miRNAs in Heart Development and Disease
Abstract
1. Introduction
1.1. Non-Coding RNA Biogenesis, Function, and Molecular Evolution
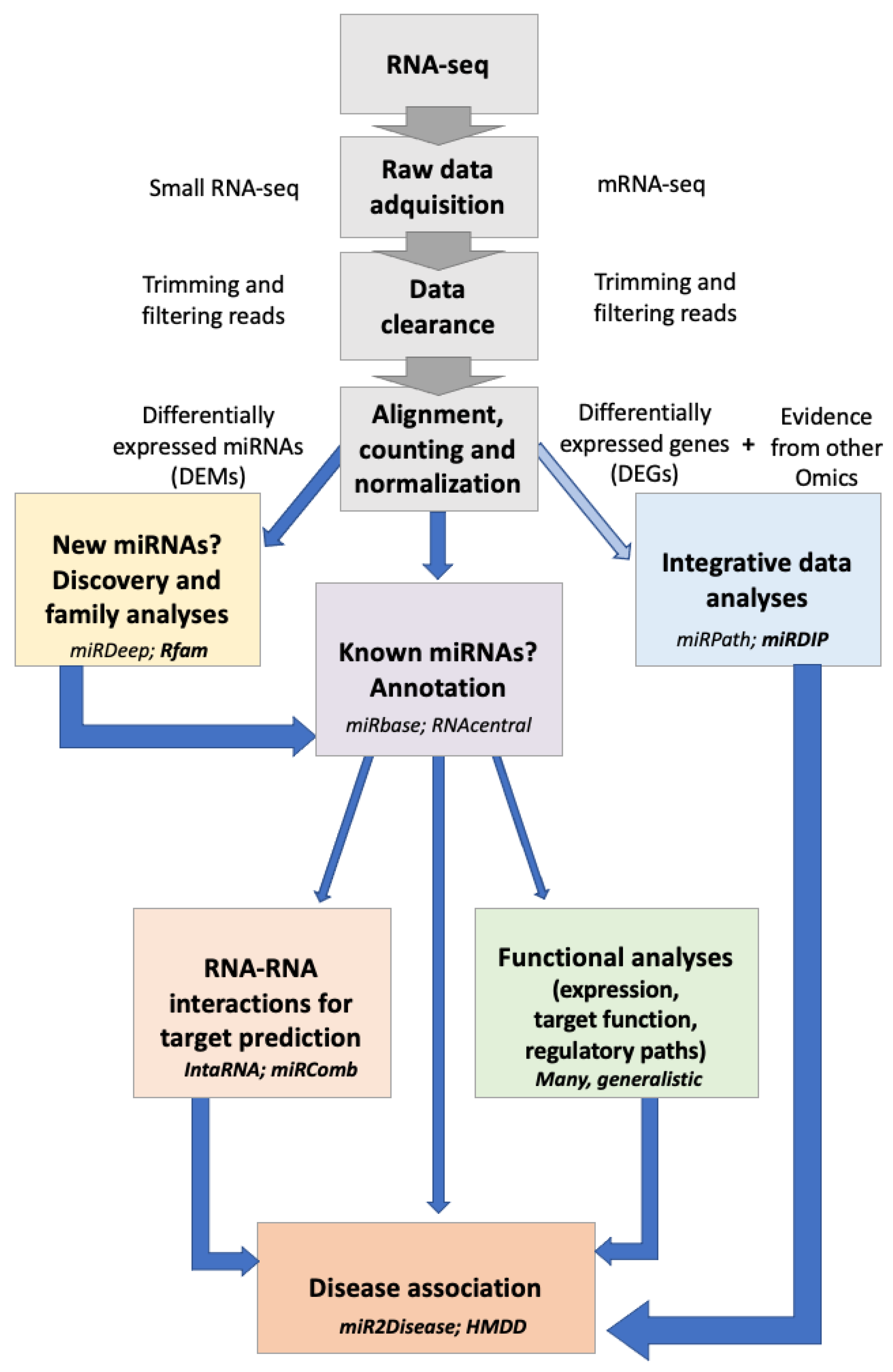
1.2. Bioinformatic Tools of Non-Coding RNAs
1.3. An Introduction to Cardiac Development and Disease
2. The Role of microRNAs in Heart Development
3. The Role of microRNAs in Congenital Heart Diseases
4. The Role of microRNAs in Atrial Fibrillation
5. The Role of miRNAs in Heart Failure and Fibrosis
6. The Role of miRNAs in Coronary Artery Disease
7. The Role of miRNAs in Myocardial Infarction
8. Novel Bioinformatic Tools for the Study of ncRNAs
8.1. Bioinformatics Tools for ncRNA Family Identification
8.2. Bioinformatics Tools for RNA–RNA Interaction Prediction
8.3. Bioinformatic Tools for Functional Analysis
8.4. Bioinformatics Tools for Disease Association
8.5. Bioinformatics Tools for Integrative Data Analysis
9. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, D.-Z. microRNAs in cardiovascular development. J. Mol. Cell. Cardiol. 2012, 52, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Sun, W.; Guo, Z.; Zhang, J.; Yu, H.; Liu, B. Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 2020, 254, 116900. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Gaiti, F.; Calcino, A.D.; Tanurdžić, M.; Degnan, B.M. Origin and evolution of the metazoan non-coding regulatory genome. Dev. Biol. 2017, 427, 193–202. [Google Scholar] [CrossRef]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief. Bioinform. 2019, 20, 1836–1852. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Lai, J.; Lehman, M.L.; Nelson, C.C. miRDeep*: An integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2013, 41, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-H.; Fan, B.; Rothschild, M.F.; Hu, Z.-L.; Li, K.; Zhao, S.-H. MiRFinder: An improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinform. 2007, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.; Shankar, R. miReader: Discovering Novel miRNAs in Species without Sequenced Genome. PLoS ONE 2013, 8, e66857. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Findeiß, S.; Washietl, S.; Hofacker, I.L.; Stadler, P.F. RNAz 2.0: Improved noncoding RNA detection. Pac. Symp. Biocomput. 2010, 2010, 69–79. [Google Scholar]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Fan, Y.; Xia, J. miRNet-Functional Analysis and Visual Exploration of miRNA-Target Interactions in a Network Context. Methods Mol. Biol. 2018, 1819, 215–233. [Google Scholar] [CrossRef]
- Petrov, A.I.; Kay, S.J.E.; Kalvari, I.; Howe, K.L.; Gray, K.A.; Bruford, E.A.; Kersey, P.J.; Cochrane, G.; Finn, R.D.; Bateman, A.; et al. RNAcentral: A comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017, 45, D128–D134. [Google Scholar] [CrossRef] [PubMed]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: MicroRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Vergoulis, T.; Vlachos, I.S.; Alexiou, P.; Georgakilas, G.; Maragkakis, M.; Reczko, M.; Gerangelos, S.; Koziris, N.; Dalamagas, T.; Hatzigeorgiou, A.G. TarBase 6.0: Capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012, 40, D222–D229. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef]
- Kiriakidou, M.; Nelson, P.T.; Kouranov, A.; Fitziev, P.; Bouyioukos, C.; Mourelatos, Z.; Hatzigeorgiou, A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004, 18, 1165–1178. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwilkowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Bernhart, S.H.; Lorenz, R. The ViennaRNA web services. Methods Mol. Biol. 2015, 1269, 307–326. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Weinstein, E.G.; Abdelhakim, A.; Yekta, S.; Rhoades, M.W.; Burge, C.B.; Bartel, D.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003, 17, 991–1008. [Google Scholar] [CrossRef]
- Lai, E.C.; Tomancak, P.; Williams, R.W.; Rubin, G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003, 4, R42. [Google Scholar] [CrossRef]
- Sylva, M.; van den Hoff, M.J.B.; Moorman, A.F.M. Development of the human heart. Am. J. Med. Genet. A 2014, 164, 1347–1371. [Google Scholar] [CrossRef] [PubMed]
- Buijtendijk, M.F.J.; Barnett, P.; van den Hoff, M.J.B. Development of the human heart. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.A. The heart and development. Semin. Perinatol. 1996, 20, 482–509. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pomares, J.M.; de la Pompa, J.L.; Franco, D.; Henderson, D.; Ho, S.Y.; Houyel, L.; Kelly, R.G.; Sedmera, D.; Sheppard, M.; Sperling, S.; et al. Congenital coronary artery anomalies: A bridge from embryology to anatomy and pathophysiology—A position statement of the development, anatomy, and pathology ESC Working Group. Cardiovasc. Res. 2016, 109, 204–216. [Google Scholar] [CrossRef] [PubMed]
- van Weerd, J.H.; Christoffels, V.M. The formation and function of the cardiac conduction system. Development 2016, 143, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.H.; Yanni, J.; Boyett, M.R.; Chandler, N.J.; Dobrzynski, H. The anatomy of the cardiac conduction system. Clin. Anat. 2009, 22, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Munshi, N.V. Development of the Cardiac Conduction System. Cold Spring Harb. Perspect. Biol. 2020, 12, a037408. [Google Scholar] [CrossRef] [PubMed]
- Gelb, B.D. Genetic Discovery for Congenital Heart Defects. In Etiology and Morphogenesis of Congenital Heart Disease; Nakanishi, T., Markwald, R.R., Baldwin, H., Keller, B.B., Srivastava, D., Yamagishi, H., Eds.; SpringerOpen: London, UK, 2016; pp. 355–360. [Google Scholar] [CrossRef]
- Russell, M.W.; Chung, W.K.; Kaltman, J.R.; Miller, T.A. Advances in the Understanding of the Genetic Determinants of Congenital Heart Disease and Their Impact on Clinical Outcomes. J. Am. Heart Assoc. 2018, 7, e006906. [Google Scholar] [CrossRef] [PubMed]
