Identification of miR-30c-5p microRNA in Serum as a Candidate Biomarker to Diagnose Endometriosis
Abstract
:1. Introduction
2. Results
2.1. Study Participant Characteristics
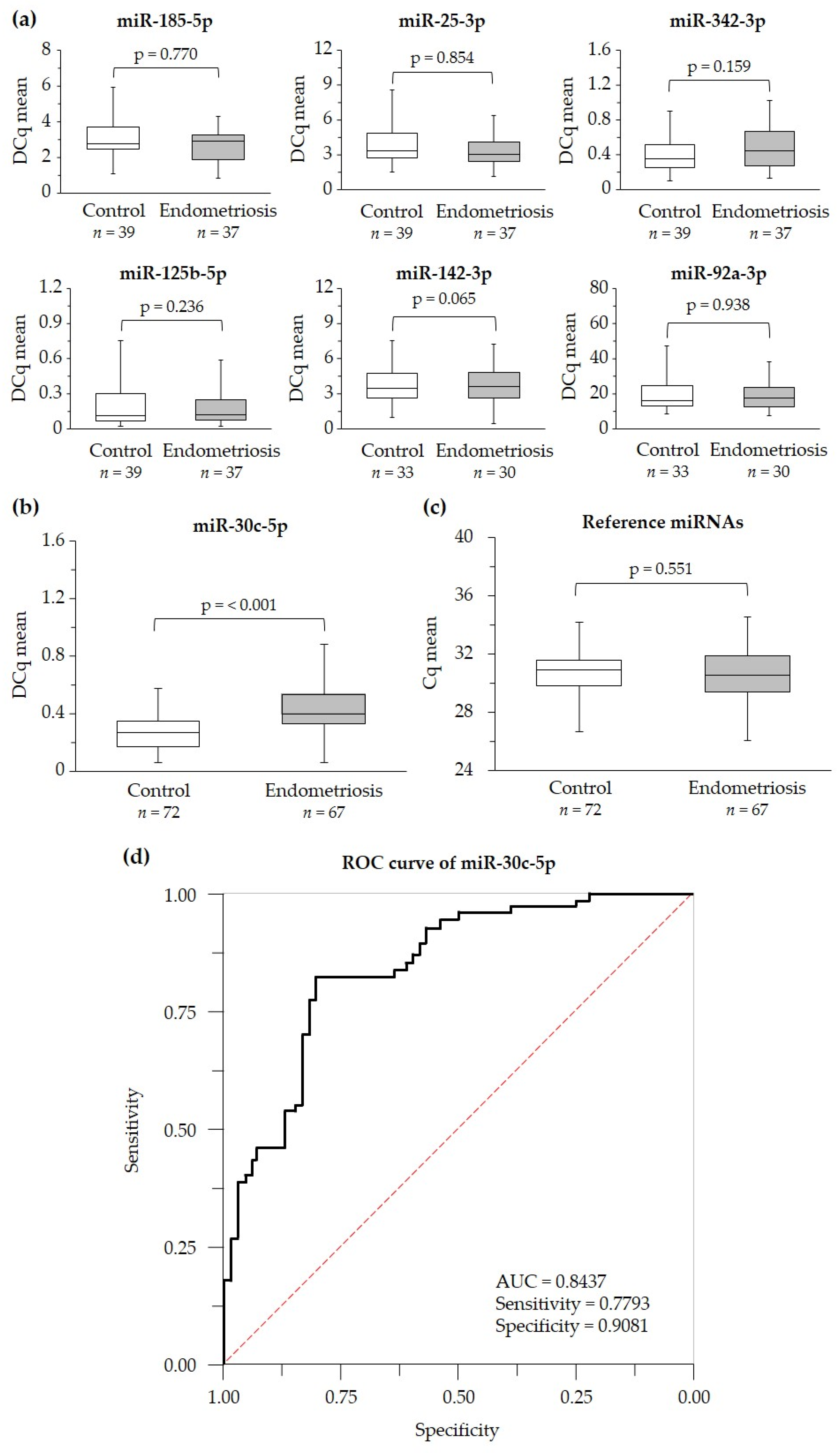
2.2. Identification of Serum Levels of Circulating miRNAs in Case and Control Groups
2.3. Suitability of miR-30c-5p as Potential Endometriosis Biomarker
2.4. Serum miR-30c-5p Levels in Endometriosis Stages
2.5. The Impact of Hemolysis on Potential Diagnosis of miR-30c-5p
2.6. Prediction of Target Genes and Functional Enrichment
3. Discussion
4. Materials and Methods
4.1. Study Population and Design
4.2. Serum Sample Collection
4.3. miRNA qPCR Profiling
4.4. RNA Extraction and cDNA Synthesis
4.5. Selection of miRNA Biomarker Candidates of Endometriosis and Reference miRNAs
4.6. miRNA Real-Time qPCR Validation
4.7. Hemolysis Control
4.8. miRNA Target Gene Prediction and Functional Enrichment Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


| miRNA ID | Sequence (5′ > 3′) |
|---|---|
| miR-185-5p | UGGAGAGAAAGGCAGUUCCUGA |
| let-7c-5p | UGAGGUAGUAGGUUGUAUGGUU |
| miR-30c-5p | UGUAAACAUCCUACACUCUCAGC |
| miR-210-3p | CUGUGCGUGUGACAGCGGCUGA |
| miR-25-3p | CAUUGCACUUGUCUCGGUCUGA |
| miR-342-3p | UCUCACACAGAAAUCGCACCCGU |
| miR-125b-5p | UCCCUGAGACCCUAACUUGUGA |
| miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-92a-3p | UAUUGCACUUGUCCCGGCCUGU |
| miR-30e-5p | UGUAAACAUCCUUGACUGGAAG |
| miR-15b-5p | UAGCAGCACAUCAUGGUUUACA |
| miR-451a | AAACCGUUACCAUUACUGAGUU |
| miR-23a-3p | AUCACAUUGCCAGGGAUUUCC |
References
- Saunders, P.T.; Horne, A.W. Endometriosis: Etiology, pathobiology, and therapeutic prospects. Cell 2021, 184, 2807–2824. [Google Scholar] [CrossRef] [PubMed]
- Lamceva, J.; Uljanovs, R.; Strumfa, I. The main theories on the pathogenesis of endometriosis. Int. J. Mol. Sci. 2023, 24, 4254. [Google Scholar] [CrossRef]
- Burney, R.O.; Giudice, L.C. Pathogenesis and pathophysiology of endometriosis. Fertil. Steril. 2012, 98, 511–519. [Google Scholar] [CrossRef]
- Gruber, T.M.; Mechsner, S. Pathogenesis of endometriosis: The origin of pain and subfertility. Cells 2021, 10, 1381. [Google Scholar] [CrossRef] [PubMed]
- Mahini, S.M.; Mortazavi, G.; Samare-Najaf, M.; Azadbakht, M.K.; Jamali, N. Non-invasive diagnosis of endometriosis: Immunologic and genetic markers. Clin. Chim. Acta 2022, 538, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Fassbender, A.; Vodolazkaia, A.; Saunders, P.; Lebovic, D.; Waelkens, E.; De Moor, B.; D’Hooghe, T. Biomarkers of endometriosis. Fertil. Steril. 2013, 99, 1135–1145. [Google Scholar] [CrossRef]
- Mol, B.W.; Bayram, N.; Lijmer, J.G.; Wiegerinck, M.A.; Bongers, M.Y.; Van Der Veen, F.; Bossuyt, P.M. The performance of CA-125 measurement in the detection of endometriosis: A meta-analysis. Fertil. Steril. 1998, 70, 1101–1108. [Google Scholar] [CrossRef]
- May, K.; Conduit-Hulbert, S.; Villar, J.; Kirtley, S.; Kennedy, S.; Becker, C. Peripheral biomarkers of endometriosis: A systematic review. Hum. Reprod. Update 2010, 16, 651–674. [Google Scholar] [CrossRef]
