Fluorescence-Based Protein Stability Monitoring—A Review
Abstract
1. Introduction
1.1. Usability of Protein Stability Assays
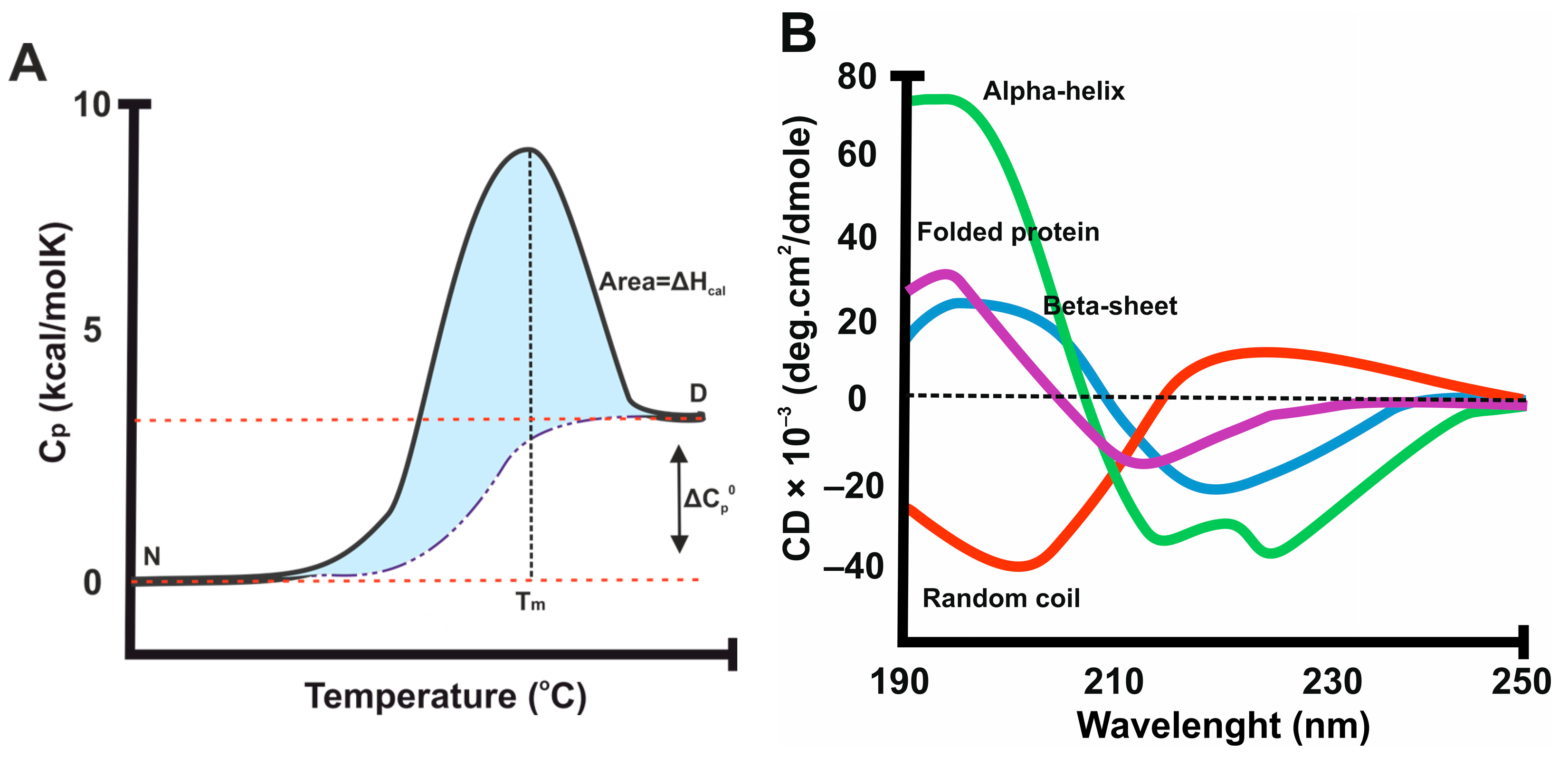
1.2. Stability Parameters
1.3. Interaction Monitoring
1.4. Non-Luminescent Methods
2. Luminescence-Based Thermal Shift Assays (TSAs)
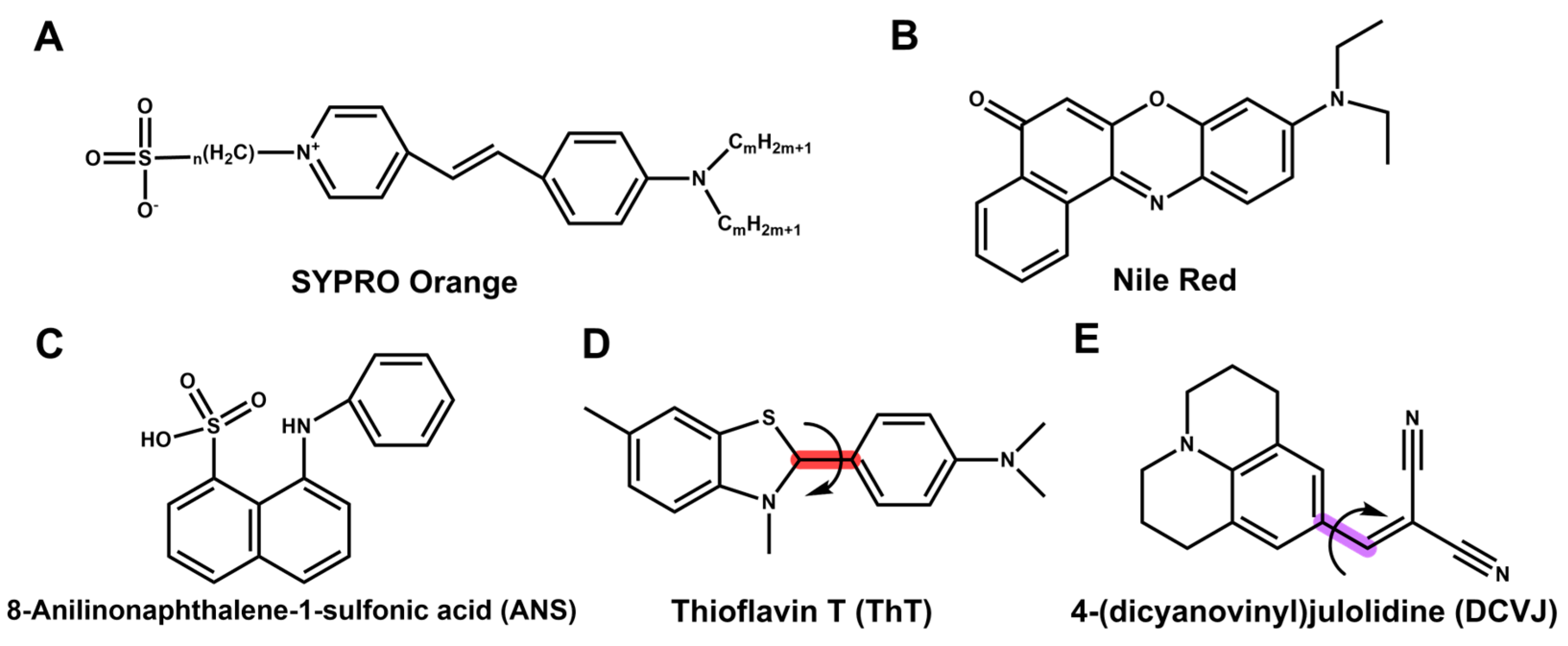
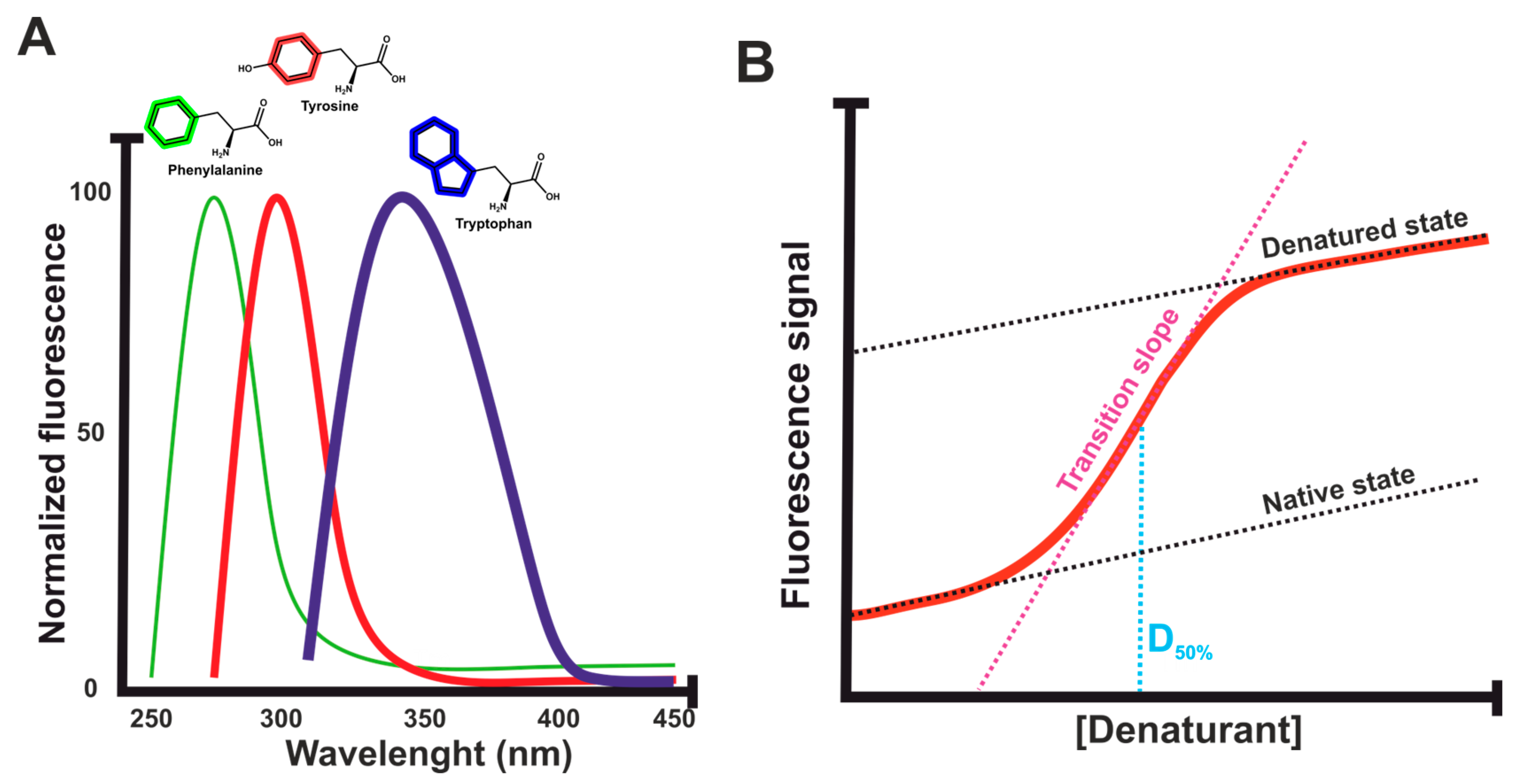
2.1. Fluorescent Dyes vs. Intrinsic Fluorescence
2.2. Equipment Demands
2.3. Buffer Components and Interferences
2.4. Functionality Comparison to Other Methods
2.5. Data Analysis
3. Isothermal Chemical Denaturation (ICD) Assays
3.1. Fluorescent Dyes vs. Intrinsic Fluorescence
3.2. Denaturants and Their Function
3.3. ICD Comparison to TSA
4. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Strutz, W. Exploring protein stability by NanoDSF. Biophys. J. 2016, 110, 393a. [Google Scholar] [CrossRef]
- Vuorinen, E.; Valtonen, S.; Eskonen, V.; Kariniemi, T.; Jakovleva, J.; Kopra, K.; Harma, H. Sensitive label-free thermal stability assay for protein denaturation and protein–Ligand interaction studies. Anal. Chem. 2020, 92, 3512–3516. [Google Scholar] [CrossRef] [PubMed]
- Shire, S.J. Formulation and manufacturability of biologics. Curr. Opin. Biotechnol. 2009, 20, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Korotchkina, L.G.; Ramani, K.; Balu-Iyer, S.V. Folding considerations for therapeutic protein formulations. Prog. Mol. Biol. Transl. Sci. 2008, 83, 255–270. [Google Scholar] [CrossRef]
- Carter, P.J. Introduction to current and future protein therapeutics: A protein engineering perspective. Exp. Cell Res. 2011, 317, 1261–1269. [Google Scholar] [CrossRef]
- Samra, H.S.; He, F. Advancements in high throughput biophysical technologies: Applications for characterization and screening during early formulation development of monoclonal antibodies. Mol. Pharm. 2012, 9, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Mathias, S.; Kube, S.; Otte, K.; Garidel, P.; Gamer, M.; Blech, M.; Fischer, S.; Karow-Zwick, A.R. Rational optimization of a monoclonal antibody improves the aggregation propensity and enhances the CMC properties along the entire pharmaceutical process chain. mAbs 2020, 12, 1787121. [Google Scholar] [CrossRef]
- Kopra, K.; Härmä, H. Quenching resonance energy transfer (QRET): A single-label technique for inhibitor screening and interaction studies. New Biotechnol. 2015, 32, 575–580. [Google Scholar] [CrossRef]
- Snapp, E.L.; Hegde, R.S. Rational design and evaluation of FRET experiments to measure protein proximities in cells. Curr. Protoc. Cell Biol. 2006, 32, 17.19.11–17.19.20. [Google Scholar] [CrossRef]
- Broussard, J.A.; Green, K.J. Research techniques made simple: Methodology and applications of Förster resonance energy transfer (FRET) microscopy. J. Investig. Dermatol. 2017, 137, e185–e191. [Google Scholar] [CrossRef]
- Piston, D.W.; Kremers, G.-J. Fluorescent protein FRET: The good, the bad and the ugly. Trends Biochem. Sci. 2007, 32, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Petrauskas, V.; Kazlauskas, E.; Gedgaudas, M.; Baranauskienė, L.; Zubrienė, A.; Matulis, D. Thermal shift assay for protein–ligand dissociation constant determination. TrAC Trends Anal. Chem. 2023, 170, 117417. [Google Scholar] [CrossRef]
- Senisterra, G.A.; Markin, E.; Yamazaki, K.; Hui, R.; Vedadi, M.; Awrey, D.E. Screening for ligands using a generic and high-throughput light-scattering-based assay. J. Biomol. Screen. 2006, 11, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Mehta, G.; Srivastava, S. Label-free detection techniques for protein microarrays: Prospects, merits and challenges. Proteomics 2010, 10, 731–748. [Google Scholar] [CrossRef]
- Miklos, A.C.; Li, C.; Pielak, G.J. Using NMR-detected backbone amide 1H exchange to assess macromolecular crowding effects on globular-protein stability. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2009; Volume 466, pp. 1–18. [Google Scholar] [CrossRef]