- Boyd, R.; McMullen, H.; Beqaj, H.; Kalfa, D. Environmental Exposures and Congenital Heart Disease. Pediatrics 2022, 149, e2021052151. [Google Scholar] [CrossRef] [PubMed]
- van der Bom, T.; Zomer, A.C.; Zwinderman, A.H.; Meijboom, F.J.; Bouma, B.J.; Mulder, B.J.M. The changing epidemiology of congenital heart disease. Nat. Rev. Cardiol. 2011, 8, 50–60. [Google Scholar] [CrossRef]
- Shao, C.; Wang, J.; Tian, J.; Tang, Y. Coronary Artery Disease: From Mechanism to Clinical Practice. Adv. Exp. Med. Biol. 2020, 1177, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, R.E.; Jordan, E. Dilated Cardiomyopathy Overview; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; GeneReviews®: Seattle, WA, USA, 1993. [Google Scholar]
- Tuohy, C.V.; Kaul, S.; Song, H.K.; Nazer, B.; Heitner, S.B. Hypertrophic cardiomyopathy: The future of treatment. Eur. J. Heart Fail. 2020, 22, 228–240. [Google Scholar] [CrossRef]
- Teekakirikul, P.; Zhu, W.; Huang, H.C.; Fung, E. Hypertrophic Cardiomyopathy: An Overview of Genetics and Management. Biomolecules 2019, 9, 878. [Google Scholar] [CrossRef]
- Coffey, S.; Roberts-Thomson, R.; Brown, A.; Carapetis, J.; Chen, M.; Enriquez-Sarano, M.; Zühlke, L.; Prendergast, B.D. Global epidemiology of valvular heart disease. Nat. Rev. Cardiol. 2021, 18, 853–864. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Q.; Gao, N.; Wu, F.; Lan, F.; Zhang, F.; Jin, L.; Huang, Z.; Ge, J.; Wang, H.; et al. MircroRNA-10b Promotes Human Embryonic Stem Cell-Derived Cardiomyocyte Proliferation via Novel Target Gene LATS1. Mol. Ther. Nucleic Acids 2020, 19, 437–445. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Min, H.; Kim, H.; Choi, Y.M.; Liu, H.C.; Ku, S.-Y. Differential MicroRNA Expression Profile of Human Embryonic Stem Cell-Derived Cardiac Lineage Cells. Tissue Eng. Regen. Med. 2017, 14, 163–169. [Google Scholar] [CrossRef]
- Wagh, V.; Pomorski, A.; Wilschut, K.J.; Piombo, S.; Bernstein, H.S. MicroRNA-363 negatively regulates the left ventricular determining transcription factor HAND1 in human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. Ther. 2014, 5, 75. [Google Scholar] [CrossRef]
- Poon, E.N.-Y.; Hao, B.; Guan, D.; Jun Li, M.; Lu, J.; Yang, Y.; Wu, B.; Wu, S.C.; Webb, S.E.; Liang, Y.; et al. Integrated transcriptomic and regulatory network analyses identify microRNA-200c as a novel repressor of human pluripotent stem cell-derived cardiomyocyte differentiation and maturation. Cardiovasc. Res. 2018, 114, 894–906. [Google Scholar] [CrossRef]
- Kuppusamy, K.T.; Jones, D.C.; Sperber, H.; Madan, A.; Fischer, K.A.; Rodriguez, M.L.; Pabon, L.; Zhu, W.Z.; Tulloch, N.L.; Yang, X.; et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, E2785–E2794. [Google Scholar] [CrossRef] [PubMed]
- Synnergren, J.; Améen, C.; Lindahl, A.; Olsson, B.; Sartipy, P. Expression of microRNAs and their target mRNAs in human stem cell-derived cardiomyocyte clusters and in heart tissue. Physiol. Genom. 2011, 43, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.-D.; Rushing, S.N.; Lieu, D.K.; Chan, C.W.; Kong, C.-W.; Geng, L.; Wilson, K.D.; Chiamvimonvat, N.; Boheler, K.R.; Wu, J.C.; et al. Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLoS ONE 2011, 6, e27417. [Google Scholar] [CrossRef]
- Zhao, Y.; Ransom, J.F.; Li, A.; Vedantham, V.; von Drehle, M.; Muth, A.N.; Tsuchihashi, T.; McManus, M.T.; Schwartz, R.J.; Srivastava, D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 2007, 129, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Velasco, E.; Garcia-Padilla, C.; Del Mar Muñoz-Gallardo, M.; Martinez-Amaro, F.J.; Caño-Carrillo, S.; Castillo-Casas, J.M.; Sanchez-Fernandez, C.; Aranega, A.E.; Franco, D. Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis. Int. J. Mol. Sci. 2022, 23, 2839. [Google Scholar] [CrossRef]
- Lopez-Sanchez, C.; Franco, D.; Bonet, F.; Garcia-Lopez, V.; Aranega, A.; Garcia-Martinez, V. Reciprocal repression between Fgf8 and miR-133 regulates cardiac induction through Bmp2 signaling. Data Br. 2015, 5, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanchez, C.; Franco, D.; Bonet, F.; Garcia-Lopez, V.; Aranega, A.; Garcia-Martinez, V. Negative Fgf8-Bmp2 feed-back is regulated by miR-130 during early cardiac specification. Dev. Biol. 2015, 406, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Coppola, A.; Romito, A.; Borel, C.; Gehrig, C.; Gagnebin, M.; Falconnet, E.; Izzo, A.; Altucci, L.; Banfi, S.; Antonarakis, S.E.; et al. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. 2014, 12, 323–337. [Google Scholar] [CrossRef]
- Wang, J.; Greene, S.B.; Bonilla-Claudio, M.; Tao, Y.; Zhang, J.; Bai, Y.; Huang, Z.; Black, B.L.; Wang, F.; Martin, J.F.; et al. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev. Cell 2010, 19, 903–912. [Google Scholar] [CrossRef]
- Alzein, M.; Lozano-Velasco, E.; Hernández-Torres, F.; García-Padilla, C.; Domínguez, J.N.; Aránega, A.; Franco, D. Differential Spatio-Temporal Regulation of T-Box Gene Expression by microRNAs during Cardiac Development. J. Cardiovasc. Dev. Dis. 2021, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Fazilaty, H.; Rago, L.; Kass Youssef, K.; Ocaña, O.H.; Garcia-Asencio, F.; Arcas, A.; Galceran, J.; Nieto, M.A. A gene regulatory network to control EMT programs in development and disease. Nat. Commun. 2019, 10, 5115. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Lu, M.M.; Massera, D.; Epstein, J.A. MicroRNA-processing enzyme Dicer is required in epicardium for coronary vasculature development. J. Biol. Chem. 2011, 286, 41036–41045. [Google Scholar] [CrossRef] [PubMed]
- Brønnum, H.; Andersen, D.C.; Schneider, M.; Sandberg, M.B.; Eskildsen, T.; Nielsen, S.B.; Kalluri, R.; Sheikh, S.P. miR-21 promotes fibrogenic epithelial-to-mesenchymal transition of epicardial mesothelial cells involving Programmed Cell Death 4 and Sprouty-1. PLoS ONE 2013, 8, e56280. [Google Scholar] [CrossRef] [PubMed]
