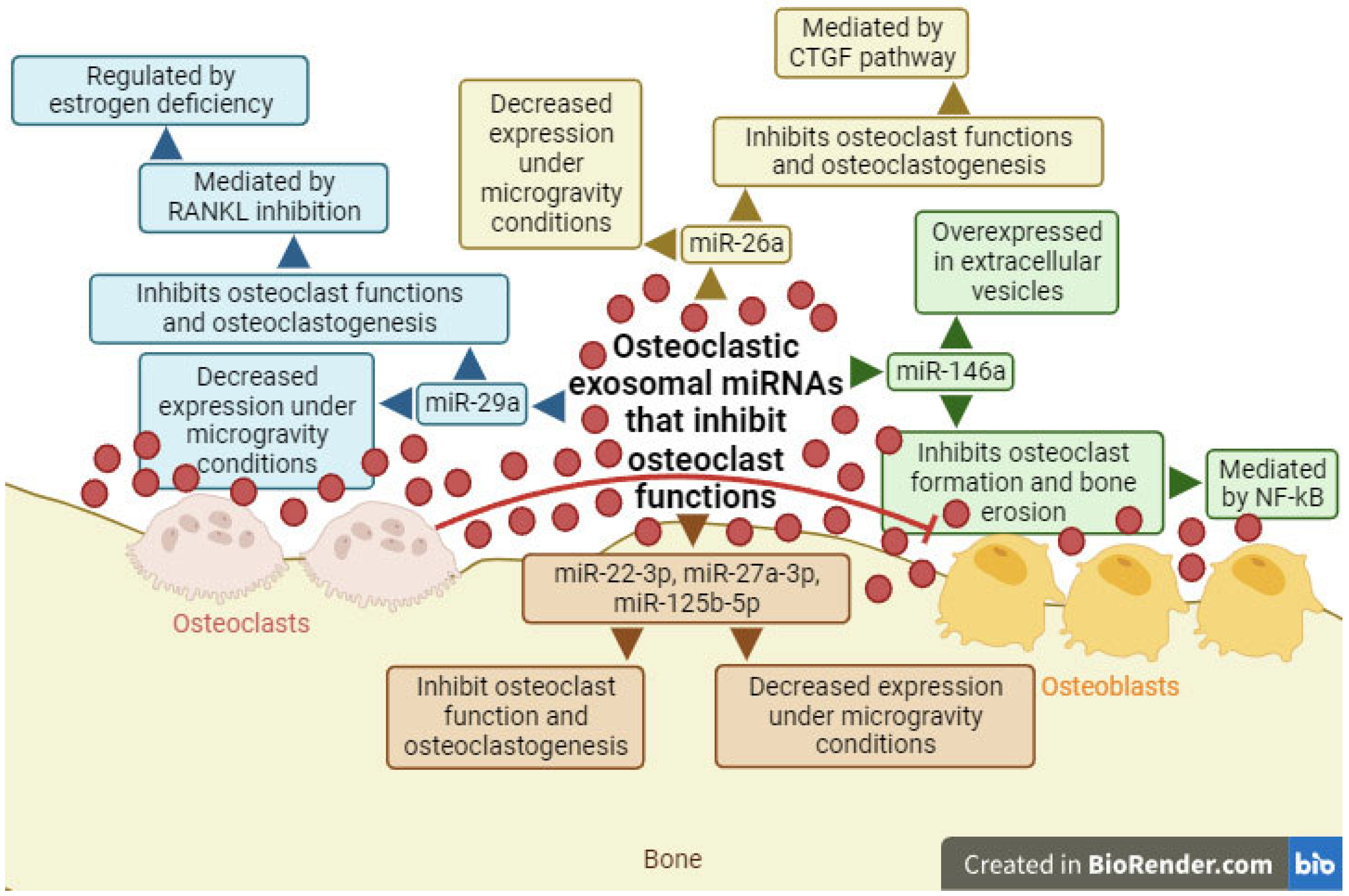
2.3. Exosomal miRNAs with the Ability to Inhibit Osteoclast Differentiation
3. Possible Implications in Patients with RA
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Almutairi, K.; Nossent, J.; Preen, D.; Keen, H.; Inderjeeth, C. The global prevalence of rheumatoid arthritis: A meta-analysis based on a systematic review. Rheumatol. Int. 2021, 41, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Goemaere, S.; Ackerman, C.; Goethals, K.; De Keyser, F.; Van Der Straeten, C.; Verbruggen, G.; Mielants, H.; Veys, E. Onset of symptoms of rheumatoid arthritis in relation to age, sex and menopausal transition. J. Rheumatol. 1990, 17, 1620–1622. [Google Scholar] [CrossRef] [PubMed]
- van Vollenhoven, R.F. Sex differences in rheumatoid arthritis: More than meets the eye…. BMC Med. 2009, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Sudoł-Szopińska, I.; Kontny, E.; Maśliński, W.; Prochorec-Sobieszek, M.; Kwiatkowska, B.; Zaniewicz-Kaniewska, K.; Warczyńska, A. The pathogenesis of rheumatoid arthritis in radiological studies. Part I: Formation of inflammatory infiltrates within the synovial membrane. J. Ultrason. 2012, 12, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Elshabrawy, H.A.; Chen, Z.; Volin, M.V.; Ravella, S.; Virupannavar, S.; Shahrara, S. The pathogenic role of angiogenesis in rheumatoid arthritis. Angiogenesis 2015, 18, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.; Gallicchio, V.S. Stem Cell Therapy as a Treatment Method for Rheumatoid Arthritis. Stem Cell Regen. Med. 2020, 4, 1–10. [Google Scholar] [CrossRef]
- Smolen, J.S.; Landewé, R.B.M.; Bergstra, S.A.; Kerschbaumer, A.; Sepriano, A.; Aletaha, D.; Caporali, R.; Edwards, C.J.; Hyrich, K.L.; Pope, J.E.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2022 update. Ann. Rheum. Dis. 2023, 82, 3–18. [Google Scholar] [CrossRef]
- Larid, G.; Pancarte, M.; Offer, G.; Clavel, C.; Martin, M.; Pradel, V.; Auger, I.; Lafforgue, P.; Roudier, J.; Serre, G.; et al. In Rheumatoid Arthritis Patients, HLA-DRB1*04:01 and Rheumatoid Nodules Are Associated with ACPA to a Particular Fibrin Epitope. Front. Immunol. 2021, 12, 692041. [Google Scholar] [CrossRef]
- Yélamos, J.; García-Lozano, J.R.; Moreno, I.; Aguilera, I.; González, M.F.; Garcia, A.; Núñez-Roldán, A.; Sanchez, B.; Inmunología, S.; Reumatologia, S. Association of HLA-DR4-Dw15 (DRB1*0405) and DR10 with rheumatoid arthritis in a Spanish population. Arthritis Rheum. 1993, 36, 811–814. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Luo, Q.; Gao, Y.; Zhang, L.; Rao, J.; Guo, Y.; Huang, Z.; Li, J. Decreased ALKBH5, FTO, and YTHDF2 in Peripheral Blood Are as Risk Factors for Rheumatoid Arthritis. BioMed Res. Int. 2020, 2020, 5735279. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Tong, Y.; Wu, L.; Niu, H.; Li, Y.; Su, L.C.; Wu, Y.; Bozec, A.; Zaiss, M.M.; Qing, P.; et al. Alteration of gut microbiota in high-risk individuals for rheumatoid arthritis is associated with disturbed metabolome and initiates arthritis by triggering mucosal immunity imbalance. Arthritis Rheumatol. 2023, 75, 1736–1748. [Google Scholar] [CrossRef] [PubMed]
- Maresz, K.J.; Hellvard, A.; Sroka, A.; Adamowicz, K.; Bielecka, E.; Koziel, J.; Gawron, K.; Mizgalska, D.; Marcinska, K.A.; Benedyk, M.; et al. Porphyromonas gingivalis Facilitates the Development and Progression of Destructive Arthritis through Its Unique Bacterial Peptidylarginine Deiminase (PAD). PLoS Pathog. 2013, 9, e1003627. [Google Scholar] [CrossRef] [PubMed]
- Hedström, A.K.; Stawiarz, L.; Klareskog, L.; Alfredsson, L. Smoking and susceptibility to rheumatoid arthritis in a Swedish population-based case–control study. Eur. J. Epidemiol. 2018, 33, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Klareskog, L.; Stolt, P.; Lundberg, K.; Källberg, H.; Bengtsson, C.; Grunewald, J.; Rönnelid, J.; Harris, H.E.; Ulfgren, A.-K.; Rantapää-Dahlqvist, S.; et al. A new model for an etiology of rheumatoid arthritis. Smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006, 54, 38–46. [Google Scholar] [CrossRef]
- Gazitt, T.; Lood, C.; Elkon, K.B. Citrullination in Rheumatoid Arthritis—A Process Promoted by Neutrophil Lysis? Rambam Maimonides Med. J. 2016, 7, e0027. [Google Scholar] [CrossRef]
- Kanmert, D.; Kastbom, A.; Almroth, G.; Skogh, T.; Enander, K.; Wetterö, J. IgG rheumatoid factors against the four human Fc-gamma subclasses in early rheumatoid arthritis (The Swedish TIRA Project). Scand. J. Immunol. 2011, 75, 115–119. [Google Scholar] [CrossRef]
- Rantapää-Dahlqvist, S.; De Jong, B.A.W.; Berglin, E.; Hallmans, G.; Wadell, G.; Stenlund, H.; Sundin, U.; Van Venrooij, W.J. Antibodies Against Cyclic Citrullinated Peptide and IgA Rheumatoid Factor Predict the Development of Rheumatoid Arthritis. Arthritis Rheum. 2003, 48, 2741–2749. [Google Scholar] [CrossRef]
- Sempere-Ortells, J.M.; Pérez-García, V.; Marín-Alberca, G.; Peris-Pertusa, A.; Benito, J.M.; Marco, F.M.; Zubcoff, J.J.; Navarro-Blasco, F.J. Quantification and phenotype of regulatory T cells in rheumatoid arthritis according to Disease Activity Score-28. Autoimmunity 2009, 42, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Carvalheiro, H.; Duarte, C.; Silva-Cardoso, S.; Da Silva, J.A.P.; Souto-Carneiro, M.M. CD8+ T cell profiles in patients with rheumatoid arthritis and their relationship to disease activity. Arthritis Rheumatol. 2015, 67, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.H.; Levescot, A.; Nelson-Maney, N.; Blaustein, R.B.; Winden, K.D.; Morris, A.; Wactor, A.; Balu, S.; Grieshaber-Bouyer, R.; Wei, K.; et al. Arthritis flares mediated by tissue-resident memory T cells in the joint. Cell Rep. 2021, 37, 1–22. [Google Scholar] [CrossRef]