- Momenbeitollahi, N.; Cloet, T.; Li, H. Pushing the detection limits: Strategies towards highly sensitive optical-based protein detection. Anal. Bioanal. Chem. 2021, 413, 5995–6011. [Google Scholar] [CrossRef]
- Tolvanen, T.A. Current advances in CETSA. Front. Mol. Biosci. 2022, 9, 866764. [Google Scholar] [CrossRef]
- Molina, D.M.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef]
- Mateus, A.; Kurzawa, N.; Becher, I.; Sridharan, S.; Helm, D.; Stein, F.; Typas, A.; Savitski, M.M. Thermal proteome profiling for interrogating protein interactions. Mol. Syst. Biol. 2020, 16, e9232. [Google Scholar] [CrossRef]
- Tsuboyama, K.; Dauparas, J.; Chen, J.; Laine, E.; Mohseni Behbahani, Y.; Weinstein, J.J.; Mangan, N.M.; Ovchinnikov, S.; Rocklin, G.J. Mega-scale experimental analysis of protein folding stability in biology and design. Nature 2023, 620, 434–444. [Google Scholar] [CrossRef]
- Sauer, P.; Bantscheff, M. Thermal Proteome Profiling for Drug Target Identification and Probing of Protein States. In Mass Spectrometry-Based Proteomics. Methods in Molecular Biology; Gevaert, K., Ed.; Humana: New York, NY, USA, 2023; Volume 2718, pp. 73–98. [Google Scholar] [CrossRef]
- Alexandrov, A.I.; Mileni, M.; Chien, E.Y.; Hanson, M.A.; Stevens, R.C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 2008, 16, 351–359. [Google Scholar] [CrossRef]
- Errasti-Murugarren, E.; Bartoccioni, P.; Palacín, M. Membrane protein stabilization strategies for structural and functional studies. Membranes 2021, 11, 155. [Google Scholar] [CrossRef] [PubMed]
- Bruce, D.; Cardew, E.; Freitag-Pohl, S.; Pohl, E. How to stabilize protein: Stability screens for thermal shift assays and nano differential scanning fluorimetry in the virus-X project. JoVE J. Vis. Exp. 2019, 14, e58666. [Google Scholar] [CrossRef]
- Kwan, T.O.; Kolek, S.A.; Danson, A.E.; Reis, R.I.; Camacho, I.S.; Shaw Stewart, P.D.; Moraes, I. Measuring Protein Aggregation and Stability Using High-Throughput Biophysical Approaches. Front. Mol. Biosci. 2022, 9, 890862. [Google Scholar] [CrossRef] [PubMed]
- Kopra, K.; Valtonen, S.; Mahran, R.; Kapp, J.N.; Hassan, N.; Gillette, W.; Dennis, B.; Li, L.; Westover, K.D.; Plückthun, A. Thermal Shift Assay for Small GTPase Stability Screening: Evaluation and Suitability. Int. J. Mol. Sci. 2022, 23, 7095. [Google Scholar] [CrossRef]
- Wen, J.; Lord, H.; Knutson, N.; Wikström, M. Nano differential scanning fluorimetry for comparability studies of therapeutic proteins. Anal. Biochem. 2020, 593, 113581. [Google Scholar] [CrossRef]
- Spink, C.H. Differential scanning calorimetry. Methods Cell Biol. 2008, 84, 115–141. [Google Scholar] [CrossRef]
- Fang, X.; Zheng, Y.; Duan, Y.; Liu, Y.; Zhong, W. Recent advances in design of fluorescence-based assays for high-throughput screening. Anal. Chem. 2018, 91, 482–504. [Google Scholar] [CrossRef]
- Attene-Ramos, M.S.; Austin, C.; Xia, M. High throughput screening. In Encyclopedia of Toxicology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar] [CrossRef]
- Gao, K.; Oerlemans, R.; Groves, M.R. Theory and applications of differential scanning fluorimetry in early-stage drug discovery. Biophys. Rev. 2020, 12, 85–104. [Google Scholar] [CrossRef]
- Ericsson, U.B.; Hallberg, B.M.; DeTitta, G.T.; Dekker, N.; Nordlund, P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 2006, 357, 289–298. [Google Scholar] [CrossRef]
- Magliery, T.J.; Lavinder, J.J.; Sullivan, B.J. Protein stability by number: High-throughput and statistical approaches to one of protein science’s most difficult problems. Curr. Opin. Chem. Biol. 2011, 15, 443–451. [Google Scholar] [CrossRef]
- Boozer, C.; Kim, G.; Cong, S.; Guan, H.; Londergan, T. Looking towards label-free biomolecular interaction analysis in a high-throughput format: A review of new surface plasmon resonance technologies. Curr. Opin. Biotechnol. 2006, 17, 400–405. [Google Scholar] [CrossRef]
- Blümich, B. Introduction to compact NMR: A review of methods. TrAC Trends Anal. Chem. 2016, 83, 2–11. [Google Scholar] [CrossRef]
- Palmer III, A.G. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004, 104, 3623–3640. [Google Scholar] [CrossRef]