- Pontemezzo, E.; Foglio, E.; Vernucci, E.; Magenta, A.; D’Agostino, M.; Sileno, S.; Astanina, E.; Bussolino, F.; Pellegrini, L.; Germani, A.; et al. miR-200c-3p Regulates Epitelial-to-Mesenchymal Transition in Epicardial Mesothelial Cells by Targeting Epicardial Follistatin-Related Protein 1. Int. J. Mol. Sci. 2021, 22, 4971. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, A.; Expósito, A.; Muñoz, M.D.M.; de Manuel, M.J.; Cámara-Morales, A.; Serrano-Osorio, F.; García-Padilla, C.; Hernández-Torres, F.; Domínguez, J.N.; Aránega, A.; et al. MiR-195 enhances cardiomyogenic differentiation of the proepicardium/septum transversum by Smurf1 and Foxp1 modulation. Sci. Rep. 2020, 10, 9334. [Google Scholar] [CrossRef]
- Yu, K.; Ji, Y.; Wang, H.; Xuan, Q.K.; Li, B.B.; Xiao, J.J.; Sun, W.; Kong, X.Q. Association of miR-196a2, miR-27a, and miR-499 polymorphisms with isolated congenital heart disease in a Chinese population. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Song, Y.; Higgins, H.; Guo, J.; Harrison, K.; Schultz, E.N.; Hales, B.J.; Moses, E.K.; Goldblatt, J.; Pachter, N.; Zhang, G. Clinical significance of circulating microRNAs as markers in detecting and predicting congenital heart defects in children. J. Transl. Med. 2018, 16, 42. [Google Scholar] [CrossRef]
- Chen, W.; Li, S. Circulating microRNA as a Novel Biomarker for Pulmonary Arterial Hypertension Due to Congenital Heart Disease. Pediatr. Cardiol. 2017, 38, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Z.; Song, X.; Liu, L.; Su, G.; Cui, Y. Bioinformatic Analysis of Genes and MicroRNAs Associated with Atrioventricular Septal Defect in Down Syndrome Patients. Int. Heart J. 2016, 57, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Ji, L.; Liu, L.; Liu, Y.; Hou, H.; Yu, K.; Sun, Q.; Zhao, Z. Characterization of circulating microRNA expression in patients with a ventricular septal defect. PLoS ONE 2014, 9, e106318. [Google Scholar] [CrossRef]
- Dueñas, A.; Expósito, A.; Aranega, A.; Franco, D. The Role of Non-Coding RNA in Congenital Heart Diseases. J. Cardiovasc. Dev. Dis. 2019, 6, 15. [Google Scholar] [CrossRef]
- van Rooij, E.; Olson, E.N. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Investig. 2007, 117, 2369–2376. [Google Scholar] [CrossRef]
- Danielson, L.S.; Park, D.S.; Rotllan, N.; Chamorro-Jorganes, A.; Guijarro, M.V.; Fernandez-Hernando, C.; Fishman, G.I.; Phoon, C.K.; Hernando, E. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013, 27, 1460–1467. [Google Scholar] [CrossRef]
- Sucharov, C.C.; Sucharov, J.; Karimpour-Fard, A.; Nunley, K.; Stauffer, B.L.; Miyamoto, S.D. Micro-RNA expression in hypoplastic left heart syndrome. J. Card. Fail. 2015, 21, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.T.M.; Ng, E.K.O.; Chow, P.; Kwong, A.; Cheung, Y. Circulating microRNA expression profile and systemic right ventricular function in adults after atrial switch operation for complete transposition of the great arteries. BMC Cardiovasc. Disord. 2013, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Tutarel, O.; Dangwal, S.; Bretthauer, J.; Westhoff-Bleck, M.; Roentgen, P.; Anker, S.D.; Bauersachs, J.; Thum, T. Circulating miR-423_5p fails as a biomarker for systemic ventricular function in adults after atrial repair for transposition of the great arteries. Int. J. Cardiol. 2013, 167, 63–66. [Google Scholar] [CrossRef]
- Low, K.J.; Buxton, C.C.; Newbury-Ecob, R.A. Tetralogy of Fallot, microcephaly, short stature and brachymesophalangy is associated with hemizygous loss of noncoding MIR17HG and coding GPC5. Clin. Dysmorphol. 2015, 24, 113–114. [Google Scholar] [CrossRef]
- Huang, J.-B.; Mei, J.; Jiang, L.-Y.; Jiang, Z.-L.; Liu, H.; Zhang, J.-W.; Ding, F.B. MiR-196a2 rs11614913 T>C Polymorphism Is Associated with an Increased Risk of Tetralogy of Fallot in a Chinese Population. Acta Cardiol. Sin. 2015, 31, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ma, X.-J.; Wang, H.-J.; Li, W.-C.; Chen, L.; Ma, D.; Huang, G.Y. Expression of Cx43-related microRNAs in patients with tetralogy of Fallot. World J. Pediatr. 2014, 10, 138–144. [Google Scholar] [CrossRef]
- O’Brien, J.E.J.; Kibiryeva, N.; Zhou, X.-G.; Marshall, J.A.; Lofland, G.K.; Artman, M.; Chen, J.; Bittel, D.C. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ. Cardiovasc. Genet. 2012, 5, 279–286. [Google Scholar] [CrossRef]
- Zhang, J.; Chang, J.-J.; Xu, F.; Ma, X.-J.; Wu, Y.; Li, W.-C.; Wang, H.J.; Huang, G.Y.; Ma, D. MicroRNA deregulation in right ventricular outflow tract myocardium in nonsyndromic tetralogy of fallot. Can. J. Cardiol. 2013, 29, 1695–1703. [Google Scholar] [CrossRef]
- Bittel, D.C.; Kibiryeva, N.; Marshall, J.A.; O’Brien, J.E. MicroRNA-421 Dysregulation Is Associated with Tetralogy of Fallot. Cells 2014, 3, 713–723. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, K.; Li, Y.; Shi, K.; Liu, Y.; Yang, Y.F.; Fang, Y.; Mao, M. Screening miRNA and their target genes related to tetralogy of Fallot with microarray. Cardiol. Young 2014, 24, 442–446. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Z.; Wu, C.; Pan, Z.; Xiang, L.; Liu, H.; Jin, X.; Tong, K.; Fan, S.; Jin, X. Potential association of long noncoding RNA HA117 with tetralogy of Fallot. Genes Dis. 2018, 5, 185–190. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Meese, E.; Keller, A.; Abdul-Khaliq, H.; Rädle-Hurst, T. Analysis of circulating microRNAs in patients with repaired Tetralogy of Fallot with and without heart failure. J. Transl. Med. 2017, 15, 156. [Google Scholar] [CrossRef]
- Lai, C.T.M.; Ng, E.K.O.; Chow, P.-C.; Kwong, A.; Cheung, Y.-F. Circulating MicroRNA in patients with repaired tetralogy of Fallot. Eur. J. Clin. Investig. 2017, 47, 574–582. [Google Scholar] [CrossRef]