- Kanjana, K.; Chevaisrakul, P.; Matangkasombut, P.; Paisooksantivatana, K.; Lumjiaktase, P. Inhibitory activity of FOXP3+ regulatory T cells reveals high specificity for displaying immune tolerance in remission state rheumatoid arthritis. Sci. Rep. 2020, 10, 19789. [Google Scholar] [CrossRef] [PubMed]
- Fresneda Alarcon, M.; McLaren, Z.; Wright, H.L. Neutrophils in the Pathogenesis of Rheumatoid Arthritis and Systemic Lupus Erythematosus: Same Foe Different M.O. Front. Immunol. 2021, 12, 649693. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shang, X.; Qi, X.; Ba, D.; Lv, J.; Zhou, X.; Wang, H.; Shaxika, N.; Wang, J.; Ma, X. Clinical Significance of M1/M2 Macrophages and Related Cytokines in Patients with Spinal Tuberculosis. Dis. Markers 2020, 2020, 2509454. [Google Scholar] [CrossRef] [PubMed]
- Fukui, S.; Iwamoto, N.; Takatani, A.; Igawa, T.; Shimizu, T.; Umeda, M.; Nishino, A.; Horai, Y.; Hirai, Y.; Koga, T.; et al. M1 and M2 Monocytes in rheumatoid arthritis: A contribution of imbalance of M1/M2 monocytes to osteoclastogenesis. Front. Immunol. 2018, 8, 1958. [Google Scholar] [CrossRef] [PubMed]
- Al-Saadany, H.M.; Hussein, M.S.; Gaber, R.A.; Zaytoun, H.A. Th-17 cells and serum IL-17 in rheumatoid arthritis patients: Correlation with disease activity and severity. Egypt. Rheumatol. 2016, 38, 1–7. [Google Scholar] [CrossRef]
- Hao, Y.; Xie, L.; Xia, J.; Liu, Z.; Yang, B.; Zhang, M. Plasma interleukin-21 levels and genetic variants are associated with susceptibility to rheumatoid arthritis. BMC Musculoskelet. Disord. 2021, 22, 246. [Google Scholar] [CrossRef]
- Lioté, F.; Boval-Boizard, B.; Weill, D.; Kuntz, D.; Wautier, J.-L. Blood monocyte activation in rheumatoid arthritis: Increased monocyte adhesiveness, integrin expression, and cytokine release. Clin. Exp. Immunol. 1996, 106, 13–19. [Google Scholar] [CrossRef]
- Melis, L.; Vandooren, B.; Kruithof, E.; Jacques, P.; De Vos, M.; Mielants, H.; Verbruggen, G.; De Keyser, F.; Elewaut, D. Systemic levels of IL-23 are strongly associated with disease activity in rheumatoid arthritis but not spondyloarthritis. Ann. Rheum. Dis. 2010, 69, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.-T.; Sun, Y.-H.; Zong, S.-H.; Xiang, Y.-B. Serum levels of IL-6 and TNF-α may correlate with activity and severity of rheumatoid arthritis. Med. Sci. Monit. 2015, 21, 4030–4038. [Google Scholar] [CrossRef] [PubMed]
- Pap, T.; Van Der Laan, W.H.; Aupperle, K.R.; Gay, R.E.; Verheijen, J.H.; Firestein, G.S.; Gay, S.; Neidhart, M. Modulation of fibroblast-mediated cartilage degradation by articular chondrocytes in rheumatoid arthritis. Arthritis Rheum. 2000, 43, 2531–2536. [Google Scholar] [CrossRef] [PubMed]
- Chabaud, M.; Lubberts, E.; Joosten, L.; Van Den Berg, W.; Miossec, P. IL-17 derived from juxta-articular bone and synovium contributes to joint degradation in rheumatoid arthritis. Arthritis Res. 2001, 3, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Johnatty, R.N.; Taub, D.D.; Reeder, S.P.; Turcovski-Corrales, S.M.; Cottam, D.W.; Stephenson, T.J.; Rees, R.C. Cytokine and chemokine regulation of proMMP-9 and TIMP-1 production by human peripheral blood lymphocytes. J. Immunol. 1997, 158, 2327–2333. [Google Scholar] [CrossRef] [PubMed]
- Hembry, R.M.; Bagga, M.R.; Reynolds, J.J.; Hamblen, D.L. Immunolocalisation studies on six matrix metalloproteinases and their inhibitors, TIMP-1 and TIMP-2, in synovia from patients with osteo- and rheumatoid arthritis. Ann. Rheum. Dis. 1995, 54, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, Y.; Nakamura, H.; Obata, K.; Yamada, H.; Hayakawa, T.; Fujikawa, K.; Okada, Y. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in synovial fluids from patients with rheumatoid arthritis or osteoarthritis. Ann. Rheum. Dis. 2000, 59, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Goldring, S.R.; Goldring, M.B. The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin. Orthop. Relat. Res. 2004, 427, S27–S36. [Google Scholar] [CrossRef]
- Chen, M.S.; Lin, C.Y.; Chiu, Y.H.; Chen, C.P.; Tsai, P.J.; Wang, H.S. IL-1β-induced matrix metalloprotease-1 promotes mesenchymal stem cell migration via PAR1 and G-protein-coupled signaling pathway. Stem Cells Int. 2018, 2018, 3524759. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Kuo, C.T.; Lin, C.C.; Hsieh, H.L.; Yang, C.M. IL-1β induces expression of matrix metalloproteinase-9 and cell migration via a c-Src-dependent, growth factor receptor transactivation in A549 cells. Br. J. Pharmacol. 2010, 160, 1595–1610. [Google Scholar] [CrossRef]
- Roodman, G.D.; Ibbotson, K.J.; MacDonald, B.R.; Kuehl, T.J.; Mundy, G.R. 1,25-dihydroxyvitamin D3 causes formation of multinucleated cells with several osteoclast characteristics in cultures of primate marrow. Proc. Natl. Acad. Sci. USA 1985, 82, 8213–8217. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Cai, X.; Ren, F.; Ye, Y.; Wang, F.; Zheng, C.; Qian, Y.; Zhang, M. The Macrophage-Osteoclast Axis in Osteoimmunity and Osteo-Related Diseases. Front. Immunol. 2021, 12, 664871. [Google Scholar] [CrossRef] [PubMed]
- Sims, N.A.; Martin, T.J. Osteoclasts Provide Coupling Signals to Osteoblast Lineage Cells through Multiple Mechanisms. Annu. Rev. Physiol. 2020, 82, 507–529. [Google Scholar] [CrossRef] [PubMed]
- Swales, C.; Athanasou, N.A.; Knowles, H.J. Angiopoietin-like 4 Is over-expressed in rheumatoid arthritis patients: Association with pathological bone resorption. PLoS ONE 2014, 9, e109524. [Google Scholar] [CrossRef] [PubMed]