- Sturtevant, J.M. Biochemical applications of differential scanning calorimetry. Annu. Rev. Phys. Chem. 1987, 38, 463–488. [Google Scholar] [CrossRef]
- Wang, W.; Nema, S.; Teagarden, D. Protein aggregation—Pathways and influencing factors. Int. J. Pharm. 2010, 390, 89–99. [Google Scholar] [CrossRef]
- Housmans, J.A.; Wu, G.; Schymkowitz, J.; Rousseau, F. A guide to studying protein aggregation. FEBS J. 2023, 290, 554–583. [Google Scholar] [CrossRef]
- Gummadi, S.N. What is the role of thermodynamics on protein stability? Biotechnol. Bioprocess Eng. 2003, 8, 9–18. [Google Scholar] [CrossRef]
- Bruylants, G.; Wouters, J.; Michaux, C. Differential scanning calorimetry in life science: Thermodynamics, stability, molecular recognition and application in drug design. Curr. Med. Chem. 2005, 12, 2011–2020. [Google Scholar] [CrossRef] [PubMed]
- Ihnat, P.M.; Zhang, J.; Xu, J.; Wu, K.; Carrillo, R.J. Chapter 6: High-throughput conformational and colloidal stability screening of biologic molecules. In Development of Biopharmaceutical Drug-Device Products; Jameel, F., Skoug, J., Nesbitt, R., Eds.; AAPS Advances in the Pharmaceutical Sciences Series; Springer: Cham, Switzerland, 2020; Volume 35, pp. 117–138. [Google Scholar] [CrossRef]
- Hau, J.C.; Fontana, P.; Zimmermann, C.; De Pover, A.; Erdmann, D.; Chène, P. Leveraging the contribution of thermodynamics in drug discovery with the help of fluorescence-based thermal shift assays. J. Biomol. Screen. 2011, 16, 552–556. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moreau, M.J.; Morin, I.; Schaeffer, P.M. Quantitative determination of protein stability and ligand binding using a green fluorescent protein reporter system. Mol. BioSyst. 2010, 6, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.M.; Taylor, C.W. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 2011, 6, 365–387. [Google Scholar] [CrossRef]
- Wu, T.; Yu, J.; Gale-Day, Z.; Woo, A.; Suresh, A.; Hornsby, M.; Gestwicki, J.E. Three essential resources to improve differential scanning fluorimetry (DSF) experiments. BioRxiv 2020. [Google Scholar] [CrossRef]
- Sun, C.; Li, Y.; Yates, E.A.; Fernig, D.G. SimpleDSFviewer: A tool to analyze and view differential scanning fluorimetry data for characterizing protein thermal stability and interactions. Protein Sci. 2020, 29, 19–27. [Google Scholar] [CrossRef]
- Jameson, D.M.; Seifried, S.E. Quantification of protein–protein interactions using fluorescence polarization. Methods 1999, 19, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S.; Yendluri, M.; Poddar, S.; Li, A.; Mallick, K.; Mallik, S.; Ghosh, B. Recent Advancements in Computational Drug Design Algorithms through Machine Learning and Optimization. Kinases Phosphatases 2023, 1, 117–140. [Google Scholar] [CrossRef]
- Bernetti, M.; Cavalli, A.; Mollica, L. Protein–ligand (un) binding kinetics as a new paradigm for drug discovery at the crossroad between experiments and modelling. MedChemComm 2017, 8, 534–550. [Google Scholar] [CrossRef] [PubMed]
- Kuzu, G.; Keskin, O.; Nussinov, R.; Gursoy, A. PRISM-EM: Template interface-based modelling of multi-protein complexes guided by cryo-electron microscopy density maps. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 1137–1148. [Google Scholar] [CrossRef]
- Cala, O.; Guillière, F.; Krimm, I. NMR-based analysis of protein–ligand interactions. Anal. Bioanal. Chem. 2014, 406, 943–956. [Google Scholar] [CrossRef]