- Saxena, A.; Tabin, C.J. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc. Natl. Acad. Sci. USA 2010, 107, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-P.; Chen, J.-F.; Regan, J.N.; Maguire, C.T.; Tang, R.-H.; Dong, X.R.; Majesky, M.W.; Wang, D.Z. Loss of microRNAs in neural crest leads to cardiovascular syndromes resembling human congenital heart defects. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 2575–2586. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Velasco, E.; Franco, D.; Aranega, A.; Daimi, H. Genetics and Epigenetics of Atrial Fibrillation. Int. J. Mol. Sci. 2020, 21, 5717. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.; Aranega, A.; Dominguez, J.N. Non-coding RNAs and Atrial Fibrillation. Adv. Exp. Med. Biol. 2020, 1229, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Girmatsion, Z.; Biliczki, P.; Bonauer, A.; Wimmer-Greinecker, G.; Scherer, M.; Moritz, A.; Moritz, A.; Bukowska, A.; Goette, A.; Nattel, S.; et al. Changes in microRNA-1 expression and IK1 up-regulation in human atrial fibrillation. Heart Rhythm. 2009, 6, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-D.; Hong, Y.-F.; Yusufuaji, Y.; Tang, B.-P.; Zhou, X.-H.; Xu, G.-J.; Li, J.X.; Sun, L.; Zhang, J.H.; Xin, Q.; et al. Altered expression of hyperpolarization-activated cyclic nucleotide-gated channels and microRNA-1 and -133 in patients with age-associated atrial fibrillation. Mol. Med. Rep. 2015, 12, 3243–3248. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zheng, S.; Xie, X.; Zhang, Y.; Wang, W.; Wang, Z.; Zhang, Y.; Wang, J.; Gao, M.; Hou, Y. MicroRNA-1 accelerates the shortening of atrial effective refractory period by regulating KCNE1 and KCNB2 expression: An atrial tachypacing rabbit model. PLoS ONE 2013, 8, e85639. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xiao, Z.; Guo, H.; Fang, X.; Liang, J.; Zhu, J.; Yang, J.; Li, H.; Pan, R.; Yuan, S.; et al. Novel role of the clustered miR-23b-3p and miR-27b-3p in enhanced expression of fibrosis-associated genes by targeting TGFBR3 in atrial fibroblasts. J. Cell. Mol. Med. 2019, 23, 3246–3256. [Google Scholar] [CrossRef]
- Harada, M.; Luo, X.; Qi, X.Y.; Tadevosyan, A.; Maguy, A.; Ordog, B.; Ledoux, J.; Kato, T.; Naud, P.; Voigt, N.; et al. Transient receptor potential canonical-3 channel-dependent fibroblast regulation in atrial fibrillation. Circulation 2012, 126, 2051–2064. [Google Scholar] [CrossRef] [PubMed]
- Dawson, K.; Wakili, R.; Ordög, B.; Clauss, S.; Chen, Y.; Iwasaki, Y.; Voigt, N.; Qi, X.Y.; Sinner, M.F.; Dobrev, D.; et al. MicroRNA29: A mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 2013, 127, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Cardin, S.; Guasch, E.; Luo, X.; Naud, P.; Le Quang, K.; Shi, Y.; Tardif, J.C.; Comtois, P.; Nattel, S. Role for MicroRNA-21 in atrial profibrillatory fibrotic remodeling associated with experimental postinfarction heart failure. Circ. Arrhythm. Electrophysiol. 2012, 5, 1027–1035. [Google Scholar] [CrossRef]
- Cao, W.; Shi, P.; Ge, J.-J. miR-21 enhances cardiac fibrotic remodeling and fibroblast proliferation via CADM1/STAT3 pathway. BMC Cardiovasc. Disord. 2017, 17, 88. [Google Scholar] [CrossRef]
- Yuan, C.-T.; Li, X.-X.; Cheng, Q.-J.; Wang, Y.-H.; Wang, J.-H.; Liu, C.-L. MiR-30a regulates the atrial fibrillation-induced myocardial fibrosis by targeting snail 1. Int. J. Clin. Exp. Pathol. 2015, 8, 15527–15536. [Google Scholar]
- Xu, J.; Wu, H.; Chen, S.; Qi, B.; Zhou, G.; Cai, L.; Zhao, L.; Wei, Y.; Liu, S. MicroRNA-30c suppresses the pro-fibrogenic effects of cardiac fibroblasts induced by TGF-β1 and prevents atrial fibrosis by targeting TGFβRII. J. Cell. Mol. Med. 2018, 22, 3045–3057. [Google Scholar] [CrossRef]
- Tsoporis, J.N.; Fazio, A.; Rizos, I.K.; Izhar, S.; Proteau, G.; Salpeas, V.; Rigopoulos, A.; Sakadakis, E.; Toumpoulis, I.K.; Parker, T.G. Increased right atrial appendage apoptosis is associated with differential regulation of candidate MicroRNAs 1 and 133A in patients who developed atrial fibrillation after cardiac surgery. J. Mol. Cell Cardiol. 2018, 121, 25–32. [Google Scholar] [CrossRef]
- Zhang, X.; Jing, W. Upregulation of miR-122 is associated with cardiomyocyte apoptosis in atrial fibrillation. Mol. Med. Rep. 2018, 18, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Tang, Y.; Peng, J.; He, J.; Zou, Q.; Yan, S.; Zheng, Z.; Pan, H. miR-520d suppresses rapid pacing-induced apoptosis of atrial myocytes through mediation of ADAM10. J. Mol. Histol. 2021, 52, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.-B.; Li, K.; Wang, G.; Gao, G.-M.; Du, J.-X. MiR-23 enhances cardiac fibroblast proliferation and suppresses fibroblast apoptosis via targeting TGF-β1 in atrial fibrillation. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4419–4424. [Google Scholar] [CrossRef] [PubMed]
- Sacks, D.; Baxter, B.; Campbell, B.C.V.; Carpenter, J.S.; Cognard, C.; Dippel, D.; Eesa, M.; Fischer, U.; Hausegger, K.; Hirsch, J.A.; et al. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int. J. Stroke 2018, 13, 612–632. [Google Scholar] [CrossRef]
- Redfield, M.M. Heart Failure with Preserved Ejection Fraction. N. Engl. J. Med. 2016, 375, 1868–1877. [Google Scholar] [CrossRef]
- Murphy, S.P.; Ibrahim, N.E.; Januzzi, J.L.J. Heart Failure with Reduced Ejection Fraction: A Review. J. Am. Med. Assoc. 2020, 324, 488–504. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Hsu, J.J.; Ziaeian, B.; Fonarow, G.C. Heart Failure with Mid-range Ejection Fraction. Curr. Heart Fail. Rep. 2020, 17, 1–8. [Google Scholar] [CrossRef]
- Ben-Nun, D.; Buja, L.M.; Fuentes, F. Prevention of heart failure with preserved ejection fraction (HFpEF): Reexamining microRNA-21 inhibition in the era of oligonucleotide-based therapeutics. Cardiovasc. Pathol. 2020, 49, 107243. [Google Scholar] [CrossRef]