- Ritchlin, C.T.; Haas-Smith, S.A.; Li, P.; Hicks, D.G.; Schwarz, E.M. Mechanisms of TNF-α- and RANKL-mediated osteoclastogenesis and bone resorption in psoriatic arthritis. J. Clin. Investig. 2003, 111, 821–831. [Google Scholar] [CrossRef] [PubMed]
- O’ Gradaigh, D.; Ireland, D.; Bord, S.; Compston, J.E. Joint erosion in rheumatoid arthritis: Interactions between tumour necrosis factor α, interleukin 1, and receptor activator of nuclear factor κB ligand (RANKL) regulate osteoclasts. Ann. Rheum. Dis. 2004, 63, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Nakashima, T. Recent advances in osteoclast biology. Histochem. Cell Biol. 2018, 149, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Gao, J.; Wang, L.; Liu, J.; Zhang, L. Regulation of differentiation and generation of osteoclasts in rheumatoid arthritis. Front. Immunol. 2022, 13, 1034050. [Google Scholar] [CrossRef]
- Lehenkari, P.; Hentunen, T.A.; Laitala-Leinonen, T.; Tuukkanen, J.; Väänänen, H.K. Carbonic anhydrase II plays a major role in osteoclast differentiation and bone resorption by effecting the steady state intracellular pH and Ca2+. Exp. Cell Res. 1998, 242, 128–137. [Google Scholar] [CrossRef]
- Lakkakorpi, P.T.; Horton, M.A.; Helfrich, M.H.; Karhukorpi, E.K.; Väänanen, H.K. Vitronectin receptor has a role in bone resorption but does not mediate tight sealing zone attachment of osteoclasts to the bone surface. J. Cell Biol. 1991, 115, 1179–1186. [Google Scholar] [CrossRef]
- Keller, J.; Catala-Lehnen, P.; Huebner, A.K.; Jeschke, A.; Heckt, T.; Lueth, A.; Krause, M.; Koehne, T.; Albers, J.; Schulze, J.; et al. Calcitonin controls bone formation by inhibiting the release of sphingosine 1-phosphate from osteoclasts. Nat. Commun. 2014, 5, 5215. [Google Scholar] [CrossRef] [PubMed]
- Kotake, S.; Udagawa, N.; Hakoda, M.; Mogi, M.; Yano, K.; Tsuda, E.; Takahashi, K.; Furuya, T.; Ishiyama, S.; Kim, K.-J.; et al. Activated human T cells directly induce osteoclastogenesis from human monocytes. Possible role of T cells in bone destruction in rheumatoid arthritis patients. Arthritis Rheum. 2001, 44, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, R.; Rasool, M. Interleukin 17 regulates SHP-2 and IL-17RA/STAT-3 dependent Cyr61, IL-23 and GM-CSF expression and RANKL mediated osteoclastogenesis by fibroblast-like synoviocytes in rheumatoid arthritis. Mol. Immunol. 2017, 91, 134–144. [Google Scholar] [CrossRef] [PubMed]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, H.; Shima, N.; Nakagawa, N.; Mochizuki, S.I.; Yano, K.; Fujise, N.; Sato, Y.; Goto, M.; Yamaguchi, K.; Kuriyama, M.; et al. Identity of osteoclastogenesis inhibitory factor (OCIF) and osteoprotegerin (OPG): A mechanism by which OPG/OCIF inhibits osteoclastogenesis in vitro. Endocrinology 1998, 139, 1329–1337. [Google Scholar] [CrossRef]
- Meednu, N.; Zhang, H.; Owen, T.; Sun, W.; Wang, V.; Cistrone, C.; Rangel-Moreno, J.; Xing, L.; Anolik, J.H. Production of RANKL by memory B cells. A link between B cells and bone erosion in rheumatoid arthritis. Arthritis Rheumatol. 2016, 68, 805–816. [Google Scholar] [CrossRef]
- Fessler, J.; Husic, R.; Schwetz, V.; Lerchbaum, E.; Aberer, F.; Fasching, P.; Ficjan, A.; Obermayer-Pietsch, B.; Duftner, C.; Graninger, W.; et al. Senescent T-cells promote bone loss in rheumatoid arthritis. Front. Immunol. 2018, 9, 95. [Google Scholar] [CrossRef]
- Polzer, K.; Joosten, L.; Gasser, J.; Distler, J.H.; Ruiz, G.; Baum, W.; Redlich, K.; Bobacz, K.; Smolen, J.S.; Van Den Berg, W.; et al. Interleukin-1 is essential for systemic inflammatory bone loss. Ann. Rheum. Dis. 2010, 69, 284–290. [Google Scholar] [CrossRef]
- Miossec, P.; Chomarat, P.; Dechanet, J.; Moreau, J.-F.; Roux, J.-P.; Delmas, P.; Banchereau, J. Interleukin-4 inhibits bone resorption through an effect on osteoclasts and proinflammatory cytokines in an ex vivo model of bone resorption in rheumatoid arthritis. Arthritis Rheum. 1994, 37, 1715–1722. [Google Scholar] [CrossRef]
- Abdel Meguid, M.H.; Hamad, Y.H.; Swilam, R.S.; Barakat, M.S. Relation of interleukin-6 in rheumatoid arthritis patients to systemic bone loss and structural bone damage. Rheumatol. Int. 2013, 33, 697–703. [Google Scholar] [CrossRef]
- Kaji, H.; Sugimoto, T.; Kanatani, M.; Fukase, M.; Kumegawa, M.; Chihara, K. Prostaglandin E, stimulates osteoclast-like cell formation and bone-resorbing activity via osteoblasts: Role of cAMP-dependent protein kinase. J. Bone Miner. Res. 1996, 11, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Kogawa, M.; Findlay, D.M.; Anderson, P.H.; Ormsby, R.; Vincent, C.; Morris, H.A.; Atkins, G.J. Osteoclastic metabolism of 25(OH)-vitamin D3: A potential mechanism for optimization of bone resorption. Endocrinology 2010, 151, 4613–4625. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.; Owens, J.M.; Chambers, T.J. Induction of osteoclast formation by parathyroid hormone depends on an action on stromal cells. J. Endocrinol. 1998, 158, 341–350. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Niida, A.; Hiroko, T.; Kasai, M.; Furukawa, Y.; Nakamura, Y.; Suzuki, Y.; Sugano, S.; Akiyama, T. DKK1, a negative regulator of Wnt signaling, is a target of the β-catenin/TCF pathway. Oncogene 2004, 23, 8520–8526. [Google Scholar] [CrossRef] [PubMed]
