Novel Epigenetic Modifiers of Histones Presenting Potent Inhibitory Effects on Adenoid Cystic Carcinoma Stemness and Invasive Properties
Abstract
1. Introduction
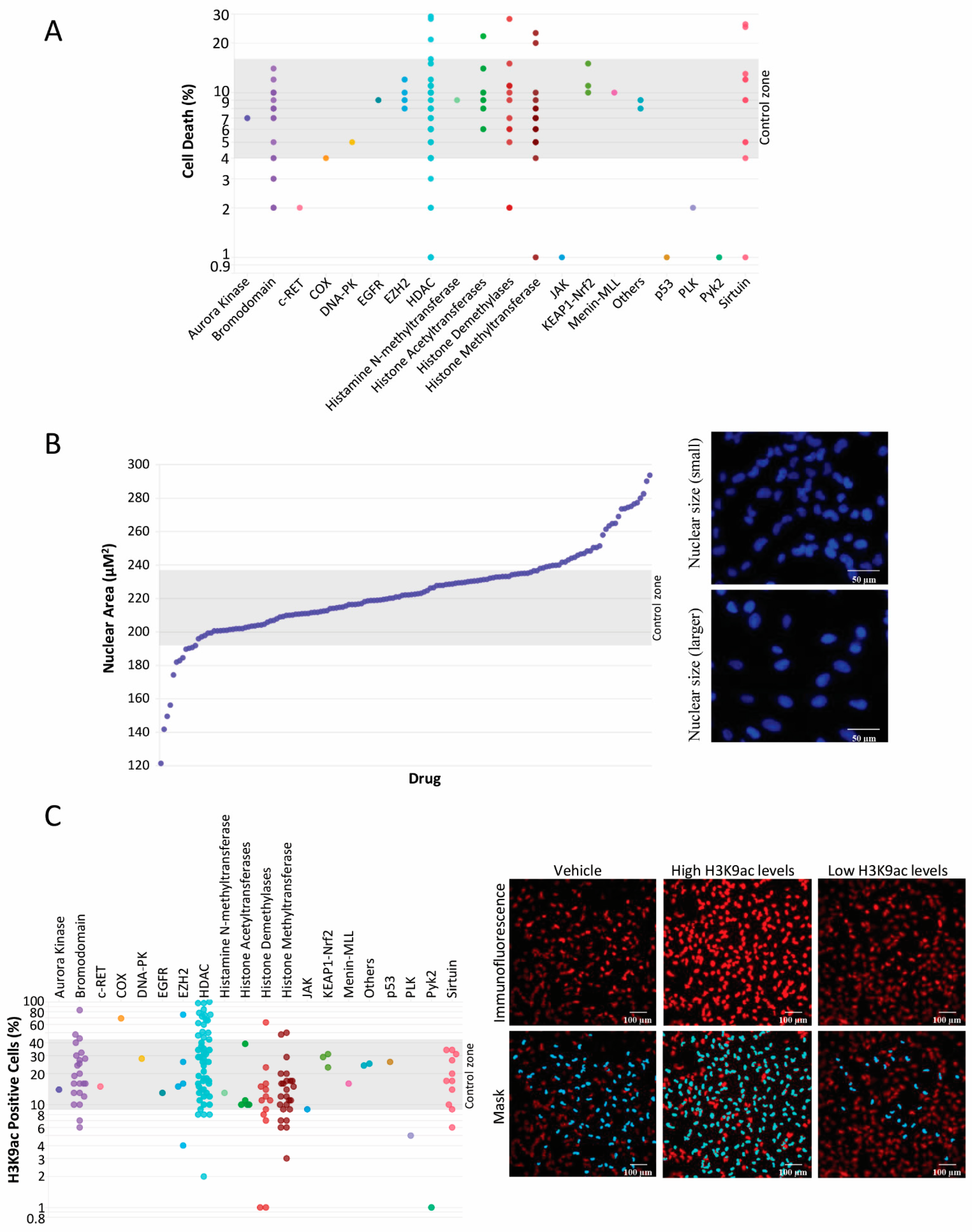
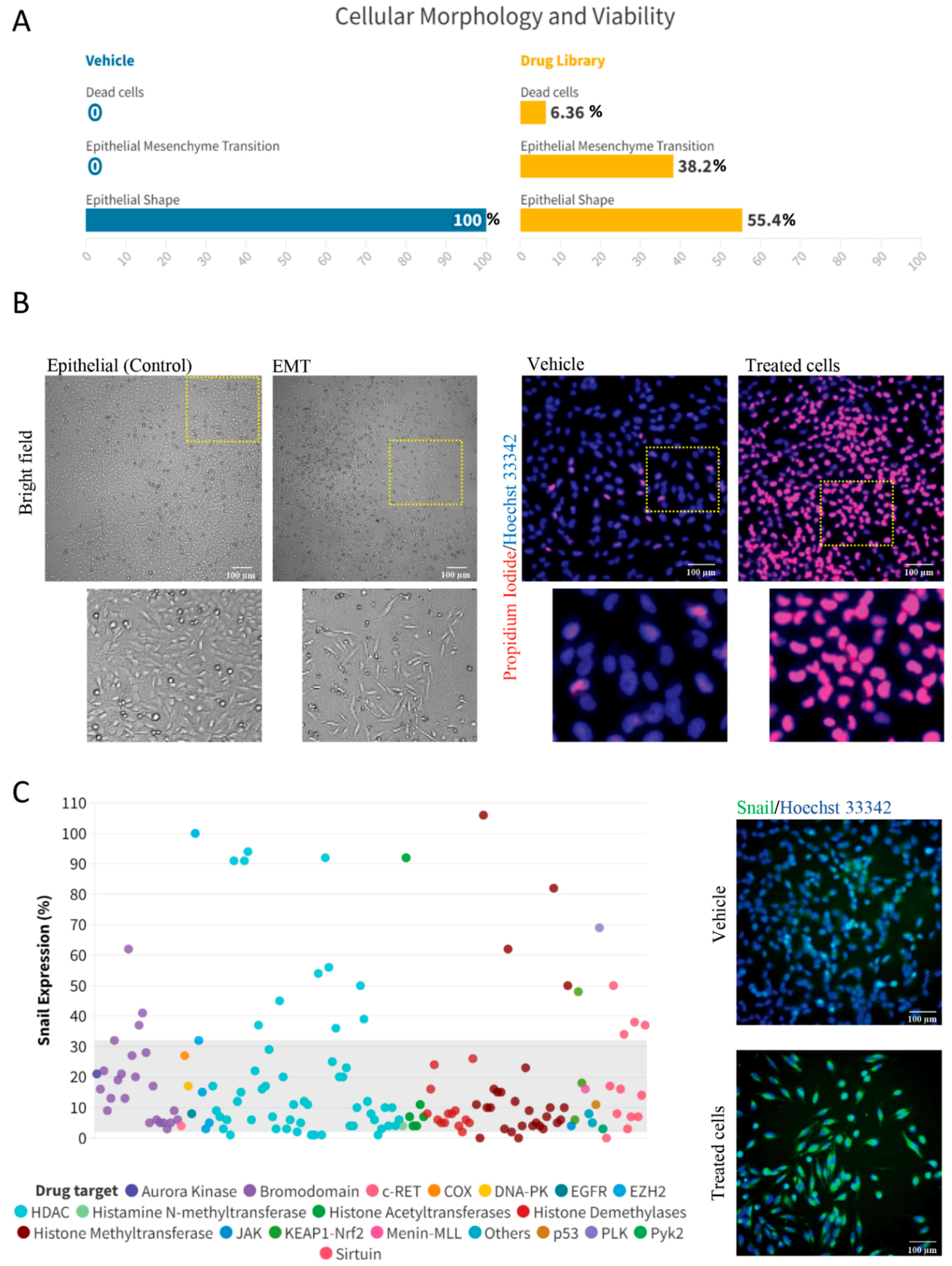
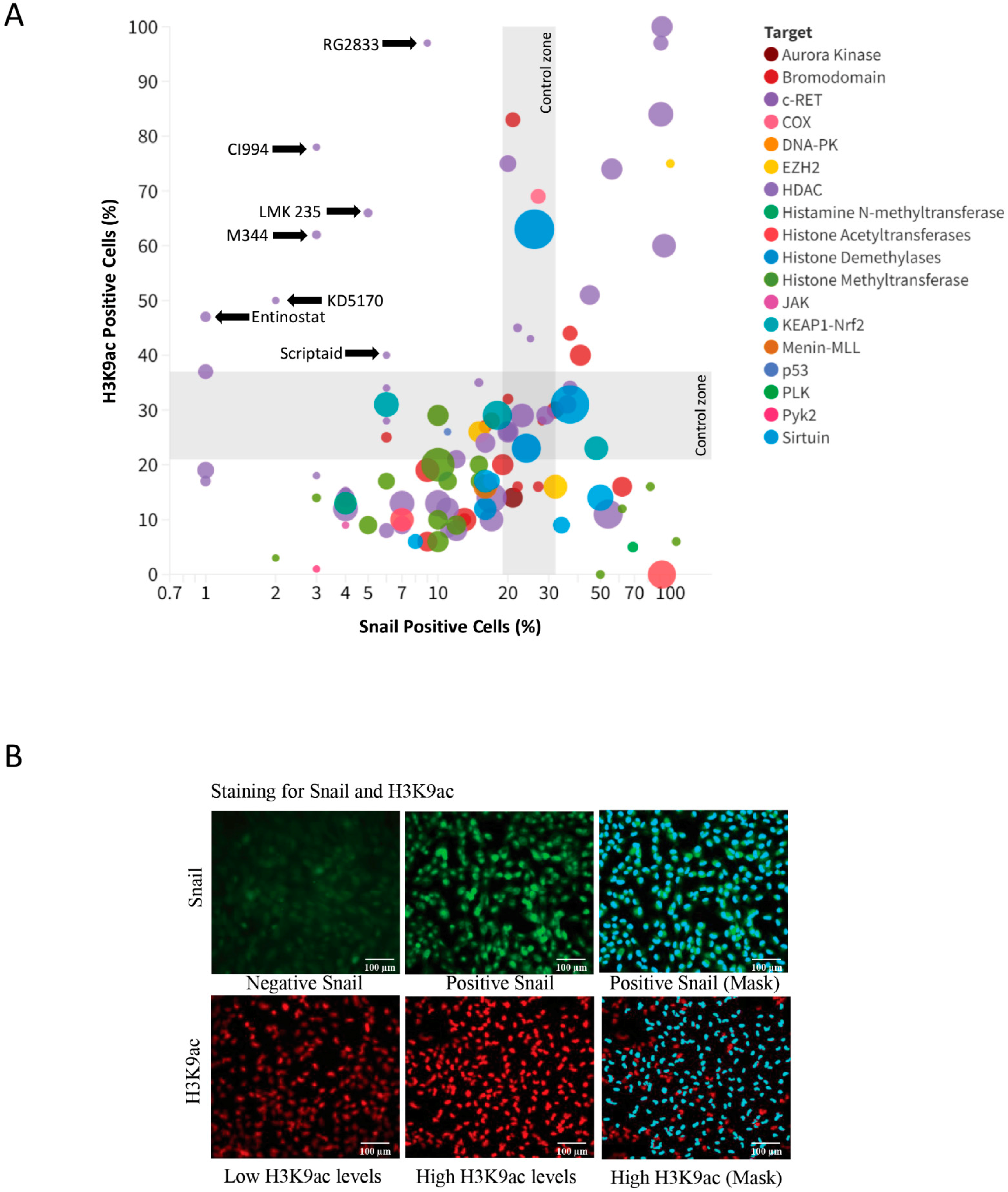
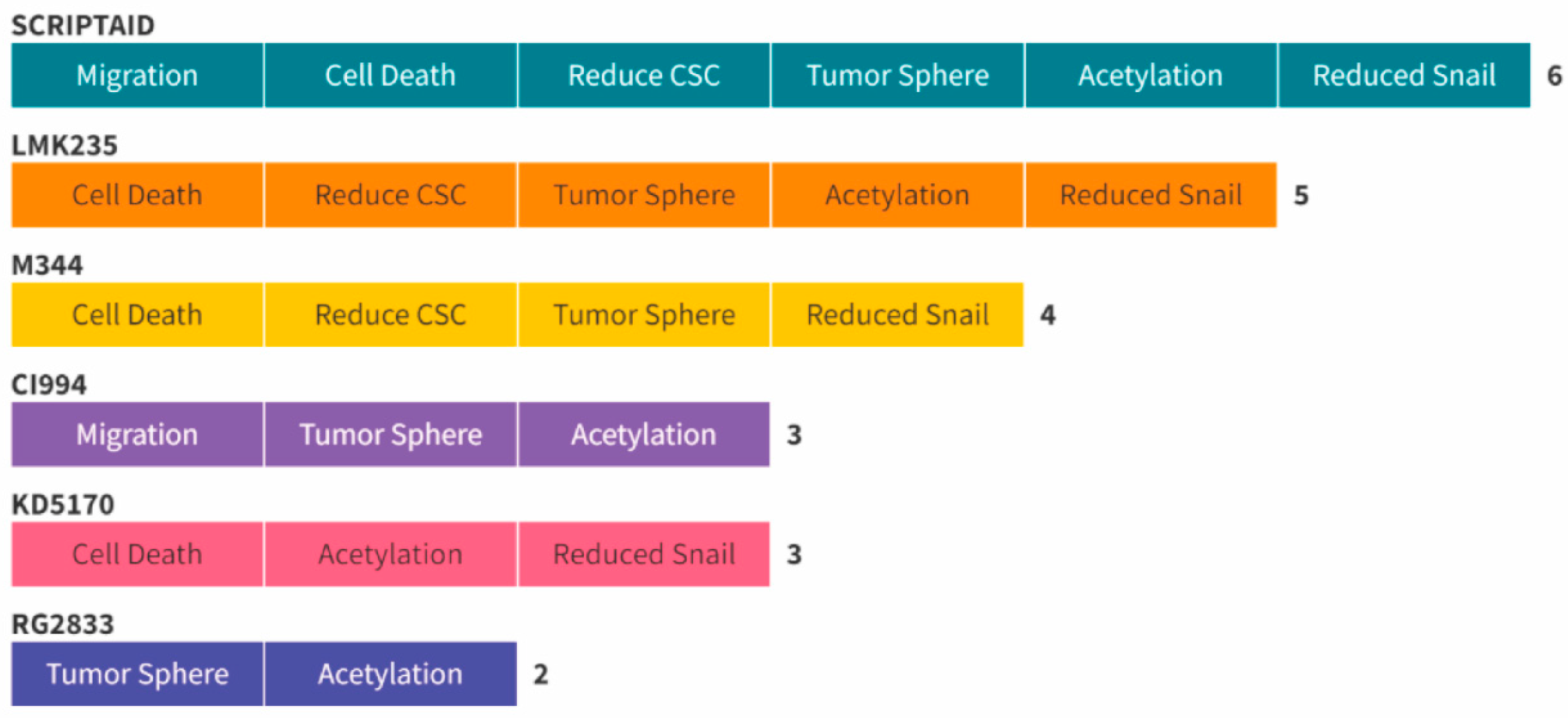
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Line and Culture Conditions
4.2. Histone Modifiers Library and High-Throughput Screening (HTS)
4.3. Immunofluorescence
4.4. Flow Cytometry
4.5. Scratch Assay
4.6. Tumorsphere Assay
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jaso, J.; Malhotra, R. Adenoid cystic carcinoma. Arch. Pathol. Lab. Med. 2011, 135, 511–515. [Google Scholar] [CrossRef]
- Dillon, P.M.; Chakraborty, S.; Moskaluk, C.A.; Joshi, P.J.; Thomas, C.Y. Adenoid cystic carcinoma: A review of recent advances, molecular targets, and clinical trials. Head Neck 2016, 38, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.O.; Guimaraes, D.M.; Martins, M.D.; Martins, M.A.T.; Warner, K.A.; Nor, J.E.; Castilho, R.M.; Squarize, C.H. Unlocking the chromatin of adenoid cystic carcinomas using HDAC inhibitors sensitize cancer stem cells to cisplatin and induces tumor senescence. Stem. Cell Res. 2017, 21, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Cantu, G. Adenoid cystic carcinoma. An indolent but aggressive tumour. Part A: From aetiopathogenesis to diagnosis. Acta Otorhinolaryngol. Ital. 2021, 41, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Cantu, G. Adenoid cystic carcinoma. An indolent but aggressive tumour. Part B: Treatment and prognosis. Acta Otorhinolaryngol. Ital. 2021, 41, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.C.; Perez-de-Oliveira, M.E.; Pedroso, C.M.; Leite, A.A.; Santos-Silva, A.R.; Lopes, M.A.; Junior, G.D.; Martins, M.D.; Wagner, V.P.; Kowalski, L.P.; et al. Systemic therapies for salivary gland carcinomas: An overview of published clinical trials. Med. Oral Patol. Oral Cir. Bucal. 2023, 26264, online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lv, L.; Yang, K. Chemotherapy targeting cancer stem cells. Am. J. Cancer Res. 2015, 5, 880–893. [Google Scholar]
- Giudice, F.S.; Pinto, D.S., Jr.; Nor, J.E.; Squarize, C.H.; Castilho, R.M. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS ONE 2013, 8, e58672. [Google Scholar] [CrossRef]
- Belulescu, I.C.; Margaritescu, C.; Dumitrescu, C.I.; Munteanu, M.C.; Daguci, L.; Margaritescu, O.C.; Matei, M. The immunophenotype of epithelial to mesenchymal transition inducing transcription factors in salivary gland adenoid cystic carcinomas. Rom. J. Morphol. Embryol. 2020, 61, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Bialkowska, A.B.; Yang, V.W. High-throughput screening strategies for targeted identification of therapeutic compounds in colorectal cancer. Future Oncol. 2012, 8, 259–272. [Google Scholar] [CrossRef]
- Li, Z.; Tamari, K.; Seo, Y.; Minami, K.; Takahashi, Y.; Tatekawa, S.; Otani, K.; Suzuki, O.; Isohashi, F.; Ogawa, K. Dihydroouabain, a novel radiosensitizer for cervical cancer identified by automated high-throughput screening. Radiother. Oncol. 2020, 148, 21–29. [Google Scholar] [CrossRef]