- Schlichting, I. X-ray crystallography of protein-ligand interactions. In Methods in Molecular Biology; Humana: Totowa, NJ, USA, 2005; pp. 155–165. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Vakser, I.A. Low-resolution structural modeling of protein interactome. Curr. Opin. Struct. Biol. 2013, 23, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Perozzo, R.; Folkers, G.; Scapozza, L. Thermodynamics of protein–ligand interactions: History, presence, and future aspects. J. Recept. Signal Transduct. 2004, 24, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Celej, M.S.; Dassie, S.A.; González, M.; Bianconi, M.L.; Fidelio, G.D. Differential scanning calorimetry as a tool to estimate binding parameters in multiligand binding proteins. Anal. Biochem. 2006, 350, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Grøftehauge, M.K.; Hajizadeh, N.R.; Swann, M.J.; Pohl, E. Protein–ligand interactions investigated by thermal shift assays (TSA) and dual polarization interferometry (DPI). Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, V.M.; Kaufman, G.K.; Urbach, A.R.; Gitlin, I.; Gudiksen, K.L.; Weibel, D.B.; Whitesides, G.M. Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein—Ligand binding. Chem. Rev. 2008, 108, 946–1051. [Google Scholar] [CrossRef] [PubMed]
- Chivers, C.E.; Koner, A.L.; Lowe, E.D.; Howarth, M. How the biotin–streptavidin interaction was made even stronger: Investigation via crystallography and a chimaeric tetramer. Biochem. J. 2011, 435, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Gambini, L.; Baggio, C.; Udompholkul, P.; Jossart, J.; Salem, A.F.; Perry, J.J.P.; Pellecchia, M. Covalent inhibitors of protein–protein interactions targeting lysine, tyrosine, or histidine residues. J. Med. Chem. 2019, 62, 5616–5627. [Google Scholar] [CrossRef] [PubMed]
- Malakoutikhah, M.; Mahran, R.; Gooran, N.; Masoumi, A.; Lundell, K.; Liljeblad, A.; Guiley, K.; Dai, S.; Zheng, Q.; Zhu, L. Nanomolar Protein Thermal Profiling with Modified Cyanine Dyes. Anal. Chem. 2023, 95, 18344–18351. [Google Scholar] [CrossRef]
- Valtonen, S.; Vuorinen, E.; Kariniemi, T.; Eskonen, V.; Le Quesne, J.; Bushell, M.; Harma, H.; Kopra, K. Nanomolar protein–protein interaction monitoring with a label-free Protein-Probe technique. Anal. Chem. 2020, 92, 15781–15788. [Google Scholar] [CrossRef]
- Kopra, K.; Hassan, N.; Vuorinen, E.; Valtonen, S.; Mahran, R.; Habib, H.; Jalkanen, P.; Susi, P.; Hytönen, V.; Hankaniemi, M. Rapid high-throughput compatible label-free virus particle quantification method based on time-resolved luminescence. Anal. Bioanal. Chem. 2022, 414, 4509–4518. [Google Scholar] [CrossRef]
- Capelle, M.A.; Gurny, R.; Arvinte, T. High throughput screening of protein formulation stability: Practical considerations. Eur. J. Pharm. Biopharm. 2007, 65, 131–148. [Google Scholar] [CrossRef]
- Parkins, D.A.; Lashmar, U.T. The formulation of biopharmaceutical products. Pharm. Sci. Technol. Today 2000, 3, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Mach, H.; Arvinte, T. Addressing new analytical challenges in protein formulation development. Eur. J. Pharm. Biopharm. 2011, 78, 196–207. [Google Scholar] [CrossRef]
- Brader, M.L. UV-absorbance, fluorescence and FT-IR spectroscopy in biopharmaceutical development. In Biophysical Characterization of Proteins in Developing Biopharmaceuticals; Elsevier: Amsterdam, The Netherlands, 2020; pp. 97–121. [Google Scholar]
- Durowoju, I.B.; Bhandal, K.S.; Hu, J.; Carpick, B.; Kirkitadze, M. Differential scanning calorimetry—A method for assessing the thermal stability and conformation of protein antigen. JoVE J. Vis. Exp. 2017, 121, e55262. [Google Scholar] [CrossRef]
- Holdgate, G.; Embrey, K.; Milbradt, A.; Davies, G. Biophysical methods in early drug discovery. ADMET DMPK 2019, 7, 222–241. [Google Scholar] [CrossRef]
- Mitra, D. Use of isothermal titration calorimetry to study various systems. Mater. Today Proc. 2020, 23, 284–300. [Google Scholar] [CrossRef]
- Jelesarov, I.; Bosshard, H.R. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recognit. 1999, 12, 3–18. [Google Scholar] [CrossRef]
- Gabbott, P. A practical introduction to differential scanning calorimetry. In Principles and Applications of Thermal Analysis; Wiley-Blackwell: Hoboken, NJ, USA, 2008; pp. 1–50. [Google Scholar] [CrossRef]
- Differential Scanning Calorimetry (DSC) to Measure Heat Flow. Available online: https://www.innovatechlabs.com/materials-analysis-dsc/#:~:text=Unique%20capabilities%3A%20DSC%20can%20measure,time%20takes%20approximately%201%20hour (accessed on 27 December 2023).