- Ruaro, B.; Casabella, A.; Paolino, S.; Pizzorni, C.; Ghio, M.; Seriolo, C.; Molfetta, L.; Odetti, P.; Smith, V.; Cutolo, M. Dickkopf-1 (Dkk-1) serum levels in systemic sclerosis and rheumatoid arthritis patients: Correlation with the Trabecular Bone Score (TBS). Clin. Rheumatol. 2018, 37, 3057–3062. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yin, Y.; Yao, L.; Lin, Z.; Sun, S.; Zhang, J.; Li, X. TNF-α treatment increases DKK1 protein levels in primary osteoblasts via upregulation of DKK1 mRNA levels and downregulation of miR-335-5p. Mol. Med. Rep. 2020, 22, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Lee, K.A.; Yoon, H.B.; Yoo, S.A.; Park, Y.W.; Chung, Y.; Kim, W.U.; Kang, C.Y. The Wnt inhibitor secreted Frizzled-Related Protein 1 (sFRP1) promotes human Th17 differentiation. Eur. J. Immunol. 2012, 42, 2564–2573. [Google Scholar] [CrossRef]
- Schett, G. Cells of the synovium in rheumatoid arthritis. Osteoclasts. Arthritis Res. Ther. 2007, 9, 203. [Google Scholar] [CrossRef]
- Rhee, D.K.; Marcelino, J.; Baker, M.; Gong, Y.; Smits, P.; Lefebvre, V.; Jay, G.D.; Stewart, M.; Wang, H.; Warman, M.L.; et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J. Clin. Investig. 2005, 115, 622–631. [Google Scholar] [CrossRef]
- Chia, S.-L.; Sawaji, Y.; Burleigh, A.; McLean, C.; Inglis, J.; Saklatvala, J.; Vincent, T. Fibroblast growth factor 2 is an intrinsic chondroprotective agent that suppresses ADAMTS-5 and delays cartilage degradation in murine osteoarthritis. Arthritis Rheum. 2009, 60, 2019–2027. [Google Scholar] [CrossRef]
- Majumdar, M.K.; Askew, R.; Schelling, S.; Stedman, N.; Blanchet, T.; Hopkins, B.; Morris, E.A.; Glasson, S.S. Double-knockout of ADAMTS-4 and ADAMTS-5 in mice results in physiologically normal animals and prevents the progression of osteoarthritis. Arthritis Rheum. 2007, 56, 3670–3674. [Google Scholar] [CrossRef] [PubMed]
- Haeusler, G.; Walter, I.; Helmreich, M.; Egerbacher, M. Localization of matrix metalloproteinases, (MMPs) their tissue inhibitors, and vascular endothelial growth factor (VEGF) in growth plates of children and adolescents indicates a role for MMPs in human postnatal growth and skeletal maturation. Calcif. Tissue Int. 2005, 76, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; Van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Nederveen, J.P.; Warnier, G.; Di Carlo, A.; Nilsson, M.I.; Tarnopolsky, M.A. Extracellular Vesicles and Exosomes: Insights from Exercise Science. Front. Physiol. 2021, 11, 604274. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.-H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Elsharkawi, F.; Elsabah, M.; Shabayek, M.; Khaled, H. Urine and serum exosomes as novel biomarkers in detection of bladder cancer. Asian Pac. J. Cancer Prev. 2019, 20, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.-L.; Cao, Y.-H.; Ni, H.; Xu, M.; Liu, D.; Liu, H.; Chen, P.; Liu, B.-C. MicroRNA-29c in urinary exosome/microvesicle as a biomarker of renal fibrosis. Am. J. Physiol. Ren. Physiol. 2013, 305, F1220–F1227. [Google Scholar] [CrossRef]
- Koga, Y.; Yasunaga, M.; Moriya, Y.; Akasu, T.; Fujita, S.; Yamamoto, S.; Matsumura, Y. Exosome can prevent RNase from degrading microRNA in feces. J. Gastrointest. Oncol. 2011, 2, 215–222. [Google Scholar] [CrossRef]
- Valenzuela, M.M.A.; Ferguson Bennit, H.R.; Gonda, A.; Diaz Osterman, C.J.; Hibma, A.; Khan, S.; Wall, N.R. Exosomes secreted from human cancer cell lines contain inhibitors of apoptosis (IAP). Cancer Microenviron. 2015, 8, 65–73. [Google Scholar] [CrossRef]
- Blenkiron, C.; Simonov, D.; Muthukaruppan, A.; Tsai, P.; Dauros, P.; Green, S.; Hong, J.; Print, C.G.; Swift, S.; Phillips, A.R. Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA. PLoS ONE 2016, 11, e0160440. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Omar, O.; Vazirisani, F.; Thomsen, P.; Ekström, K. Mesenchymal stem cell-derived exosomes have altered microRNA profiles and induce osteogenic differentiation depending on the stage of differentiation. PLoS ONE 2018, 13, e0193059. [Google Scholar] [CrossRef] [PubMed]
- Wollert, T.; Wunder, C.; Lippincott-Schwartz, J.; Hurley, J.H. Membrane scission by the ESCRT-III complex. Nature 2009, 458, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, e0193059. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Rong, Y.; Chuang, Y.-S.; Peng, D.; Emr, S.D. Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol. Cell 2015, 57, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 20424. [Google Scholar] [CrossRef]
- Admyre, C.; Grunewald, J.; Thyberg, J.; Gripenbäck, S.; Tornling, G.; Eklund, A.; Scheynius, A.; Gabrielsson, S. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur. Respir. J. 2003, 22, 578–583. [Google Scholar] [CrossRef]
- Cwiklinski, K.; de la Torre-Escudero, E.; Trelis, M.; Bernal, D.; Dufresne, P.J.; Brennan, G.P.; O’Neill, S.; Tort, J.; Paterson, S.; Marcilla, A.; et al. The extracellular vesicles of the helminth pathogen, Fasciola hepatica: Biogenesis pathways and cargo molecules involved in parasite pathogenesis. Mol. Cell. Proteom. 2015, 14, 3258–3273. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Statello, L.; Maugeri, M.; Garre, E.; Nawaz, M.; Wahlgren, J.; Papadimitriou, A.; Lundqvist, C.; Lindfors, L.; Collén, A.; Sunnerhagen, P.; et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE 2018, 13, e0195969. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 2016, 5, e19276. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Murai, C.; Shibata, S.; Munakata, Y.; Ishii, T.; Ishii, K.; Saitoh, T.; Sawai, T.; Sugamura, K.; Sasaki, T. Human parvovirus B19 as a causative agent for rheumatoid arthritis. Proc. Natl. Acad. Sci. USA 1998, 95, 8227–8232. [Google Scholar] [CrossRef] [PubMed]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Fan, W.; Ma, L.; Geng, X. miR-708-5p promotes fibroblast-like synoviocytes’ cell apoptosis and ameliorates rheumatoid arthritis by the inhibition of Wnt3a/β-catenin pathway. Drug Des. Devel. Ther. 2018, 12, 3439–3447. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Zhang, J.; He, H.; Li, J.; Wu, Y.; Shen, Z. MiR-525-3p mediates antiviral defense to rotavirus infection by targeting nonstructural protein 1. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 3212–3225. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yem, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.-T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Görlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Ketting, R.F.; Fischer, S.E.J.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H.A. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes. Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Ladewig, E.; Zhou, L.; Lai, E.C. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes. Dev. 2013, 27, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Szczyrba, J.; Jung, V.; Beitzinger, M.; Nolte, E.; Wach, S.; Hart, M.; Sapich, S.; Wiesehöfer, M.; Taubert, H.; Wennemuth, G.; et al. Analysis of argonaute complex bound mRNAs in DU145 prostate carcinoma cells reveals new miRNA target genes. Prostate Cancer 2017, 2017, 4893921. [Google Scholar] [CrossRef]
- Quévillon Huberdeau, M.; Zeitler, D.M.; Hauptmann, J.; Bruckmann, A.; Fressigné, L.; Danner, J.; Piquet, S.; Strieder, N.; Engelmann, J.C.; Jannot, G.; et al. Phosphorylation of argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. EMBO J. 2017, 36, 2088–2106. [Google Scholar] [CrossRef]
- Ouboussad, L.; Hunt, L.; Hensor, E.M.A.; Nam, J.L.; Barnes, N.A.; Emery, P.; McDermott, M.F.; Buch, M.H. Profiling microRNAs in individuals at risk of progression to rheumatoid arthritis. Arthritis Res. Ther. 2017, 19, 288. [Google Scholar] [CrossRef]
- Yue, J.; Lau, T.C.K.; Griffith, J.F.; Xu, J.; Xiao, F.; Shi, L.; Wang, D.; Wong, P.C.H.; Li, E.K.; Tam, L.P.; et al. Circulating miR-99b-5p as a novel predictor of erosion progression on high-resolution peripheral quantitative computed tomography in early rheumatoid arthritis: A prospective cohort study. Int. J. Rheum. Dis. 2019, 22, 1724–1733. [Google Scholar] [CrossRef]
- Cunningham, C.C.; Wade, S.; Floudas, A.; Orr, C.; McGarry, T.; Wade, S.; Cregan, S.; Fearon, U.; Veale, D.J. Serum miRNA Signature in Rheumatoid Arthritis and “At-Risk Individuals”. Front. Immunol. 2021, 12, 633201. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ai, H.; Zou, Y.; Xu, J. Osteoclast-derived extracellular miR-106a-5p promotes osteogenic differentiation and facilitates bone defect healing. Cell. Signal. 2023, 102, 110549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, G.; Meng, Y.; Lin, H.; Lu, Y. MAG-2 promotes invasion, mobility and adherence capability of lung cancer cells by MMP-2, CD44 and intracellular calcium in vitro. Oncol. Rep. 2009, 21, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Anh, D.J.; Dimai, H.P.; Hall, S.L.; Farley, J.R. Skeletal alkaline phosphatase activity is primarily released from human osteoblasts in an insoluble form, and the net release is inhibited by calcium and skeletal growth factors. Calcif. Tissue Int. 1998, 62, 332–340. [Google Scholar] [CrossRef] [PubMed]
- van Straalen, J.P.; Sanders, E.; Prummel, M.F.; Sanders, G.T.B. Bone-alkaline phosphatase as indicator of bone formation. Clin. Chim. Acta 1991, 201, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Huyan, T.; Du, Y.; Peng, H.; Hu, X.; Wu, C.; Li, Q.; Dong, D.; Li, J.; Shang, P. Evaluation of osteoclast-derived exosomal miRNA under simulated microgravity conditions using next-generation sequencing. Acta Astronaut. 2019, 161, 75–86. [Google Scholar] [CrossRef]
- Chen, J.; Liu, M.; Luo, X.; Peng, L.; Zhao, Z.; He, C.; He, Y. Exosomal miRNA-486-5p derived from rheumatoid arthritis fibroblast-like synoviocytes induces osteoblast differentiation through the Tob1/BMP/Smad pathway. Biomater. Sci. 2020, 8, 3430–3442. [Google Scholar] [CrossRef]
- Liu, H.; Dong, Y.; Feng, X.; Li, L.; Jiao, Y.; Bai, S.; Feng, Z.; Yu, H.; Li, X.; Zhao, Y. MiR-34a promotes bone regeneration in irradiated bone defects by enhancing osteoblastic differentiation of mesenchymal stromal cells in rats. Stem Cell Res. Ther. 2019, 10, 180. [Google Scholar] [CrossRef]
- Ma, S.; Wang, D.D.; Ma, C.Y.; Zhang, Y.D. microRNA-96 promotes osteoblast differentiation and bone formation in ankylosing spondylitis mice through activating the Wnt signaling pathway by binding to SOST. J. Cell. Biochem. 2019, 120, 15429–15442. [Google Scholar] [CrossRef]
- Qi, X.B.; Jia, B.; Wang, W.; Xu, G.H.; Guo, J.C.; Li, X.; Liu, J.N. Role of miR-199a-5p in osteoblast differentiation by targeting TET2. Gene 2020, 726, 144193. [Google Scholar] [CrossRef]
- Wang, Y.X.; Peng, Z.L.; Sun, Z.W.; Pan, Y.J.; Ai, H.; Mai, Z.H. MiR-20a promotes osteogenic differentiation in bone marrow-derived mesenchymal stem/stromal cells and bone repair of the maxillary sinus defect model in rabbits. Front. Bioeng. Biotechnol. 2023, 11, 1127908. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Li, M.; Feng, X.; Zhu, E.; Wang, B. miR-142a-5p promoted osteoblast differentiation via targeting nuclear factor IA. J. Cell. Physiol. 2021, 236, 1810–1821. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Yin, X.; Zhang, S.; Ai, H.; Luo, F.; Xu, J.; Dou, C.; Dong, S.; Ma, Q. Osteoclast-derived small extracellular vesicles induce osteogenic differentiation via inhibiting ARHGAP1. Mol. Ther. Nucleic Acids 2021, 23, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.X.; Xie, P.; Li, Y.S.; Wen, T.; Yang, X.C. Osteoclast-derived miR-23a-5p-containing exosomes inhibit osteogenic differentiation by regulating Runx2. Cell. Signal. 2020, 70, 109504. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Chen, M.; Li, C.; Xu, M.; Liu, Y.; Cong, M.; Sang, N.; Liu, S. Long non-coding RNA MT1DP shunts the cellular defense to cytotoxicity through crosstalk with MT1H and RhoC in cadmium stress. Cell Discov. 2018, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Holliday, L.S.; McHugh, K.P.; Zuo, J.; Aguirre, J.I.; Neubert, J.K.; Rody, W.J., Jr. Exosomes: Novel regulators of bone remodelling and potential therapeutic agents for orthodontics. Orthod. Craniofacial Res. 2017, 20, 95–99. [Google Scholar] [CrossRef]