- Henikoff, S.; Smith, M.M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 2015, 7, a019364. [Google Scholar] [CrossRef]
- Marino-Ramirez, L.; Kann, M.G.; Shoemaker, B.A.; Landsman, D. Histone structure and nucleosome stability. Expert Rev. Proteom. 2005, 2, 719–729. [Google Scholar] [CrossRef]
- Sugiura, M.; Sato, H.; Kanesaka, M.; Imamura, Y.; Sakamoto, S.; Ichikawa, T.; Kaneda, A. Epigenetic modifications in prostate cancer. Int. J. Urol. 2021, 28, 140–149. [Google Scholar] [CrossRef]
- Cohen, I.; Poreba, E.; Kamieniarz, K.; Schneider, R. Histone modifiers in cancer: Friends or foes? Genes Cancer 2011, 2, 631–647. [Google Scholar] [CrossRef]
- Lee, J.S.; Smith, E.; Shilatifard, A. The language of histone crosstalk. Cell 2010, 142, 682–685. [Google Scholar] [CrossRef]
- Huang, H.; Sabari, B.R.; Garcia, B.A.; Allis, C.D.; Zhao, Y. SnapShot: Histone modifications. Cell 2014, 159, 458–458.e1. [Google Scholar] [CrossRef]
- Jiang, J.; Tang, Y.; Zhu, G.; Zheng, M.; Yang, J.; Liang, X. Correlation between transcription factor Snail1 expression and prognosis in adenoid cystic carcinoma of salivary gland. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2010, 110, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu. Rev. Cell Dev. Biol. 2011, 27, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Minucci, S.; Pelicci, P.G. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer 2006, 6, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, L.D.; Webber, L.P.; Gaio, E.J.; Junior, D.S.; Goncalves, P.; Wick, M.J.; Burr, N.S.; Squarize, C.H.; Castilho, R.M. Using PDX animal