- Yuan, J.; Chen, H.; Ge, D.; Xu, Y.; Xu, H.; Yang, Y.; Gu, M.; Zhou, Y.; Zhu, J.; Ge, T.; et al. Mir-21 Promotes Cardiac Fibrosis After Myocardial Infarction via Targeting Smad7. Cell Physiol. Biochem. 2017, 42, 2207–2219. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Volkmann, I.; Jazbutyte, V.; Dangwal, S.; Park, D.-H.; Thum, T. Transforming growth factor-β-induced endothelial-to-mesenchymal transition is partly mediated by microRNA-21. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, X.; Zhang, S.; Lin, Y.; Yang, J.; Zhang, C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J. Mol. Cell. Cardiol. 2009, 47, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Ma, W.; Hao, B.; Hu, F.; Yan, L.; Yan, X.; Wang, Y.; Chen, Z.; Wang, Z. microRNA-21 promotes cardiac fibrosis and development of heart failure with preserved left ventricular ejection fraction by up-regulating Bcl-2. Int. J. Clin. Exp. Pathol. 2014, 7, 565–574. [Google Scholar] [PubMed]
- Inácio, J.M.; Cristo, F.; Pinheiro, M.; Vasques-Nóvoa, F.; Saraiva, F.; Nunes, M.M.; Rosas, G.; Reis, A.; Coimbra, R.; Oliveira, J.L.; et al. Myocardial RNA Sequencing Reveals New Potential Therapeutic Targets in Heart Failure with Preserved Ejection Fraction. Biomedicines 2023, 11, 2131. [Google Scholar] [CrossRef] [PubMed]
- van Almen, G.C.; Verhesen, W.; van Leeuwen, R.E.W.; van de Vrie, M.; Eurlings, C.; Schellings, M.W.M.; Swinnen, M.; Cleutjens, J.P.; van Zandvoort, M.A.; Heymans, S.; et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell 2011, 10, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Deckx, S.; Heggermont, W.; Carai, P.; Rienks, M.; Dresselaers, T.; Himmelreich, U.; van Leeuwen, R.; Lommen, W.; van der Velden, J.; Gonzalez, A.; et al. Osteoglycin prevents the development of age-related diastolic dysfunction during pressure overload by reducing cardiac fibrosis and inflammation. Matrix Biol. 2018, 66, 110–124. [Google Scholar] [CrossRef]
- van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef]
- Calderon-Dominguez, M.; Belmonte, T.; Quezada-Feijoo, M.; Ramos, M.; Calderon-Dominguez, J.; Campuzano, O.; Mangas, A.; Toro, R. Plasma microrna expression profile for reduced ejection fraction in dilated cardiomyopathy. Sci. Rep. 2021, 11, 7517. [Google Scholar] [CrossRef]
- Duisters, R.F.; Tijsen, A.J.; Schroen, B.; Leenders, J.J.; Lentink, V.; van der Made, I.; Herias, V.; van Leeuwen, R.E.; Schellings, M.W.; Barenbrug, P.; et al. miR-133 and miR-30 regulate connective tissue growth factor: Implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 2009, 104, 170–178. [Google Scholar] [CrossRef]
- Ramírez, C.M.; Goedeke, L.; Rotllan, N.; Yoon, J.-H.; Cirera-Salinas, D.; Mattison, J.A.; Suárez, Y.; de Cabo, R.; Gorospe, M.; Fernández-Hernando, C. MicroRNA 33 regulates glucose metabolism. Mol. Cell. Biol. 2013, 33, 2891–2902. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef]
- Bhatia, H.; Verma, G.; Datta, M. miR-107 orchestrates ER stress induction and lipid accumulation by post-transcriptional regulation of fatty acid synthase in hepatocytes. Biochim. Biophys. Acta 2014, 1839, 334–343. [Google Scholar] [CrossRef]
- Essandoh, K.; Li, Y.; Huo, J.; Fan, G.-C. MiRNA-Mediated Macrophage Polarization and Its Potential Role in the Regulation of Inflammatory Response. Shock 2016, 46, 122–131. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Esteves, J.V.; Enguita, F.J.; Machado, U.F. MicroRNAs-Mediated Regulation of Skeletal Muscle GLUT4 Expression and Translocation in Insulin Resistance. J. Diabetes Res. 2017, 2017, 7267910. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Most, P.; Peppel, K. Induction of microRNA-138 by pro-inflammatory cytokines causes endothelial cell dysfunction. FEBS Lett. 2014, 588, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Chen, S.; Gordon, A.D.; Chakrabarti, S. miR-146a mediates inflammatory changes and fibrosis in the heart in diabetes. J. Mol. Cell Cardiol. 2017, 105, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Heymans, S.; Corsten, M.F.; Verhesen, W.; Carai, P.; van Leeuwen, R.E.W.; Custers, K.; Peters, T.; Hazebroek, M.; Stöger, L.; Wijnands, E.; et al. Macrophage microRNA-155 promotes cardiac hypertrophy and failure. Circulation 2013, 128, 1420–1432. [Google Scholar] [CrossRef]
- Chu, B.; Wu, T.; Miao, L.; Mei, Y.; Wu, M. MiR-181a regulates lipid metabolism via IDH1. Sci. Rep. 2015, 5, 8801. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Song, Y.; Liu, L.; Hou, N.; An, X.; Zhan, D.; Li, Y.; Zhou, L.; Li, P.; Yu, L.; et al. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death Differ. 2017, 24, 1205–1213. [Google Scholar] [CrossRef]
- Geiger, J.; Dalgaard, L.T. Interplay of mitochondrial metabolism and microRNAs. Cell Mol. Life Sci. 2017, 74, 631–646. [Google Scholar] [CrossRef]
- Soni, M.S.; Rabaglia, M.E.; Bhatnagar, S.; Shang, J.; Ilkayeva, O.; Mynatt, R.; Zhou, Y.P.; Schadt, E.E.; Thornberry, N.A.; Muoio, D.M.; et al. Downregulation of carnitine acyl-carnitine translocase by miRNAs 132 and 212 amplifies glucose-stimulated insulin secretion. Diabetes 2014, 63, 3805–3814. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Melton, D.W.; Gelfond, J.A.L.; McManus, L.M.; Shireman, P.K. MiR-351 transiently increases during muscle regeneration and promotes progenitor cell proliferation and survival upon differentiation. Physiol. Genom. 2012, 44, 1042–1051. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Drosatos, K.; Hiyama, Y.; Goldberg, I.J.; Zannis, V.I. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J. Lipid Res. 2010, 51, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Carrer, M.; Liu, N.; Grueter, C.E.; Williams, A.H.; Frisard, M.I.; Hulver, M.W.; Bassel-Duby, R.; Olson, E.N. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc. Natl. Acad. Sci. USA 2012, 109, 15330–15335. [Google Scholar] [CrossRef]