- Sun, W.; Zhao, C.; Li, Y.; Wang, L.; Nie, G.; Peng, J.; Wang, A.; Zhang, P.; Tian, W.; Li, Q.; et al. Osteoclast-derived microRNA-containing exosomes selectively inhibit osteoblast activity. Cell Discov. 2016, 2, 16015. [Google Scholar] [CrossRef]
- Li, D.; Liu, J.; Guo, B.; Liang, C.; Dang, L.; Lu, C.; He, X.; Cheung, H.Y.S.; Xu, L.; Lu, C.; et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat. Commun. 2016, 7, 10872. [Google Scholar] [CrossRef]
- Zeng, Q.; Wang, Y.; Gao, J.; Yan, Z.; Li, Z.; Zou, X.; Li, Y.; Wang, J.; Guo, Y. miR-29b-3p regulated osteoblast differentiation via regulating IGF-1 secretion of mechanically stimulated osteocytes. Cell. Mol. Biol. Lett. 2019, 24, 11. [Google Scholar] [CrossRef]
- Huynh, N.; Vonmoss, L.; Smith, D.; Rahman, I.; Felemban, M.F.; Zuo, J.; Rody, W.J.; McHugh, K.P.; Holliday, L.S. Characterization of regulatory extracellular vesicles from osteoclasts. J. Dent. Res. 2016, 95, 673–679. [Google Scholar] [CrossRef]
- Stephens, E.; Roy, M.; Bisson, M.; Nguyen, H.D.; Scott, M.S.; Boire, G.; Bouchard, L.; Roux, S. Osteoclast signaling-targeting miR-146a-3p and miR-155-5p are downregulated in Paget’s disease of bone. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165852. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Yang, M.; Hu, W.; Cai, S. Overexpression of miRNA-22-3p attenuates osteoporosis by targeting MAPK14. Exp. Ther. Med. 2021, 22, 692. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, J.H.; Kim, I.; Lee, J.; Seong, S.; Park, Y.; Kim, N. MicroRNA-26a Regulates RANKL-Induced Osteoclast Formation. Mol. Cells 2015, 38, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Lian, W.S.; Ko, J.Y.; Chen, Y.S.; Ke, H.J.; Hsieh, C.K.; Kuo, C.W.; Wang, S.Y.; Huang, B.W.; Tseng, J.G.; Wang, F.S. MicroRNA-29a represses osteoclast formation and protects against osteoporosis by regulating PCAF-mediated RANKL and CXCL12. Cell Death Dis. 2019, 10, 705. [Google Scholar] [CrossRef]
- Minamizaki, T.; Nakao, Y.; Irie, Y.; Ahmed, F.; Itoh, S.; Sarmin, N.; Yoshioka, H.; Nobukiyo, A.; Fujimoto, C.; Niida, S.; et al. The matrix vesicle cargo miR-125b accumulates in the bone matrix, inhibiting bone resorption in mice. Commun. Biol. 2020, 3, 30. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Maruyama, E.O.; Martinez, J.; Lopes, J.; Hsu, T.; Wu, W.; Hsu, W.; Maruyama, T. miRNA-27a is essential for bone remodeling by modulating p62-mediated osteoclast signaling. Elife 2023, 12, e79768. [Google Scholar] [CrossRef]
- Song, M.S.; Rossi, J.J. The anti-miR21 antagomir, a therapeutic tool for colorectal cancer, has a potential synergistic effect by perturbing an angiogenesis-associated miR30. Front. Genet. 2014, 4, 301. [Google Scholar] [CrossRef]
- Sujitha, S.; Rasool, M. Berberine coated mannosylated liposomes curtail RANKL stimulated osteoclastogenesis through the modulation of GSK3β pathway via upregulating miR-23a. Int. Immunopharmacol. 2019, 74, 105703. [Google Scholar] [CrossRef]
- Sujitha, S.; Dinesh, P.; Rasool, M. Berberine modulates ASK1 signaling mediated through TLR4/TRAF2 via upregulation of miR-23a. Toxicol. Appl. Pharmacol. 2018, 359, 34–46. [Google Scholar] [CrossRef]
- Cao, Y.; Tang, S.; Nie, X.; Zhou, Z.; Ruan, G.; Han, W.; Zhu, Z.; Ding, C. Decreased miR-214–3p activates NF-κB pathway and aggravates osteoarthritis progression. eBioMedicine 2021, 65, 103283. [Google Scholar] [CrossRef]
- Ting, K.K.; Zhao, Y.; Shen, W.; Coleman, P.; Yam, M.; Chan-Ling, T.; Li, J.; Moller, T.; Gillies, M.; Vadas, M.A.; et al. Therapeutic regulation of VE-cadherin with a novel oligonucleotide drug for diabetic eye complications using retinopathy mouse models. Diabetologia 2019, 62, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Young, J.A.; Ting, K.K.; Li, J.; Moller, T.; Dunn, L.; Lu, Y.; Lay, A.J.; Moses, J.; Prado-Lourenço, L.; Khachigian, L.M.; et al. Regulation of vascular leak and recovery from ischemic injury by general and VE-cadherin-restricted mirna antagonists of mir-27. Blood 2013, 122, 2911–2919. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.Y.; Yang, K.P.; Li, Z.Z. MiR-22 restrains proliferation of rheumatoid arthritis by targeting IL6R and may be concerned with the suppression of NF-κB pathway. Kaohsiung J. Med. Sci. 2020, 36, 20–26. [Google Scholar] [CrossRef]
- Fan, P.; He, L.; Hu, N.; Luo, J.; Zhang, J.; Mo, L.F.; Wang, Y.H.; Pu, D.; Lv, X.H.; Hao, Z.M.; et al. Effect of 1,25-(OH)2D3 on proliferation of fibroblast-like synoviocytes and expressions of pro-inflammatory cytokines through regulating MicroRNA-22 in a rat model of rheumatoid arthritis. Cell. Physiol. Biochem. 2017, 42, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Cao, S. Role of microRNA-26a in cartilage injury and chondrocyte proliferation and apoptosis in rheumatoid arthritis rats by regulating expression of CTGF. J. Cell. Physiol. 2020, 235, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Zhu, W.; Xu, J.; Wang, B.; Hou, W.; Zhang, R.; Zhong, N.; Ning, Q.; Han, Y.; Yu, H.; et al. MicroRNA-26a negatively regulates toll-like receptor 3 expression of rat macrophages and ameliorates pristane induced arthritis in rats. Arthritis Res. Ther. 2014, 16, R9. [Google Scholar] [CrossRef]