models to identify and stratify adenoid cystic carcinoma patients presenting an enhanced response to HDAC inhibitors. Am. J. Cancer Res. 2023, 13, 143–160. [Google Scholar] [PubMed]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Sahara, S.; Warner, K.A.; Herzog, A.E.; Zhang, Z.; Nor, J.E. Therapeutic inhibition of Bmi-1 ablates chemoresistant cancer stem cells in adenoid cystic carcinoma. Oral Oncol. 2023, 142, 106437. [Google Scholar] [CrossRef] [PubMed]
- Clay, M.R.; Tabor, M.; Owen, J.H.; Carey, T.E.; Bradford, C.R.; Wolf, G.T.; Wicha, M.S.; Prince, M.E. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck 2010, 32, 1195–1201. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, J.; Chai, K.; Ying, X.; Zhou, B.P. The Role of Snail in EMT and Tumorigenesis. Curr. Cancer Drug Targets 2013, 13, 963–972. [Google Scholar] [CrossRef]
- Nishioka, R.; Itoh, S.; Gui, T.; Gai, Z.; Oikawa, K.; Kawai, M.; Tani, M.; Yamaue, H.; Muragaki, Y. SNAIL induces epithelial-to-mesenchymal transition in a human pancreatic cancer cell line (BxPC3) and promotes distant metastasis and invasiveness in vivo. Exp. Mol. Pathol. 2010, 89, 149–157. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Zhou, B.P.; Deng, J.; Xia, W.; Xu, J.; Li, Y.M.; Gunduz, M.; Hung, M.C. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004, 6, 931–940. [Google Scholar] [CrossRef]
- Lloyd, S.; Yu, J.B.; Wilson, L.D.; Decker, R.H. Determinants and patterns of survival in adenoid cystic carcinoma of the head and neck, including an analysis of adjuvant radiation therapy. Am. J. Clin. Oncol. 2011, 34, 76–81. [Google Scholar] [CrossRef]