- Watson, C.J.; Gupta, S.K.; O’Connell, E.; Thum, S.; Glezeva, N.; Fendrich, J.; Gallagher, J.; Ledwidge, M.; Grote-Levi, L.; McDonald, K.; et al. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur. J. Heart Fail. 2015, 17, 405–415. [Google Scholar] [CrossRef]
- Freeman, A.M.; Raman, S.V.; Aggarwal, M.; Maron, D.J.; Bhatt, D.L.; Parwani, P.; Osborne, J.; Earls, J.P.; Min, J.K.; Bax, J.J.; et al. Integrating Coronary Atherosclerosis Burden and Progression with Coronary Artery Disease Risk Factors to Guide Therapeutic Decision Making. Am. J. Med. 2023, 136, 260–269.e7. [Google Scholar] [CrossRef]
- Henein, M.Y.; Vancheri, S.; Longo, G.; Vancheri, F. The Role of Inflammation in Cardiovascular Disease. Int. J. Mol. Sci. 2022, 23, 12906. [Google Scholar] [CrossRef] [PubMed]
- Surina, S.; Fontanella, R.A.; Scisciola, L.; Marfella, R.; Paolisso, G.; Barbieri, M. miR-21 in Human Cardiomyopathies. Front. Cardiovasc. Med. 2021, 8, 767064. [Google Scholar] [CrossRef]
- Wei, Y.; Nazari-Jahantigh, M.; Neth, P.; Weber, C.; Schober, A. MicroRNA-126, -145, and -155: A therapeutic triad in atherosclerosis? Arter. Thromb. Vasc. Biol. 2013, 33, 449–454. [Google Scholar] [CrossRef]
- Li, P.; Wei, J.; Li, X.; Cheng, Y.; Chen, W.; Cui, Y.; Simoncini, T.; Gu, Z.; Yang, J.; Fu, X. 17β-Estradiol Enhances Vascular Endothelial Ets-1/miR-126-3p Expression: The Possible Mechanism for Attenuation of Atherosclerosis. J. Clin. Endocrinol. Metab. 2017, 102, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.A.; Yamakuchi, M.; Ferlito, M.; Mendell, J.T.; Lowenstein, C.J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA 2008, 105, 1516–1521. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Narang, R.; Sreenivas, V.; Rastogi, V.; Bhatia, J.; Saluja, D.; Srivastava, K. Circulatory miR-133b and miR-21 as Novel Biomarkers in Early Prediction and Diagnosis of Coronary Artery Disease. Genes 2020, 11, 164. [Google Scholar] [CrossRef] [PubMed]
- Bruen, R.; Fitzsimons, S.; Belton, O. miR-155 in the Resolution of Atherosclerosis. Front. Pharmacol. 2019, 10, 463. [Google Scholar] [CrossRef] [PubMed]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Simoons, M.L.; Chaitman, B.R.; White, H.D.; Katus, H.A.; Lindahl, B.; Morrow, D.A.; Clemmensen, P.M.; et al. Third universal definition of myocardial infarction. Circulation 2012, 126, 2020–2035. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Liu, M.; Sun, R.; Zheng, Y.; Zhang, P. Myocardial Infarction: Symptoms and Treatments. Cell Biochem. Biophys. 2015, 72, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Tan, N.; Yang, J.; Liu, X.; Cao, X.; He, P.; Dong, X.; Qin, S.; Zhang, C. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin. Sci. 2010, 119, 87–95. [Google Scholar] [CrossRef]
- Huang, X.; Wang, J. miR-1 Mediated AMPK Pathway on Cardiomyocyte Apoptosis in Hypertensive Rats. Cell. Mol. Biol. 2022, 68, 135–140. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Chen, W.; Xie, L.; Zhao, Z.-A.; Yang, J.; Chen, Y.; Lei, W.; Shen, Z. MicroRNA-133 overexpression promotes the therapeutic efficacy of mesenchymal stem cells on acute myocardial infarction. Stem Cell Res. Ther. 2017, 8, 268. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, Y.; Zhang, J.-F.; Fu, W.-M. MicroRNA-133 mediates cardiac diseases: Mechanisms and clinical implications. Exp. Cell Res. 2017, 354, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Yang, C.; Han, Z. Circulating miR-499 as a potential biomarker for acute myocardial infarction. Ann. Transl. Med. 2016, 4, 135. [Google Scholar] [CrossRef]
- Abkhooie, L.; Sarabi, M.M.; Kahroba, H.; Eyvazi, S.; Montazersaheb, S.; Tarhriz, V.; Hejazi, M.S. Potential Roles of MyomiRs in Cardiac Development and Related Diseases. Curr. Cardiol. Rev. 2021, 17, e010621188335. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.-F.; Chu, T.; Ruan, Z.; Zhang, M.; Zhou, M.; Zhang, Q.; Zhang, R.; Wu, L. Inflammation-Related MicroRNAs Are Associated with Plaque Stability Calculated by IVUS in Coronary Heart Disease Patients. J. Interv. Cardiol. 2019, 2019, 9723129. [Google Scholar] [CrossRef]
- Bronze-da-Rocha, E. MicroRNAs expression profiles in cardiovascular diseases. Biomed. Res. Int. 2014, 2014, 985408. [Google Scholar] [CrossRef]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.-F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.; Srivastava, D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef]
- Kunz, M.; Xiao, K.; Liang, C.; Viereck, J.; Pachel, C.; Frantz, S.; Thum, T.; Dandekar, T. Bioinformatics of cardiovascular miRNA biology. J. Mol. Cell. Cardiol. 2015, 89, 3–10. [Google Scholar] [CrossRef]
- Lukasik, A.; Wójcikowski, M.; Zielenkiewicz, P. Tools4miRs—One place to gather all the tools for miRNA analysis. Bioinformatics 2016, 32, 2722–2724. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Burge, S.W.; Bateman, A.; Daub, J.; Eberhardt, R.Y.; Eddy, S.R.; Floden, E.W.; Gadner, P.P.; Jones, T.A.; Tate, J. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015, 43, D130–D137. [Google Scholar] [CrossRef]
- Eddy, S.R. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinform. 2002, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Richter, A.S.; Backofen, R. IntaRNA: Efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 2008, 24, 2849–2856. [Google Scholar] [CrossRef] [PubMed]
- Vila-Casadesús, M.; Gironella, M.; Lozano, J.J. MiRComb: An R Package to Analyse miRNA-mRNA Interactions. Examples across Five Digestive Cancers. PLoS ONE 2016, 11, e0151127. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Alanis-Lobato, G.; Andrade-Navarro, M.A.; Schaefer, M.H. HIPPIE v2.0: Enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res. 2017, 45, D408–D414. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Wang, J.; Duncan, D.; Shi, Z.; Zhang, B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013, 41, W77–W83. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Cho, J.-W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Kostoulas, N.; Vergoulis, T.; Georgakilas, G.; Reczko, M.; Maragkakis, M.; Paraskevopoulou, M.D.; Prionidis, K.; Dalamagas, T.; Hatzigeorgiou, A.G.; et al. DIANA miRPath v.2.0: Investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012, 40, W498–W504. [Google Scholar] [CrossRef]