- Bullock, C.H.; McAlpine, S.M.; Roberts, S.E.; Derfalvi, B. MicroRNA-27a-3p enhances the inflammatory phenotype of Juvenile Idiopathic Arthritis fibroblast-like synoviocytes. Pediatr. Rheumatol. 2023, 21, 53. [Google Scholar] [CrossRef]
- Hou, C.; Wang, D.; Zhang, L. MicroRNA-34a-3p inhibits proliferation of rheumatoid arthritis fibroblast-like synoviocytes. Mol. Med. Rep. 2019, 20, 2563–2570. [Google Scholar] [CrossRef]
- Kong, X.H.; Shi, S.F.; Hu, H.J.; Wang, J.X. MicroRNA-20a suppresses RANKL-modulated osteoclastogenesis and prevents bone erosion in mice with rheumatoid arthritis through the TLR4/p38 pathway. J. Biol. Regul. Homeost. Agents 2021, 35, 921–931. [Google Scholar] [CrossRef]
- Philippe, L.; Alsaleh, G.; Pichot, A.; Ostermann, E.; Zuber, G.; Frisch, B.; Sibilia, J.; Pfeffer, S.; Bahram, S.; Wachsmann, D.; et al. MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann. Rheum. Dis. 2013, 72, 1071–1079. [Google Scholar] [CrossRef]
- Bao, W.; Xie, M.; Ye, Y. Age-associated B cells indicate disease activity in rheumatoid arthritis. Cell. Immunol. 2022, 377, 104533. [Google Scholar] [CrossRef]
- Zisman, D.; Safieh, M.; Simanovich, E.; Feld, J.; Kinarty, A.; Zisman, L.; Gazitt, T.; Haddad, A.; Elias, M.; Rosner, I.; et al. Tocilizumab (TCZ) Decreases Angiogenesis in Rheumatoid Arthritis Through Its Regulatory Effect on miR-146a-5p and EMMPRIN/CD147. Front. Immunol. 2021, 12, 739592. [Google Scholar] [CrossRef]
- Liu, W.; Wu, Y.H.; Zhang, L.; Xue, B.; Wang, Y.; Liu, B.; Liu, X.Y.; Zuo, F.; Yang, X.Y.; Chen, F.Y.; et al. MicroRNA-146a suppresses rheumatoid arthritis fibroblastlike synoviocytes proliferation and inflammatory responses by inhibiting the TLR4/NF-kB signaling. Oncotarget 2018, 9, 23944–23959. [Google Scholar] [CrossRef] [PubMed]
- Ammari, M.; Presumey, J.; Ponsolles, C.; Roussignol, G.; Roubert, C.; Escriou, V.; Toupet, K.; Mausset-Bonnefont, A.L.; Cren, M.; Robin, M.; et al. Delivery of miR-146a to Ly6Chigh monocytes inhibits pathogenic bone erosion in inflammatory arthritis. Theranostics 2018, 8, 5972–5985. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Kartha, R.V.; Subramanian, S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front. Genet. 2014, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhong, Z.; Miao, Q.; Zhang, Y.; Ni, B.; Zhang, M.; Tang, J. circPTPN22 as a novel biomarker and ceRNA in peripheral blood mononuclear cells of rheumatoid arthritis. Mol. Med. Rep. 2021, 24, 617. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jiang, C.; Li, J.; Li, X.; Zhao, L.; Yun, H.; Xu, W.; Fan, W.; Liu, Q.; Dong, H. Serum-derived exosomes containing NEAT1 promote the occurrence of rheumatoid arthritis through regulation of miR-144-3p/ROCK2 axis. Ther. Adv. Chronic Dis. 2021, 12, 2040622321991705. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, N.; Li, H.J.; Guo, X.Y.; Lu, L.; Guo, Y. Inhibition of lncRNA NEAT1 induces dysfunction of fibroblast-like synoviocytes in rheumatoid arthritis via miRNA-338-3p-mediated regulation of glutamine metabolism. J. Orthop. Surg. Res. 2022, 17, 401. [Google Scholar] [CrossRef]
- Bi, X.; Guo, X.H.; Mo, B.Y.; Wang, M.L.; Luo, X.Q.; Chen, Y.X.; Liu, F.; Olsen, N.; Pan, Y.F.; Zheng, S.G. LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. EBioMedicine 2019, 50, 408–420. [Google Scholar] [CrossRef]
- Peng, H.; Xing, J.; Wang, X.; Ding, X.; Tang, X.; Zou, J.; Wang, S.; Liu, Y. Circular RNA circNUP214 Modulates the T Helper 17 Cell Response in Patients with Rheumatoid Arthritis. Front. Immunol. 2022, 13, 885896. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, F.; Zhu, S.; Zhang, W.; Hou, J.; He, R.; Wang, K.; Wang, Z.; Liang, T. Exosomal long non-coding RNA TRAFD1-4:1 derived from fibroblast-like synoviocytes suppresses chondrocyte proliferation and migration by degrading cartilage extracellular matrix in rheumatoid arthritis. Exp. Cell Res. 2023, 422, 113441. [Google Scholar] [CrossRef]
- Yang, Z.; Lin, S.D.; Zhan, F.; Liu, Y.; Zhan, Y.W. LncRNA GAS5 alleviates rheumatoid arthritis through regulating miR-222-3p/Sirt1 signalling axis. Autoimmunity 2021, 54, 13–22. [Google Scholar] [CrossRef]
- Li, L.; Zhan, M.; Li, M. Circular RNA circ_0130438 suppresses TNF-α-induced proliferation, migration, invasion and inflammation in human fibroblast-like MH7A synoviocytes by regulating miR-130a-3p/KLF9 axis. Transpl. Immunol. 2022, 72, 101588. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ni, B.; Li, C.; Sun, W.; Wang, Z.; Wang, H.; Hou, X.; Yan, S.; Wang, X.; Xu, D. CircRNA_17725 Promotes Macrophage Polarization towards M2 by Targeting FAM46C to Alleviate Arthritis. Mediat. Inflamm. 2023, 2023, 6818524. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Song, M.J.; Fang, J. LncRNA OSER1-AS1 regulates the inflammation and apoptosis of rheumatoid arthritis fibroblast like synoviocytes via regulating miR-1298-5p/E2F1 axis. Bioengineered 2022, 13, 4951–4963. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, H.; Pan, L.Y.; Han, Y.Q.; Chen, Y.; Jiang, Y.; Wang, Y. LncRNA OIP5-AS1/miR-410-3p/Wnt7b axis promotes the proliferation of rheumatoid arthritis fibroblast-like synoviocytes via regulating the Wnt/β-catenin pathway. Autoimmunity 2023, 56, 2189136. [Google Scholar] [CrossRef]