- Warner, K.A.; Oklejas, A.E.; Pearson, A.T.; Zhang, Z.; Wu, W.; Divi, V.; Rodriguez-Ramirez, C.; Castilho, R.M.; Polverini, P.J.; Nor, J.E. UM-HACC-2A: MYB-NFIB fusion-positive human adenoid cystic carcinoma cell line. Oral Oncol. 2018, 87, 21–28. [Google Scholar] [CrossRef]
- Perez-Moreno, M.A.; Locascio, A.; Rodrigo, I.; Dhondt, G.; Portillo, F.; Nieto, M.A.; Cano, A. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J. Biol. Chem. 2001, 276, 27424–27431. [Google Scholar] [CrossRef]
- Peinado, H.; Portillo, F.; Cano, A. Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 2004, 48, 365–375. [Google Scholar] [CrossRef]
- Cano, A.; Perez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Zhu, Y.; Xia, T.; Chen, D.Q.; Xiong, X.; Shi, L.; Zuo, Y.; Xiao, H.; Liu, L. Promising role of protein arginine methyltransferases in overcoming anti-cancer drug resistance. Drug Resist. Updat. 2024, 72, 101016. [Google Scholar] [CrossRef]
- Silva, L.C.; Borgato, G.B.; Wagner, V.P.; Martins, M.D.; Rocha, G.Z.; Lopes, M.A.; Santos-Silva, A.R.; de Castro Junior, G.; Kowalski, L.P.; Nor, J.E.; et al. Cephaeline is an inductor of histone H3 acetylation and inhibitor of mucoepidermoid carcinoma cancer stem cells. J. Oral Pathol. Med. 2022, 51, 553–562. [Google Scholar] [CrossRef]
- Silva, L.C.; Leite, A.A.; Borgato, G.B.; Wagner, V.P.; Martins, M.D.; Loureiro, F.J.A.; Lopes, M.A.; Santos-Silva, A.R.; Sperandio, M.; de Castro Junior, G.; et al. Oral squamous cell carcinoma cancer stem cells have different drug sensitive to pharmacological NFkappaB and histone deacetylation inhibition. Am. J. Cancer Res. 2023, 13, 6038–6050. [Google Scholar]