- Haque, M.A.; Kaur, P.; Islam, A.; Hassan, M.I. Application of circular dichroism spectroscopy in studying protein folding, stability, and interaction. In Advances in Protein Molecular and Structural Biology Methods; Elsevier: Amsterdam, The Netherlands, 2022; pp. 213–224. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004, 383, 318–351. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef]
- Baldwin, A.J.; Kay, L.E. NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 2009, 5, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Zuiderweg, E.R. Mapping protein−protein interactions in solution by NMR spectroscopy. Biochemistry 2002, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R.; Gamsjaeger, R.; Mackay, J.P. The structural analysis of protein–protein interactions by NMR spectroscopy. Proteomics 2009, 9, 5224–5232. [Google Scholar] [CrossRef]
- Kovermann, M.; Rogne, P.; Wolf-Watz, M. Protein dynamics and function from solution state NMR spectroscopy. Q. Rev. Biophys. 2016, 49, e6. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.G.; Wanner, R.; Johnson, C.M.; Breitsprecher, D.; Winter, G.; Duhr, S.; Baaske, P.; Ferguson, N. Novel microscale approaches for easy, rapid determination of protein stability in academic and commercial settings. Biochim. Biophys. Acta BBA Proteins Proteom. 2014, 1844, 2241–2250. [Google Scholar] [CrossRef] [PubMed]
- Stark, J.L.; Powers, R. Application of NMR and molecular docking in structure-based drug discovery. In Topics in Current Chemistry; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–34. [Google Scholar] [CrossRef]
- Protein Thermal Shift technology Optimizing Buffer Conditions and High-Throughput Screening of Ligand-Protein Binding. 2023. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/Application-Notes/protein-thermal-shift-app-note.pdf (accessed on 27 December 2023).
- Wu, T.; Hornsby, M.; Zhu, L.; Joshua, C.Y.; Shokat, K.M.; Gestwicki, J.E. Protocol for performing and optimizing differential scanning fluorimetry experiments. STAR Protoc. 2023, 4, 102688. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, T.K.; Paul, S. Protein-misfolding diseases and chaperone-based therapeutic approaches. FEBS J. 2006, 273, 1331–1349. [Google Scholar] [CrossRef]
- Bhayani, J.A.; Ballicora, M.A. Determination of dissociation constants of protein ligands by thermal shift assay. Biochem. Biophys. Res. Commun. 2022, 590, 1–6. [Google Scholar] [CrossRef]
- Bai, N.; Roder, H.; Dickson, A.; Karanicolas, J. Isothermal analysis of ThermoFluor data can readily provide quantitative binding affinities. Sci. Rep. 2019, 9, 2650. [Google Scholar] [CrossRef]
- Christie, R.M. Fluorescent dyes. In Handbook of Textile and Industrial Dyeing; Woodhead Publishing: Sawston, UK, 2011; pp. 562–587. [Google Scholar]
- Elgert, C.; Rühle, A.; Sandner, P.; Behrends, S. Thermal shift assay: Strengths and weaknesses of the method to investigate the ligand-induced thermostabilization of soluble guanylyl cyclase. J. Pharm. Biomed. Anal. 2020, 181, 113065. [Google Scholar] [CrossRef]
- Burstein, E.A.; Vedenkina, N.S.; Ivkova, M.N. Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 1973, 18, 263–279. [Google Scholar] [CrossRef]
- Cimmperman, P.; Matulis, D. Protein Thermal Denaturation Measurements via a Fluorescent Dye; Royal Society of Chemistry: London, UK, 2011. [Google Scholar]
- Temel, D.B.; Landsman, P.; Brader, M.L. Orthogonal methods for characterizing the unfolding of therapeutic monoclonal antibodies: Differential scanning calorimetry, isothermal chemical denaturation, and intrinsic fluorescence with concomitant static light scattering. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2016; Volume 567, pp. 359–389. [Google Scholar] [CrossRef]
- Rumble, C.; Rich, K.; He, G.; Maroncelli, M. CCVJ is not a simple rotor probe. J. Phys. Chem. A 2012, 116, 10786–10792. [Google Scholar] [CrossRef]
- GloMelt™ Thermal Shift Protein Stability Kit. 2023. Available online: https://www.biotrend.com/en/other-products-186/glomelt-thermal-shift-protein-stability-514058784.html (accessed on 27 December 2023).
- Ghisaidoobe, A.B.; Chung, S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- Joshi, D.; Kumar, D.; Maini, A.K.; Sharma, R.C. Detection of biological warfare agents using ultra violet-laser induced fluorescence LIDAR. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 112, 446–456. [Google Scholar] [CrossRef]
- Hawe, A.; Sutter, M.; Jiskoot, W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 2008, 25, 1487–1499. [Google Scholar] [CrossRef]
- Di Stasio, E.; Bizzarri, P.; Misiti, F.; Pavoni, E.; Brancaccio, A. A fast and accurate procedure to collect and analyze unfolding fluorescence signal: The case of dystroglycan domains. Biophys. Chem. 2004, 107, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.A.; Jameson, D.M. Time-resolved methods in biophysics. 8. Frequency domain fluorometry: Applications to intrinsic protein fluorescence. Photochem. Photobiol. Sci. 2008, 7, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Duy, C.; Fitter, J. How aggregation and conformational scrambling of unfolded states govern fluorescence emission spectra. Biophys. J. 2006, 90, 3704–3711. [Google Scholar] [CrossRef]
- Zahid, N.I.; Abou-Zied, O.K.; Hashim, R.; Heidelberg, T. Fluorescence probing of the temperature-induced phase transition in a glycolipid self-assembly: Hexagonal ↔ micellar and cubic ↔ lamellar. Langmuir 2012, 28, 4989–4995. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yoo, H.J.; Park, E.J.; Na, D.H. Nano differential scanning fluorimetry-based thermal stability screening and optimal buffer selection for immunoglobulin G. Pharmaceuticals 2021, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Langer, A.; Lüdecke, A.; Bartoschik, T.; Cehlar, O.; Duhr, S.; Baaske, P.; Streicher, W. A New Spectral Shift-Based Method to Characterize Molecular Interactions. ASSAY Drug Dev. Technol. 2022, 20, 83–94. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, W.; Gong, G.; Xie, L.; Wang, M.-W.; Hu, Y. A new approach to produce IgG4-like bispecific antibodies. Sci. Rep. 2021, 11, 18630. [Google Scholar] [CrossRef]
- Uncle Stays Hot for Proteins with Isothermal Apps. Available online: https://www.unchainedlabs.com/wp-content/uploads/2022/06/App_Note_Uncle_Uncle-stays-hot-for-proteins_RevA_r2.pdf (accessed on 27 December 2023).