- Kafali, H.; Artuc, H.; Demir, N. Use of CA125 fluctuation during the menstrual cycle as a tool in the clinical diagnosis of endometriosis; a preliminary report. Eur. J. Obstet. Gynecol. Reprod. Biol. 2004, 116, 85–88. [Google Scholar] [CrossRef]
- Hanke, M.; Hoefig, K.; Merz, H.; Feller, A.C.; Kausch, I.; Jocham, D.; Warnecke, J.M.; Sczakiel, G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2010, 28, 655–661. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, Characterization, and Clinical Utility for Oral Cancer DetectionSalivary microRNA. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef]
- von Grothusen, C.; Frisendahl, C.; Modhukur, V.; Lalitkumar, P.G.; Peters, M.; Faridani, O.R.; Salumets, A.; Boggavarapu, N.R.; Gemzell-Danielsson, K. Uterine fluid microRNAs are dysregulated in women with recurrent implantation failure. Hum. Reprod. 2022, 37, 734–746. [Google Scholar] [CrossRef]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Li, J.; Smyth, P.; Flavin, R.; Cahill, S.; Denning, K.; Aherne, S.; Guenther, S.M.; O’Leary, J.J.; Sheils, O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Asakura, K.; Kadota, T.; Matsuzaki, J.; Yoshida, Y.; Yamamoto, Y.; Nakagawa, K.; Takizawa, S.; Aoki, Y.; Nakamura, E.; Miura, J. A miRNA-based diagnostic model predicts resectable lung cancer in humans with high accuracy. Commun. Biol. 2020, 3, 134. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C.; Wong, C.F.; Li, K.P.; Fong, A.H.; Fung, K.Y.; Au, J.S. miR-145 as a Potential Biomarker and Therapeutic Target in Patients with Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2023, 24, 10022. [Google Scholar] [CrossRef] [PubMed]
- Dar, G.M.; Agarwal, S.; Kumar, A.; Sharma, A.K.; Verma, R.; Sattar, R.S.A.; Ahmad, E.; Ali, A.; Mahajan, B.; Saluja, S.S. A non-invasive miRNA-based approach in early diagnosis and therapeutics of oral cancer. Crit. Rev. Oncol./Hematol. 2022, 180, 103850. [Google Scholar] [CrossRef] [PubMed]
- Doghish, A.S.; Moustafa, H.A.M.; Elballal, M.S.; Sallam, A.-A.M.; El-Dakroury, W.A.; Mageed, S.S.A.; Elesawy, A.E.; Abdelmaksoud, N.M.; Shahin, R.K.; Midan, H.M. The potential role of miRNAs in the pathogenesis of testicular germ cell tumors-A Focus on signaling pathways interplay. Pathol.-Res. Pract. 2023, 248, 154611. [Google Scholar] [CrossRef]
- Cheng, Y.-F.; Gu, X.-J.; Yang, T.-M.; Wei, Q.-Q.; Cao, B.; Zhang, Y.; Shang, H.-F.; Chen, Y.-P. Signature of miRNAs derived from the circulating exosomes of patients with amyotrophic lateral sclerosis. Front. Aging Neurosci. 2023, 15, 1106497. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Woo, J.; Kim, S.T.; Moon, M.; Kim, S.Y.; Cho, H.; Kim, S.; Kim, H.-K.; Park, J.