- Cho, S.; Jang, I.; Jun, Y.; Yoon, S.; Ko, M.; Kwon, Y.; Choi, I.; Chang, H.; Ryu, D.; Lee, B.; et al. MiRGator v3.0: A microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013, 41, D252–D257. [Google Scholar] [CrossRef]
- Wright, P.R.; Georg, J.; Mann, M.; Sorescu, D.A.; Richter, A.S.; Lott, S.; Kleinkauf, R.; Hess, W.R.; Backofen, R. CopraRNA and IntaRNA: Predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014, 42, W119–W123. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014, 42, D1070–D1074. [Google Scholar] [CrossRef] [PubMed]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, S.; Wan, Y.; Zhao, Y.; Shi, J.; Zhou, Y.; Cui, Q. MISIM v2.0: A web server for inferring microRNA functional similarity based on microRNA-disease associations. Nucleic Acids Res. 2019, 47, W536–W541. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ai, H.; Li, B.; Zhang, C.; Meng, F.; Ai, Y. MIMRDA: A Method Incorporating the miRNA and mRNA Expression Profiles for Predicting miRNA-Disease Associations to Identify Key miRNAs (microRNAs). Front. Genet. 2022, 13, 825318. [Google Scholar] [CrossRef]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Knott, K.E.; Wong, G. miRToolsGallery: A tag-based and rankable microRNA bioinformatics resources database portal. Database 2018, 2018, bay004. [Google Scholar] [CrossRef]
- Aghaee-Bakhtiari, S.H. miRandb: A Metadatabase of Online Resources of miRNA and miRNA Targets. Methods Mol. Biol. 2019, 1970, 15–30. [Google Scholar] [CrossRef]
- The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Henry, V.J.; Bandrowski, A.E.; Pepin, A.-S.; Gonzalez, B.J.; Desfeux, A. OMICtools: An informative directory for multi-omic data analysis. Database 2014, 2014, bau069. [Google Scholar] [CrossRef]
- Pignatelli, M.; Vilella, A.J.; Muffato, M.; Gordon, L.; White, S.; Flicek, P.; Herrero, J. ncRNA orthologies in the vertebrate lineage. Database 2016, 2016, bav127. [Google Scholar] [CrossRef]
- Wang, L.; Zhong, Q.; Feng, Y.; Tang, X.; Wang, Q.; Zou, Y.; Duan, J. Long noncoding RNA TUG1 is downregulated in sepsis and may sponge miR-27a to downregulate tumor necrosis factor-α. J. Int. Med. Res. 2020, 48, 300060520910638. [Google Scholar] [CrossRef]
- Lv, Z.; Yang, K.; Wang, Y. Long non-coding RNA breast cancer-associated transcript 54 sponges microRNA-1269b to suppress the proliferation of hemangioma-derived endothelial cells. Bioengineered 2022, 13, 6188–6195. [Google Scholar] [CrossRef]
- Vilella-Figuerola, A.; Gallinat, A.; Escate, R.; Mirabet, S.; Padró, T.; Badimon, L. Systems Biology in Chronic Heart Failure-Identification of Potential miRNA Regulators. Int. J. Mol. Sci. 2022, 23, 15226. [Google Scholar] [CrossRef] [PubMed]
- Xue, P.; Liu, Y.; Wang, H.; Huang, J.; Luo, M. miRNA-103-3p-Hlf regulates apoptosis and autophagy by targeting hepatic leukaemia factor in heart failure. ESC Hear. Fail. 2023, 10, 3038–3045. [Google Scholar] [CrossRef]
- Romano, G.L.; Platania, C.B.M.; Forte, S.; Salomone, S.; Drago, F.; Bucolo, C. MicroRNA target prediction in glaucoma. Prog. Brain Res. 2015, 220, 217–240. [Google Scholar] [CrossRef]
- Sabatino, J.; Wicik, Z.; De Rosa, S.; Eyileten, C.; Jakubik, D.; Spaccarotella, C.; Mongiardo, A.; Postula, M.; Indolfi, C. MicroRNAs fingerprint of bicuspid aortic valve. J. Mol. Cell. Cardiol. 2019, 134, 98–106. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.-S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Zheng, Y.; Gao, L. MiRNA-disease association prediction based on meta-paths. Brief. Bioinform. 2022, 23, bbab571. [Google Scholar] [CrossRef]
- Yousef, M.; Goy, G.; Bakir-Gungor, B. miRModuleNet: Detecting miRNA-mRNA Regulatory Modules. Front. Genet. 2022, 13, 767455. [Google Scholar] [CrossRef]
- Yuan, L.; Zhao, J.; Sun, T.; Shen, Z. A machine learning framework that integrates multi-omics data predicts cancer-related LncRNAs. BMC Bioinform. 2021, 22, 332. [Google Scholar] [CrossRef] [PubMed]
- Bang, D.; Gu, J.; Park, J.; Jeong, D.; Koo, B.; Yi, J.; Shin, J.; Jung, I.; Kim, S.; Lee, S. A Survey on Computational Methods for Investigation on ncRNA-Disease Association through the Mode of Action Perspective. Int. J. Mol. Sci. 2022, 23, 11498. [Google Scholar] [CrossRef] [PubMed]
- Paschoal, A.R.; Maracaja-Coutinho, V.; Setubal, J.C.; Simões, Z.L.P.; Verjovski-Almeida, S.; Durham, A.M. Non-coding transcription characterization and annotation: A guide and web resource for non-coding RNA databases. RNA Biol. 2012, 9, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Zhang, D.; Lv, L.; Shi, W.; Song, Z.; Yi, B.; Lai, B.; Chen, Q.; Yang, S.; Hua, P. Bioinformatic gene analysis for potential biomarkers and therapeutic targets of atrial fibrillation-related stroke. J. Transl. Med. 2019, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Kotlabova, K.; Doucha, J.; Hromadnikova, I. Placental-specific microRNA in maternal circulation--identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 2011, 89, 185–191. [Google Scholar] [CrossRef]
- Omran, A.; Elimam, D.; Webster, K.A.; Shehadeh, L.A.; Yin, F. MicroRNAs: A new piece in the paediatric cardiovascular disease puzzle. Cardiol. Young. 2013, 23, 642–655. [Google Scholar] [CrossRef]
- de Los Reyes-García, A.M.; Zapata-Martínez, L.; Águila, S.; Lozano, M.L.; Martínez, C.; González-Conejero, R. microRNAs as biomarkers of risk of major adverse cardiovascular events in atrial fibrillation. Front. Cardiovasc. Med. 2023, 10, 1135127. [Google Scholar] [CrossRef] [PubMed]
- Duygu, B.; de Windt, L.J.; da Costa Martins, P.A. Targeting microRNAs in heart failure. Trends Cardiovasc. Med. 2016, 26, 99–110. [Google Scholar] [CrossRef]