- Miles, A.; Janes, R.W.; Wallace, B.A. Tools and methods for circular dichroism spectroscopy of proteins: A tutorial review. Chem. Soc. Rev. 2021, 50, 8400–8413. [Google Scholar] [CrossRef]
- Spectrometer; 163 to 950 nm, Circular Dichroism, J 1500, Standard PMT, Jasco, 100/240 V. 2023. Available online: https://www.laboratory-equipment.com/j1500-circular-dichroism-spectrometer-jasco-7000-j005a.html (accessed on 27 December 2023).
- Circular Dichroism Spectrophotometers. 2023. Available online: https://olisclarity.com/circular-dichroism-models (accessed on 27 December 2023).
- Huynh, K.; Partch, C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015, 79, 28.9.1–28.9.14. [Google Scholar] [CrossRef]
- Lee, P.-H.; Huang, X.X.; Teh, B.T.; Ng, L.-M. TSA-CRAFT: A free software for automatic and robust thermal shift assay data analysis. SLAS Discov. Adv. Life Sci. RD 2019, 24, 606–612. [Google Scholar] [CrossRef]
- LightCycler® Real-Time PCR Systems-Application Manual. 2023. Available online: https://plantbio.okstate.edu/images/pdfs/Roche_RT-PCR_Manual.pdf (accessed on 27 December 2023).
- Portable Maverick qPCR Multiplex Real Time qPCR Systems from Anitoa Systems. 2023. Available online: https://www.biocompare.com/23398-PCR-Machines/13423326-Maverick-qPCR-Multiplex-Real-Time-qPCR/?pda=26969|13423326_0_1|79695|2|&dfp=true (accessed on 27 December 2023).
- Erlich, H.A. Principles and applications for DNA amplification. In PCR Technology; Stockton Press: New York, NY, USA, 1989. [Google Scholar]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The Digital MIQE Guidelines: Minimum Information for Publication of Q uantitative Digital PCR E xperiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Kuang, J.; Yan, X.; Genders, A.J.; Granata, C.; Bishop, D.J. An overview of technical considerations when using quantitative real-time PCR analysis of gene expression in human exercise research. PLoS ONE 2018, 13, e0196438. [Google Scholar] [CrossRef]
- Prometheus, the New Gold Standard for Challenging Stability Characterizations. 2023. Available online: https://resources.nanotempertech.com/prometheus/nanotemper-technologies-brochure-prometheus (accessed on 27 December 2023).
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Boivin, S.; Kozak, S.; Meijers, R. Optimization of protein purification and characterization using Thermofluor screens. Protein Expr. Purif. 2013, 91, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Y.; Wu, Z.-Q.; Yin, D.-C.; Zhou, B.-R.; Guo, Y.-Z.; Lu, H.-M.; Zhou, R.-B.; Shang, P. A strategy for selecting the pH of protein solutions to enhance crystallization. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Zbacnik, T.J.; Holcomb, R.E.; Katayama, D.S.; Murphy, B.M.; Payne, R.W.; Coccaro, R.C.; Evans, G.J.; Matsuura, J.E.; Henry, C.S.; Manning, M.C. Role of buffers in protein formulations. J. Pharm. Sci. 2017, 106, 713–733. [Google Scholar] [CrossRef]
- Ciulli, A. Biophysical screening for the discovery of small-molecule ligands. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; pp. 357–388. [Google Scholar] [CrossRef]
- Houser, J.; Kosourova, J.; Kubickova, M.; Wimmerova, M. Development of 48-condition buffer screen for protein stability assessment. Eur. Biophys. J. 2021, 50, 461–471. [Google Scholar] [CrossRef]
- Auld, D.S.; Coassin, P.A.; Coussens, N.P.; Hensley, P.; Klumpp-Thomas, C.; Michael, S.; Sittampalam, G.S.; Trask, O.J.; Wagner, B.K.; Weidner, J.R. Microplate selection and recommended practices in high-throughput screening and quantitative biology. In Assay Guidance Manual [Internet]; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2020. [Google Scholar]
- Coyle, J.; Walser, R. Applied biophysical methods in fragment-based drug discovery. SLAS DISCOVERY Adv. Sci. Drug Discov. 2020, 25, 471–490. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.C. Fragment Based Drug Design: Tools, Practical Approaches, and Examples. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Kranz, J.K.; Schalk-Hihi, C. Protein thermal shifts to identify low molecular weight fragments. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2011; Volume 493, pp. 277–298. [Google Scholar] [CrossRef]
- Ryder, A.G.; Power, S.; Glynn, T.J. Fluorescence-lifetime-based pH sensing using resorufin. In Proceedings of the Opto-Ireland 2002: Optics and Photonics Technologies and Applications, Galway, Ireland, 5–6 September 2002; pp. 827–835. [Google Scholar] [CrossRef]
- Kroeger, T.; Frieg, B.; Zhang, T.; Hansen, F.K.; Marmann, A.; Proksch, P.; Nagel-Steger, L.; Groth, G.; Smits, S.H.; Gohlke, H. EDTA aggregates induce SYPRO orange-based fluorescence in thermal shift assay. PLoS ONE 2017, 12, e0177024. [Google Scholar] [CrossRef] [PubMed]
- Ablinger, E.; Leitgeb, S.; Zimmer, A. Differential scanning fluorescence approach using a fluorescent molecular rotor to detect thermostability of proteins in surfactant-containing formulations. Int. J. Pharm. 2013, 441, 255–260. [Google Scholar] [CrossRef]
- Mezzasalma, T.M.; Kranz, J.K.; Chan, W.; Struble, G.T.; Schalk-Hihi, C.; Deckman, I.C.; Springer, B.A.; Todd, M.J. Enhancing recombinant protein quality and yield by protein stability profiling. J. Biomol. Screen. 2007, 12, 418–428. [Google Scholar] [CrossRef]
- Sviben, D.; Bertoša, B.; Hloušek-Kasun, A.; Forcic, D.; Halassy, B.; Brgles, M. Investigation of the thermal shift assay and its power to predict protein and virus stabilizing conditions. J. Pharm. Biomed. Anal. 2018, 161, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Vagenende, V.; Yap, M.G.; Trout, B.L. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry 2009, 48, 11084–11096. [Google Scholar] [CrossRef]
- Malik, K.; Matejtschuk, P.; Thelwell, C.; Burns, C.J. Differential scanning fluorimetry: Rapid screening of formulations that promote the stability of reference preparations. J. Pharm. Biomed. Anal. 2013, 77, 163–166. [Google Scholar] [CrossRef]