- Guzman, M.L.; Yang, N.; Sharma, K.K.; Balys, M.; Corbett, C.A.; Jordan, C.T.; Becker, M.W.; Steidl, U.; Abdel-Wahab, O.; Levine, R.L.; et al. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: A novel potential strategy in acute myelogenous leukemia. Mol. Cancer Ther. 2014, 13, 1979–1990. [Google Scholar] [CrossRef]
- Acasigua, G.A.; Warner, K.A.; Nor, F.; Helman, J.; Pearson, A.T.; Fossati, A.C.; Wang, S.; Nor, J.E. BH3-mimetic small molecule inhibits the growth and recurrence of adenoid cystic carcinoma. Oral Oncol. 2015, 51, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Markman, R.L.; Webber, L.P.; Nascimento Filho, C.H.V.; Reis, L.A.; Vargas, P.A.; Lopes, M.A.; Zanella, V.; Martins, M.D.; Squarize, C.H.; Castilho, R.M. Interfering with bromodomain epigenome readers as therapeutic option in mucoepidermoid carcinoma. Cell Oncol. 2019, 42, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.D.; Silveira, F.M.; Webber, L.P.; Wagner, V.P.; Martins, M.A.T.; Squarize, C.H.; Castilho, R.M. The impact of photobiomodulation therapy on the biology and behavior of head and neck squamous cell carcinomas cell lines. J. Photochem. Photobiol. B 2020, 209, 111924. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.P. Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) into Osteoclasts. Bio Protoc. 2020, 10, e3854. [Google Scholar] [CrossRef]
- Johnson, S.; Chen, H.; Lo, P.K. In vitro Tumorsphere Formation Assays. Bio Protoc. 2013, 3, e325. [Google Scholar] [CrossRef]