- Alcalde, M.; Toro, R.; Bonet, F.; Córdoba-Caballero, J.; Martínez-Barrios, E.; Ranea, J.A.; Vallverdú-Prats, M.; Brugada, R.; Meraviglia, V.; Bellin, M.; et al. Role of microRNAs in arrhythmogenic cardiomyopathy: Translation as biomarkers into clinical practice. Transl. Res. 2023, 259, 72–82. [Google Scholar] [CrossRef]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Täubel, J.; Hauke, W.; Rump, S.; Viereck, J.; Batkai, S.; Poetzsch, J.; Rode, L.; Weigt, H.; Genschel, C.; Lorch, U.; et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: Results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur. Heart J. 2021, 42, 178–188. [Google Scholar] [CrossRef] [PubMed]
| Type/Goal | Tool | Reference |
|---|---|---|
| Discovery | miRDeep | [12] |
| miRDeep2 | [13] | |
| MiRFinder | [14] | |
| miReader | [15] | |
| RNAz2 | [16] | |
| Database and annotation | miRbase | [17] |
| miRNet | [18] | |
| RNAcentral | [19] | |
| Target prediction | miRanda | [20] |
| TargetScan | [21] | |
| PicTar | [22] | |
| PITA | [23] | |
| miRDB | [24] | |
| RNAhybrid | [25] | |
| Target databases | DIANA-TarBase | [26] |
| MiRTarBase | [27] | |
| MiRWalk | [28] | |
| Pathway analysis | DIANA Tools | [29] |
| Cytoscape | [30] | |
| Structure prediction | ViennaRNA Package | [31] |
| MiRScan | [32] | |
| miRseeker | [33] |
| miRNA | Result | Reference |
|---|---|---|
| miR-18a | Targets CTGF and TSP-1 | [118] |
| miR-19a | Targets CTGF and TSP-1 | [118] |
| miR-21 | Upregulated in HFpEF. Involved in cellular processes such as the proliferation, hypertrophy, apoptosis, and fibrosis of different cell types of the heart | [112] |
| miR-22 | Targets OGN | [119] |
| miR-25 | Upregulated in HFpEF. Inhibits HAPLN1mRNA | [117] |
| miR-26a | Upregulated in HFpEF. Inhibits NPPB and HAPLN1 mRNA | [117] |
| miR-29 | Targets extracellular matrix-related genes | [120] |
| miR-29a | Correlated with myocardial fibrosis | [121] |
| miR-30 | Targets CTGF | [122] |
| miR-33a/b | Targets PCK1 and G6PC | [123] |
| miR-103 | Targets CAV1 and FASN | [124] |
| miR-107 | Targets CAV1 and FASN | [125] |
| miR-125a | Upregulated in HFrEF. Induces M2 polarization in macrophages | [126] |
| miR-126 | Targets SPRED-1 | [127] |
| miR-133 | Targets GLUT4 | [128] |
| miR-138 | Targets S100A1 | [129] |
| miR-140 | Upregulated in HFpEF. Inhibits NPPB mRNA | [117] |
| miR-146a | Targets NFKB1 | [130] |
| miR-155 | Targets SOCS1 | [131] |
| miR-181a/c | Targets IDH1 | [132] |
| miR-199a | Targets GSK3B | [133] |
| miR-210 | Targets ISCU | [134] |
| miR-212 | Targets CACT | [135] |
| miR-223 | Targets GLUT4 | [128] |
| miR-351 | Targets E2F3 | [136] |
| miR-370 | Targets miR-122 expression | [137] |
| miR-378 | Targets PPARGC1B | [138] |
| miR-4429a | Upregulated in HFpEF. Inhibits HAPLN1 mRNA | [117] |
| Functionality | Tool | Possible Applications | Reference |
|---|---|---|---|
| Family identification: | |||
| Rfam | RNA annotation | [161] | |
| Infernal | Find homologous RNAs | [162] | |
| RNA–RNA interactions: | |||
| Prediction: | IntaRNA | Target prediction | [163] |
| miRComb | Target prediction | [164] | |
| Functional analysis: | |||
| STRING | Target expression retrieval | [165] | |
| Based on expression | HiPPIE | Target interactome data retrieval | [166] |
| Cytoscape | Network visualization | [30] | |
| Gene Ontology | Enrichment analysis | ||
| Based on target function | KEGG | Enrichment analysis | |
| REACTOME | Pathway enrichment analysis | [167] | |
| Based on transcription regulation | WebGestalt | Find associated transcription factors, miRNAs, and mRNAs | [168] |
| TTRUST | Find associated transcription factors, miRNAs, and mRNAs | [169] | |
| Integrated tools | DIANA miRPath | Automatic functional interpretation based on the existing evidence (all of the above) | [170] |
| miRGator | [171] | ||
| CopraRNA | [172] | ||
| Disease association: | |||
| miR2Disease | Linking miRNA to disease | [173] | |
| HMDD | Retrieve miRNA–disease-associated omics data | [174] | |
| multiMiR | Use of disease and drug associations to find targets | [175] | |
| MISIM | Predict miRNA–disease association | [176] | |
| MIMRDA | Find top-ranked key miRNAs associated with diseases | [177] | |
| Integrative data analysis: | |||
| ncRNA-specific | miRToolsGallery (and similar) | Search, filter, and rank ncRNA-dedicated tools | [178] |
| miRDIP | Find miRNA regulatory networks using ML methodologies | [179] | |
| Generalistic | Galaxy | Build, run, and share bioinformatics pipelines using standard-use tools | [180] |
| GeneMANIA | Explore functional relationships between miRNA and protein-coding genes | [181] | |
| OMICtools | Integrate multiomics data to find novel regulatory networks and pathways | [182] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozano-Velasco, E.; Inácio, J.M.; Sousa, I.; Guimarães, A.R.; Franco, D.; Moura, G.; Belo, J.A. miRNAs in Heart Development and Disease. Int. J. Mol. Sci. 2024, 25, 1673. https://doi.org/10.3390/ijms25031673
Lozano-Velasco E, Inácio JM, Sousa I, Guimarães AR, Franco D, Moura G, Belo JA. miRNAs in Heart Development and Disease. International Journal of Molecular Sciences. 2024; 25(3):1673. https://doi.org/10.3390/ijms25031673
Chicago/Turabian StyleLozano-Velasco, Estefania, José Manuel Inácio, Inês Sousa, Ana Rita Guimarães, Diego Franco, Gabriela Moura, and José António Belo. 2024. "miRNAs in Heart Development and Disease" International Journal of Molecular Sciences 25, no. 3: 1673. https://doi.org/10.3390/ijms25031673
APA StyleLozano-Velasco, E., Inácio, J. M., Sousa, I., Guimarães, A. R., Franco, D., Moura, G., & Belo, J. A. (2024). miRNAs in Heart Development and Disease. International Journal of Molecular Sciences, 25(3), 1673. https://doi.org/10.3390/ijms25031673