- Shi, S.; Semple, A.; Cheung, J.; Shameem, M. DSF method optimization and its application in predicting protein thermal aggregation kinetics. J. Pharm. Sci. 2013, 102, 2471–2483. [Google Scholar] [CrossRef]
- Vivoli, M.; Novak, H.R.; Littlechild, J.A.; Harmer, N.J. Determination of protein-ligand interactions using differential scanning fluorimetry. JoVE J. Vis. Exp. 2014, 91, e51809. [Google Scholar] [CrossRef]
- Crowther, G.J.; He, P.; Rodenbough, P.P.; Thomas, A.P.; Kovzun, K.V.; Leibly, D.J.; Bhandari, J.; Castaneda, L.J.; Hol, W.G.; Gelb, M.H. Use of thermal melt curves to assess the quality of enzyme preparations. Anal. Biochem. 2010, 399, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, R.; Nagarajaram, H.A. Intrinsically disordered proteins: An overview. Int. J. Mol. Sci. 2022, 23, 14050. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.C.; Sanches, K.; Pinheiro-Aguiar, R.; Almeida, V.S.; Caruso, I.P. Protein surface interactions—Theoretical and experimental studies. Front. Mol. Biosci. 2021, 8, 706002. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Smith, T.; Hicks, R.H.; Doekhie, A.; Koumanov, F.; Wells, S.A.; Edler, K.J.; van den Elsen, J.; Holman, G.D.; Marchbank, K.J. Thermal stability, storage and release of proteins with tailored fit in silica. Sci. Rep. 2017, 7, 46568. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F. Protein stability [determination] problems. Front. Mol. Biosci. 2022, 9, 880358. [Google Scholar] [CrossRef] [PubMed]
- Nettleship, J.E.; Brown, J.; Groves, M.R.; Geerlof, A. Methods for protein characterization by mass spectrometry, thermal shift (ThermoFluor) assay, and multiangle or static light scattering. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2008; pp. 299–318. [Google Scholar] [CrossRef]
- Zwanzig, R. Two-state models of protein folding kinetics. Proc. Natl. Acad. Sci. USA 1997, 94, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Cimmperman, P.; Baranauskienė, L.; Jachimovičiūtė, S.; Jachno, J.; Torresan, J.; Michailovienė, V.; Matulienė, J.; Sereikaitė, J.; Bumelis, V.; Matulis, D. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys. J. 2008, 95, 3222–3231. [Google Scholar] [CrossRef]
- Strucksberg, K.; Rosenkranz, T.; Fitter, J. Reversible and irreversible unfolding of multi-domain proteins. Biochim. Biophys. Acta BBA Proteins Proteom. 2007, 1774, 1591–1603. [Google Scholar] [CrossRef]
- González, M.n.; Argaraña, C.E.; Fidelio, G.D. Extremely high thermal stability of streptavidin and avidin upon biotin binding. Biomol. Eng. 1999, 16, 67–72. [Google Scholar] [CrossRef]
- Szmola, R.; Kukor, Z.; Sahin-Tóth, M. Human mesotrypsin is a unique digestive protease specialized for the degradation of trypsin inhibitors. J. Biol. Chem. 2003, 278, 48580–48589. [Google Scholar] [CrossRef]
- Samuel, E.L.; Holmes, S.L.; Young, D.W. Processing binding data using an open-source workflow. J. Cheminformatics 2021, 13, 99. [Google Scholar] [CrossRef]
- Reys, V.; Kowalewski, J.; Gelin, M.; Lionne, C. wTSA-CRAFT: An open-access web server for rapid analysis of thermal shift assay experiments. Bioinform. Adv. 2023, 3, vbad136. [Google Scholar] [CrossRef]
- Hellman, L.M.; Yin, L.; Wang, Y.; Blevins, S.J.; Riley, T.P.; Belden, O.S.; Spear, T.T.; Nishimura, M.I.; Stern, L.J.; Baker, B.M. Differential scanning fluorimetry based assessments of the thermal and kinetic stability of peptide–MHC complexes. J. Immunol. Methods 2016, 432, 95–101. [Google Scholar] [CrossRef]
- Son, J.W.; Son, J.M.; Hur, K.H.; Lee, W.; Song, I.S.; Na, D.H. Application of isothermal chemical denaturation to early-stage formulation development of fibrinogen. Bull. Korean Chem. Soc. 2023, 44, 348–352. [Google Scholar] [CrossRef]
- Freire, E.; Schön, A.; Hutchins, B.M.; Brown, R.K. Chemical denaturation as a tool in the formulation optimization of biologics. Drug Discov. Today 2013, 18, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.B.; Flynn, R.P.; Wooten, H.R.; Noufer, H.A.; Cancel, R.A.; Zhang, J.; Subramony, J.A.; Pechenov, S.; Wang, Y. Submicron aggregation of chemically denatured monoclonal antibody. Mol. Pharm. 2018, 15, 4710–4721. [Google Scholar] [CrossRef] [PubMed]
- Augustijn, D.; Kulakova, A.; Mahapatra, S.; Harris, P.; Rinnan, Å. Isothermal chemical denaturation: Data analysis, error detection, and correction by parafac2. Anal. Chem. 2020, 92, 6958–6967. [Google Scholar] [CrossRef] [PubMed]
- Dzwolak, W.; Loksztejn, A.; Galinska-Rakoczy, A.; Adachi, R.; Goto, Y.; Rupnicki, L. Conformational indeterminism in protein misfolding: Chiral amplification on amyloidogenic pathway of insulin. J. Am. Chem. Soc. 2007, 129, 7517–7522. [Google Scholar] [CrossRef]
- Jain, D.; Mahammad, S.S.; Singh, P.P.; Kodipyaka, R. A review on parenteral delivery of peptides and proteins. Drug Dev. Ind. Pharm. 2019, 45, 1403–1420. [Google Scholar] [CrossRef] [PubMed]
- Wafer, L.; Kloczewiak, M.; Polleck, S.M.; Luo, Y. Isothermal chemical denaturation of large proteins: Path-dependence and irreversibility. Anal. Biochem. 2017, 539, 60–69. [Google Scholar] [CrossRef]
- Epps, D.E.; Sarver, R.W.; Rogers, J.M.; Herberg, J.T.; Tomich, P.K. The ligand affinity of proteins measured by isothermal denaturation kinetics. Anal. Biochem. 2001, 292, 40–50. [Google Scholar] [CrossRef]
- Sasse, J.; Gallagher, S.R. Staining proteins in gels. Curr. Protoc. Mol. Biol. 2003, 63, 10–16. [Google Scholar] [CrossRef]
- Mahran, R.; Vello, N.; Komulainen, A.; Malakoutikhah, M.; Härmä, H.; Kopra, K. Isothermal chemical denaturation assay for monitoring protein stability and inhibitor interactions. Sci. Rep. 2023, 13, 20066. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.K. Chemical Denaturation. In Molecular Life Sciences: An Encyclopedic Reference; Bell, E., Ed.; Springer: New York, NY, USA, 2021. [Google Scholar] [CrossRef]