-Y. MicroRNA super-resolution imaging in blood for Alzheimer’s disease. BMB Rep. 2023, 56, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.F.; Wen, D.; Zhao, Q.; Shen, P.Y.; Shi, H.; Zhao, Q.; Chen, Y.X.; Zhang, W. Urinary MicroRNA-30c-5p and MicroRNA-192-5p as potential biomarkers of ischemia-reperfusion-induced kidney injury. Exp. Biol. Med. 2017, 242, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Benito, B.; García-Elías, A.; Ois, Á.; Tajes, M.; Vallès, E.; Ble, M.; Bisbe, L.Y.; Giralt-Steinhauer, E.; Rodríguez-Campello, A.; Capdevila, M.C. Plasma levels of miRNA-1-3p are associated with subclinical atrial fibrillation in patients with cryptogenic stroke. Rev. Española De Cardiol. (Engl. Ed.) 2022, 75, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Düzgün, Z.; Kayikçioğlu, L.M.; Aktan, Ç.; Bara, B.; Eroğlu, F.Z.; Yağmur, B.; Çetintaş, V.B.; Bayindir, M.; Nalbantgil, S.; Vardarli, A.T. Decreased circulating microRNA-21 and microRNA-143 are associated to pulmonary hypertension. Turk. J. Med. Sci. 2023, 53, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, J.; Wan, H.; Wang, K.; Wu, J.; Cao, Y.; Hu, L.; Yu, Y.; Sun, H.; Yu, Y. Plasma extracellular vesicles microRNA-208b-3p and microRNA-143-3p as novel biomarkers for sudden cardiac death prediction in acute coronary syndrome. Mol. Omics 2023, 19, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Dabi, Y.; Suisse, S.; Puchar, A.; Delbos, L.; Poilblanc, M.; Descamps, P.; Haury, J.; Golfier, F.; Jornea, L.; Bouteiller, D. Endometriosis-associated infertility diagnosis based on saliva microRNA signatures. Reprod. BioMed. Online 2023, 46, 138–149. [Google Scholar] [CrossRef]
- Cho, S.; Mutlu, L.; Grechukhina, O.; Taylor, H.S. Circulating microRNAs as potential biomarkers for endometriosis. Fertil. Steril. 2015, 103, 1252–1260.e1. [Google Scholar] [CrossRef]
- Maged, A.M.; Deeb, W.S.; El Amir, A.; Zaki, S.S.; El Sawah, H.; Al Mohamady, M.; Metwally, A.A.; Katta, M.A. Diagnostic accuracy of serum miR-122 and miR-199a in women with endometriosis. Int. J. Gynaecol. Obs. 2018, 141, 14–19. [Google Scholar] [CrossRef]
- Moustafa, S.; Burn, M.; Mamillapalli, R.; Nematian, S.; Flores, V.; Taylor, H.S. Accurate diagnosis of endometriosis using serum microRNAs. Am. J. Obs. Gynecol. 2020, 223, 557.e1–557.e11. [Google Scholar] [CrossRef]
- Bashti, O.; Noruzinia, M.; Garshasbi, M.; Abtahi, M. miR-31 and miR-145 as Potential Non-Invasive Regulatory Biomarkers in Patients with Endometriosis. Cell J. 2018, 20, 84–89. [Google Scholar] [CrossRef]
- Papari, E.; Noruzinia, M.; Kashani, L.; Foster, W.G. Identification of candidate microRNA markers of endometriosis with the use of next-generation sequencing and quantitative real-time polymerase chain reaction. Fertil. Steril. 2020, 113, 1232–1241. [Google Scholar] [CrossRef]
- Braicu, O.-L.; Budisan, L.; Buiga, R.; Jurj, A.; Achimas-Cadariu, P.; Pop, L.A.; Braicu, C.; Irimie, A.; Berindan-Neagoe, I. miRNA expression profiling in formalin-fixed paraffin-embedded endometriosis and ovarian cancer samples. OncoTargets Ther. 2017, 10, 4225–4238. [Google Scholar] [CrossRef]
- Ohlsson Teague, E.M.C.; Van der Hoek, K.H.; Van der Hoek, M.B.; Perry, N.; Wagaarachchi, P.; Robertson, S.A.; Print, C.G.; Hull, L.M. MicroRNA-regulated pathways associated with endometriosis. Mol. Endocrinol. 2009, 23, 265–275. [Google Scholar] [CrossRef]