- Almeida, L.O.; Guimaraes, D.M.; Squarize, C.H.; Castilho, R.M. Profiling the Behavior of Distinct Populations of Head and Neck Cancer Stem Cells. Cancers 2016, 8, 7. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pina, P.S.S.; Jang, Y.; Emerick, C.; Scarini, J.F.; Sousa, S.C.O.M.; Squarize, C.H.; Castilho, R.M. Novel Epigenetic Modifiers of Histones Presenting Potent Inhibitory Effects on Adenoid Cystic Carcinoma Stemness and Invasive Properties. Int. J. Mol. Sci. 2024, 25, 1646. https://doi.org/10.3390/ijms25031646
Pina PSS, Jang Y, Emerick C, Scarini JF, Sousa SCOM, Squarize CH, Castilho RM. Novel Epigenetic Modifiers of Histones Presenting Potent Inhibitory Effects on Adenoid Cystic Carcinoma Stemness and Invasive Properties. International Journal of Molecular Sciences. 2024; 25(3):1646. https://doi.org/10.3390/ijms25031646
Chicago/Turabian StylePina, Paulo S. S., Yeejin Jang, Carolina Emerick, João Figueira Scarini, Suzana C. O. M. Sousa, Cristiane H. Squarize, and Rogerio M. Castilho. 2024. "Novel Epigenetic Modifiers of Histones Presenting Potent Inhibitory Effects on Adenoid Cystic Carcinoma Stemness and Invasive Properties" International Journal of Molecular Sciences 25, no. 3: 1646. https://doi.org/10.3390/ijms25031646
APA StylePina, P. S. S., Jang, Y., Emerick, C., Scarini, J. F., Sousa, S. C. O. M., Squarize, C. H., & Castilho, R. M. (2024). Novel Epigenetic Modifiers of Histones Presenting Potent Inhibitory Effects on Adenoid Cystic Carcinoma Stemness and Invasive Properties. International Journal of Molecular Sciences, 25(3), 1646. https://doi.org/10.3390/ijms25031646