- Paladino, A.; Vitagliano, L.; Graziano, G. The Action of Chemical Denaturants: From Globular to Intrinsically Disordered Proteins. Biology 2023, 12, 754. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Oas, T.G.; Myers, J.K. Fast and faster: A designed variant of the B-domain of protein A folds in 3 μsec. Protein Sci. 2004, 13, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Krasnovskaya, O.O.; Malinnikov, V.M.; Dashkova, N.S.; Gerasimov, V.M.; Grishina, I.V.; Kireev, I.I.; Lavrushkina, S.V.; Panchenko, P.A.; Zakharko, M.A.; Ignatov, P.A. Thiourea modified doxorubicin: A perspective pH-sensitive prodrug. Bioconjugate Chem. 2019, 30, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C. Cooperative Dual Catalysis with Thiourea Organocatalysts; Rutgers University-School of Graduate Studies: New Brunswick, NJ, USA, 2017. [Google Scholar]
- Makhatadze, G.I. Thermodynamics of protein interactions with urea and guanidinium hydrochloride. J. Phys. Chem. B 1999, 103, 4781–4785. [Google Scholar] [CrossRef]
- Schön, A.; Brown, R.K.; Hutchins, B.M.; Freire, E. Ligand binding analysis and screening by chemical denaturation shift. Anal. Biochem. 2013, 443, 52–57. [Google Scholar] [CrossRef]
- Das, A.; Mukhopadhyay, C. Urea-mediated protein denaturation: A consensus view. J. Phys. Chem. B 2009, 113, 12816–12824. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Sharma, S.; Bano, B. Comparison of guanidine hydrochloride (GdnHCl) and urea denaturation on inactivation and unfolding of human placental cystatin (HPC). Protein J. 2005, 24, 283–292. [Google Scholar] [CrossRef]
- Ibarra-Molero, B.; Sanchez-Ruiz, J.M. Irreversible Protein Denaturation. In Encyclopedia of Biophysics; Roberts, G., Watts, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Rossky, P.J. Protein denaturation by urea: Slash and bond. Proc. Natl. Acad. Sci. USA 2008, 105, 16825–16826. [Google Scholar] [CrossRef]
- Vancraenenbroeck, R.; Harel, Y.S.; Zheng, W.; Hofmann, H. Polymer effects modulate binding affinities in disordered proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 19506–19512. [Google Scholar] [CrossRef]
- Coletta, M.; Costa, H.; De Sanctis, G.; Neri, F.; Smulevich, G.; Turner, D.L.; Santos, H. pH dependence of structural and functional properties of oxidized cytochrome c” from Methylophilus methylotrophus. J. Biol. Chem. 1997, 272, 24800–24804. [Google Scholar] [CrossRef]
- Jain, R.; Kumar, R.; Kumar, S.; Chhabra, R.; Agarwal, M.C. Analysis of the pH-dependent stability and millisecond folding kinetics of horse cytochrome c. Arch. Biochem. Biophys. 2015, 585, 52–63. [Google Scholar] [CrossRef]
- Xu, Q.; Keiderling, T.A. Effect of sodium dodecyl sulfate on folding and thermal stability of acid-denatured cytochrome c: A spectroscopic approach. Protein Sci. 2004, 13, 2949–2959. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Prinz, T.K.; Stäbler, A.; Sängerlaub, S. Effect of sodium sulfite, sodium dodecyl sulfate, and urea on the molecular interactions and properties of whey protein isolate-based films. Front. Chem. 2017, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Senisterra, G.; Chau, I.; Vedadi, M. Thermal denaturation assays in chemical biology. Assay Drug Dev. Technol. 2012, 10, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Svilenov, H.; Markoja, U.; Winter, G. Isothermal chemical denaturation as a complementary tool to overcome limitations of thermal differential scanning fluorimetry in predicting physical stability of protein formulations. Eur. J. Pharm. Biopharm. 2018, 125, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Senisterra, G.A.; Soo Hong, B.; Park, H.-W.; Vedadi, M. Application of high-throughput isothermal denaturation to assess protein stability and screen for ligands. J. Biomol. Screen. 2008, 13, 337–342. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ross, P.; Weihofen, W.; Siu, F.; Xie, A.; Katakia, H.; Wright, S.K.; Hunt, I.; Brown, R.K.; Freire, E. Isothermal chemical denaturation to determine binding affinity of small molecules to G-protein coupled receptors. Anal. Biochem. 2015, 473, 41–45. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jung, F.; Frey, K.; Zimmer, D.; Mühlhaus, T. DeepSTABp: A Deep Learning Approach for the Prediction of Thermal Protein Stability. Int. J. Mol. Sci. 2023, 24, 7444. [Google Scholar] [CrossRef] [PubMed]
- Atsavapranee, B.; Stark, C.D.; Sunden, F.; Thompson, S.; Fordyce, P.M. Fundamentals to function: Quantitative and scalable approaches for measuring protein stability. Cell Syst. 2021, 12, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhao, J.; Zeng, L.; Vihinen, M. ProTstab2 for Prediction of Protein Thermal Stabilities. Int. J. Mol. Sci. 2022, 23, 10798. [Google Scholar] [CrossRef]
- Mazzeo, A.; Carpenter, P. Stability studies for biologics. In Handbook of Stability Testing in Pharmaceutical Development: Regulations, Methodologies, and Best Practices; Springer: New York, NY, USA, 2009; pp. 353–369. [Google Scholar] [CrossRef]
- Huelsmeyer, M.; Kuzman, D.; Bončina, M.; Martinez, J.; Steinbrugger, C.; Weusten, J.; Calero-Rubio, C.; Roche, W.; Niederhaus, B.; VanHaelst, Y. A universal tool for stability predictions of biotherapeutics, vaccines and in vitro diagnostic products. Sci. Rep. 2023, 13, 10077. [Google Scholar] [CrossRef]
- Valtonen, S.; Vuorinen, E.; Eskonen, E.; Malakoutikhah, M.; Kopra, K.; Harma, H. Sensitive, homogeneous, and label-free protein-probe assay for antibody aggregation and thermal stability studies. mAbs 2021, 13, 1955810. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gooran, N.; Kopra, K. Fluorescence-Based Protein Stability Monitoring—A Review. Int. J. Mol. Sci. 2024, 25, 1764. https://doi.org/10.3390/ijms25031764
Gooran N, Kopra K. Fluorescence-Based Protein Stability Monitoring—A Review. International Journal of Molecular Sciences. 2024; 25(3):1764. https://doi.org/10.3390/ijms25031764
Chicago/Turabian StyleGooran, Negin, and Kari Kopra. 2024. "Fluorescence-Based Protein Stability Monitoring—A Review" International Journal of Molecular Sciences 25, no. 3: 1764. https://doi.org/10.3390/ijms25031764
APA StyleGooran, N., & Kopra, K. (2024). Fluorescence-Based Protein Stability Monitoring—A Review. International Journal of Molecular Sciences, 25(3), 1764. https://doi.org/10.3390/ijms25031764