- Pan, Q.; Luo, X.; Toloubeydokhti, T.; Chegini, N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol. Hum. Reprod. 2007, 13, 797–806. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell communication: microRNAs as hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef] [PubMed]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Nisenblat, V.; Bossuyt, P.M.; Shaikh, R.; Farquhar, C.; Jordan, V.; Scheffers, C.S.; Mol, B.W.J.; Johnson, N.; Hull, M.L.; Gynaecology, C.; et al. Blood biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst. Rev. 1996, 2006, 5. [Google Scholar] [CrossRef] [PubMed]
- Leonova, A.; Turpin, V.E.; Agarwal, S.K.; Leonardi, M.; Foster, W.G. A critical appraisal of the circulating levels of differentially expressed microRNA in endometriosisdagger. Biol. Reprod. 2021, 105, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Blondal, T.; Jensby Nielsen, S.; Baker, A.; Andreasen, D.; Mouritzen, P.; Wrang Teilum, M.; Dahlsveen, I.K. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013, 59, S1–S6. [Google Scholar] [CrossRef]
- Misir, S.; Hepokur, C.; Oksasoglu, B.; Yildiz, C.; Yanik, A.; Aliyazicioglu, Y.; Reproduction, H. Circulating serum miR-200c and miR-34a-5p as diagnostic biomarkers for endometriosis. J. Gynecol. Obstet. Hum. Reprod. 2021, 50, 102092. [Google Scholar] [CrossRef] [PubMed]
- Zafari, N.; Tarafdari, A.M.; Izadi, P.; Noruzinia, M.; Yekaninejad, M.S.; Bahramy, A.; Mohebalian, A. A panel of plasma miRNAs 199b-3p, 224–225p and Let-7d-3p as non-invasive diagnostic biomarkers for endometriosis. Reprod. Sci. 2021, 28, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, H.; Jin, D.; Zhang, Y. Serum miR-17, IL-4, and IL-6 levels for diagnosis of endometriosis. Medicine 2018, 97, e10853. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huang, W.; Ren, C.; Zhao, M.; Jiang, X.; Fang, X.; Xia, X. Analysis of Serum microRNA Profile by Solexa Sequencing in Women With Endometriosis. Reprod. Sci. 2016, 23, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.T.; Zhao, Y.N.; Han, B.W.; Hong, S.J.; Chen, Y.Q. Circulating microRNAs identified in a genome-wide serum microRNA expression analysis as noninvasive biomarkers for endometriosis. J. Clin. Endocrinol. Metab. 2013, 98, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- McGrath, I.M.; Montgomery, G.W.; Mortlock, S. Insights from Mendelian randomization and genetic correlation analyses into the relationship between endometriosis and its comorbidities. Hum. Reprod. Update 2023, 29, 655–674. [Google Scholar] [CrossRef]
- Brosens, I. Diagnosis of endometriosis. Semin. Reprod. Endocrinol. 1997, 15, 229–233. [Google Scholar] [CrossRef]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Kroh, E.M.; Parkin, R.K.; Mitchell, P.S.; Tewari, M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010, 50, 298–301. [Google Scholar] [CrossRef]