- Li, B.; Li, N.; Zhang, L.; Li, K.; Xie, Y.; Xue, M.; Zheng, Z. Hsa-circ-0001859 Regulates ATF2 Expression by Functioning as an MIR-204/211 Sponge in Human Rheumatoid Arthritis. J. Immunol. Res. 2018, 2018, 9412387. [Google Scholar] [CrossRef]
- Yang, J.; Cheng, M.; Gu, B.; Wang, J.; Yan, S.; Xu, D. CircRNA_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via miR-6089/AKT1/NF-κB axis. Cell Death Dis. 2020, 11, 833. [Google Scholar] [CrossRef]
- Yan, S.; Wang, P.; Wang, J.; Yang, J.; Lu, H.; Jin, C.; Cheng, M.; Xu, D. Long Non-coding RNA HIX003209 Promotes Inflammation by Sponging miR-6089 via TLR4/NF-κB Signaling Pathway in Rheumatoid Arthritis. Front. Immunol. 2019, 10, 2218. [Google Scholar] [CrossRef]
- Wen, J.T.; Liu, J.; Wan, L.; Xin, L.; Guo, J.C.; Sun, Y.Q.; Wang, X.; Wang, J. Triptolide inhibits cell growth and inflammatory response of fibroblast-like synoviocytes by modulating hsa-circ-0003353/microRNA-31-5p/CDK1 axis in rheumatoid arthritis. Int. Immunopharmacol. 2022, 106, 108616. [Google Scholar] [CrossRef]
- Fan, C.; Cui, X.; Chen, S.; Huang, S.; Jiang, H. LncRNA LOC100912373 modulates PDK1 expression by sponging miR-17-5p to promote the proliferation of fibroblast-like synoviocytes in rheumatoid arthritis. Am. J. Transl. Res. 2020, 12, 7709–7723. [Google Scholar]
- Jiang, H.; Liu, J.; Fan, C.; Wang, J.; Li, W. lncRNAS56464.1 as a ceRNA promotes the proliferation of fibroblast-like synoviocytes in experimental arthritis via the Wnt signaling pathway and sponges miR-152-3p. Int. J. Mol. Med. 2021, 47, 17. [Google Scholar] [CrossRef]
- Shaikh, F.S.; Siegel, R.J.; Srivastava, A.; Fox, D.A.; Ahmed, S. Challenges and promise of targeting miRNA in rheumatic diseases: A computational approach to identify miRNA association with cell types, cytokines, and disease mechanisms. Front. Immunol. 2024, 14, 1322806. [Google Scholar] [CrossRef] [PubMed]
| Axis | ceRNA | miRNA | Molecular Target | Effects on RA | Reference |
|---|---|---|---|---|---|
| TRAFD1-4:1/miR-27a-3p/CXCL1 | TRAF1-4:1 | miR-27a-3p | CXCL1 | TRAF1-4:1 inhibited the proliferation and migration of chondrocytes | [163] |
| GAS5/miR-222-3p/Sirt1 | GAS5 | miR-222-3p | SIRT1 | GAS5 regulates FLS function by inhibiting their proliferation and inflammation and promoting their apoptosis | [164] |
| circ_0130438/miR-130a-3p/KLF9 | circ_0130438 | miR-130a-3p | KLF9 | circ_0130438 inhibits TNF-α-induced invasion, proliferation, migration and inflammation in MH7A cells | [165] |
| circRNA_17725/miR-4668-5p/FAM46C | circRNA_17725 | miR-4668-5p | FAM46C | circRNA_17725 induces macrophage proliferation at M2 | [166] |
| OSER1-AS1/miR-1298-5p/E2F1 | OSER1-AS1 | miR-1298-5p | E2F1 | OSER1-AS1 inhibits apoptosis and inflammation of the RA-FLS | [167] |
| OIP5-AS1/miR-410-3p/Wnt7b | OIP5-AS1 | miR-410-3p | Wnt7b | OIP5-AS1 inhibits FLS proliferation | [168] |
| hsa_circ_0001859/miR-204/211/ATF2 | hsa_circ_0001859 | miR-204/211 | ATF2 | Silencing of hsa_circ_0001859 reduces inflammation in SW982 cells | [169] |
| circRNA_09505/miR-6089/AKT1/NF-κB | circRNA_09505 | miR-6089 | AKT1/NF-κB | circRNA_09505 promotes the release of pro-inflammatory cytokines (TNF-α, IL-6, IL-8, IL-12 and IL-1β) from macrophages | [170] |
| HIX003209/miR-6089/TLR4 | HIX003209 | miR-6089 | TLR4 | HIX003209 promotes pro-inflammatory cytokine release (TNF-α, IL-6 and IL-1β) in macrophages | [171] |
| circ0003353/miR-31-5p/CDK1 | circ0003353 | miR-31-5p | CDK1 | Overexpression of circ0003353 promotes proliferation, cell cycle progression and inflammatory cytokine production (IL-1β and IL-6) of RA-FLS | [172] |
| LOC100912373/miR-17-5p/PDK1 | LOC100912373 | miR-17-5p | PDK1 | LOC100912373 induces FLS proliferation | [173] |
| lncRNAS56464.1/miR-152-3p/Wnt pathway | lncRNAS56464.1 | miR-152-3p | Wnt pathway | lncRNAS56464.1 promotes FLS proliferation | [174] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pascual-García, S.; Martínez-Peinado, P.; Pujalte-Satorre, C.; Navarro-Sempere, A.; Esteve-Girbés, J.; López-Jaén, A.B.; Javaloyes-Antón, J.; Cobo-Velacoracho, R.; Navarro-Blasco, F.J.; Sempere-Ortells, J.M. Exosomal Osteoclast-Derived miRNA in Rheumatoid Arthritis: From Their Pathogenesis in Bone Erosion to New Therapeutic Approaches. Int. J. Mol. Sci. 2024, 25, 1506. https://doi.org/10.3390/ijms25031506
Pascual-García S, Martínez-Peinado P, Pujalte-Satorre C, Navarro-Sempere A, Esteve-Girbés J, López-Jaén AB, Javaloyes-Antón J, Cobo-Velacoracho R, Navarro-Blasco FJ, Sempere-Ortells JM. Exosomal Osteoclast-Derived miRNA in Rheumatoid Arthritis: From Their Pathogenesis in Bone Erosion to New Therapeutic Approaches. International Journal of Molecular Sciences. 2024; 25(3):1506. https://doi.org/10.3390/ijms25031506
Chicago/Turabian StylePascual-García, Sandra, Pascual Martínez-Peinado, Carolina Pujalte-Satorre, Alicia Navarro-Sempere, Jorge Esteve-Girbés, Ana B. López-Jaén, Juan Javaloyes-Antón, Raúl Cobo-Velacoracho, Francisco J. Navarro-Blasco, and José M. Sempere-Ortells. 2024. "Exosomal Osteoclast-Derived miRNA in Rheumatoid Arthritis: From Their Pathogenesis in Bone Erosion to New Therapeutic Approaches" International Journal of Molecular Sciences 25, no. 3: 1506. https://doi.org/10.3390/ijms25031506
APA StylePascual-García, S., Martínez-Peinado, P., Pujalte-Satorre, C., Navarro-Sempere, A., Esteve-Girbés, J., López-Jaén, A. B., Javaloyes-Antón, J., Cobo-Velacoracho, R., Navarro-Blasco, F. J., & Sempere-Ortells, J. M. (2024). Exosomal Osteoclast-Derived miRNA in Rheumatoid Arthritis: From Their Pathogenesis in Bone Erosion to New Therapeutic Approaches. International Journal of Molecular Sciences, 25(3), 1506. https://doi.org/10.3390/ijms25031506