- Kang, K.; Peng, X.; Luo, J.; Gou, D. Identification of circulating miRNA biomarkers based on global quantitative real-time PCR profiling. J. Anim. Sci. Biotechnol. 2012, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Cosar, E.; Mamillapalli, R.; Ersoy, G.S.; Cho, S.; Seifer, B.; Taylor, H.S. Serum microRNAs as diagnostic markers of endometriosis: A comprehensive array-based analysis. Fertil. Steril. 2016, 106, 402–409. [Google Scholar] [CrossRef]
- Nothnick, W.B.; Falcone, T.; Joshi, N.; Fazleabas, A.T.; Graham, A. Serum miR-451a Levels Are Significantly Elevated in Women With Endometriosis and Recapitulated in Baboons (Papio anubis) with Experimentally-Induced Disease. Reprod. Sci. 2017, 24, 1195–1202. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Yuan, M.; Li, D.; Sun, C.; Wang, G. Serum exosomal microRNAs as potential circulating biomarkers for endometriosis. Dis. Markers 2020, 2020, 2456340. [Google Scholar] [CrossRef]
- Qi, R.; Weiland, M.; Gao, X.-H.; Zhou, L.; Mi, Q.-S. Identification of endogenous normalizers for serum microRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology 2012, 55, 1640–1642. [Google Scholar] [CrossRef]
- Wang, K.; Yuan, Y.; Cho, J.-H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef]
- Rekker, K.; Saare, M.; Roost, A.M.; Kaart, T.; Soritsa, D.; Karro, H.; Soritsa, A.; Simon, C.; Salumets, A.; Peters, M. Circulating miR-200-family micro-RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertil. Steril. 2015, 104, 938–946.e2. [Google Scholar] [CrossRef]
- Vanhie, A.; O, D.; Peterse, D.; Beckers, A.; Cuellar, A.; Fassbender, A.; Meuleman, C.; Mestdagh, P.; D’Hooghe, T. Plasma miRNAs as biomarkers for endometriosis. Hum. Reprod. 2019, 34, 1650–1660. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, X.; Xia, X.; Fang, X.; Zhang, T.; Huang, F. Endometrial epithelial cells-derived exosomes deliver microRNA-30c to block the BCL9/Wnt/CD44 signaling and inhibit cell invasion and migration in ovarian endometriosis. Cell Death Discov. 2022, 8, 151. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, Y.; Pan, D. miR-30c may serve a role in endometriosis by targeting plasminogen activator inhibitor-1. Exp. Ther. Med. 2017, 14, 4846–4852. [Google Scholar] [CrossRef]
- Budak, H.; Bulut, R.; Kantar, M.; Alptekin, B. MicroRNA nomenclature and the need for a revised naming prescription. Brief. Funct. Genom. 2016, 15, 65–71. [Google Scholar] [CrossRef]
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. miRNA nomenclature: A view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet. 2015, 31, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Lee, E.J.; Jiang, J.; Sarkar, A.; Yang, L.; Elton, T.S.; Chen, C. Real-time PCR quantification of precursor and mature microRNA. Methods 2008, 44, 31–38. [Google Scholar] [CrossRef]
- Razi, M.H.; Eftekhar, M.; Ghasemi, N.; Sheikhha, M.H.; Firoozabadi, A.D. Expression levels of circulatory mir-185-5p, vascular endothelial growth factor, and platelet-derived growth factor target genes in endometriosis. Int. J. Reprod. Biomed. (IJRM) 2020, 18, 347. [Google Scholar] [CrossRef]
- Abo, C.; Biquard, L.; Girardet, L.; Chouzenoux, S.; Just, P.-A.; Chapron, C.; Vaiman, D.; Borghese, B. Unbiased In Silico Analysis of Gene Expression Pinpoints Circulating miRNAs Targeting KIAA1324, a New Gene Drastically Downregulated in Ovarian Endometriosis. Biomedicines 2022, 10, 2065. [Google Scholar] [CrossRef] [PubMed]
- Bahramy, A.; Zafari, N.; Izadi, P.; Soleymani, F.; Kavousi, S.; Noruzinia, M. The role of miRNAs 340-5p, 92a-3p, and 381-3p in patients with endometriosis: A plasma and mesenchymal stem-like cell study. BioMed Res. Int. 2021, 2021, 5298006. [Google Scholar] [CrossRef] [PubMed]
- Braza-Boïls, A.; Salloum-Asfar, S.; Marí-Alexandre, J.; Arroyo, A.B.; González-Conejero, R.; Barceló-Molina, M.; García-Oms, J.; Vicente, V.; Estellés, A.; Gilabert-Estellés, J.; et al. Peritoneal fluid modifies the microRNA expression profile in endometrial and endometriotic cells from women with endometriosis. Hum. Reprod. 2015, 30, 2292–2302. [Google Scholar] [CrossRef]
- Börschel, C.S.; Stejskalova, A.; Schäfer, S.D.; Kiesel, L.; Götte, M. miR-142-3p reduces the size, migration, and contractility of endometrial and endometriotic stromal cells by targeting integrin-and Rho GTPase-related pathways that regulate cytoskeletal function. Biomedicines 2020, 8, 291. [Google Scholar] [CrossRef]
- Ma, L.; Li, Z.; Li, W.; Ai, J.; Chen, X. MicroRNA-142-3p suppresses endometriosis by regulating KLF9-mediated autophagy in vitro and in vivo. RNA Biol. 2019, 16, 1733–1748. [Google Scholar] [CrossRef]
- Zhu, J.; Xu, Z.; Wu, P.; Zeng, C.; Peng, C.; Zhou, Y.; Xue, Q.J.R.S. MicroRNA-92a-3p Inhibits Cell Proliferation and Invasion by Regulating the Transcription Factor 21/Steroidogenic Factor 1 Axis in Endometriosis. Reprod. Sci. 2023, 30, 2188–2197. [Google Scholar] [CrossRef] [PubMed]
- Byon, J.C.; Papayannopoulou, T. MicroRNAs: Allies or Foes in erythropoiesis? J. Cell. Physiol. 2012, 227, 7–13. [Google Scholar] [CrossRef]
- Masaki, S.; Ohtsuka, R.; Abe, Y.; Muta, K.; Umemura, T. Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem. Biophys. Res. Commun. 2007, 364, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.D.; O’Carroll, D. The Journal of the American Society of Hematology. The miR-144/451eGFP allele, a novel tool for resolving the erythroid potential of hematopoietic precursors. Blood 2011, 118, 2988–2992. [Google Scholar] [CrossRef]
- Rasmussen, K.D.; Simmini, S.; Abreu-Goodger, C.; Bartonicek, N.; Di Giacomo, M.; Bilbao-Cortes, D.; Horos, R.; Von Lindern, M.; Enright, A.J.; O’Carroll, D. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 2010, 207, 1351–1358. [Google Scholar] [CrossRef]
- Zhan, M.; Miller, C.P.; Papayannopoulou, T.; Stamatoyannopoulos, G.; Song, C.-Z. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp. Hematol. 2007, 35, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Bruchova, H.; Yoon, D.; Agarwal, A.M.; Mendell, J.; Prchal, J.T. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp. Hematol. 2007, 35, 1657–1667. [Google Scholar] [CrossRef]
- Leecharoenkiat, K.; Tanaka, Y.; Harada, Y.; Chaichompoo, P.; Sarakul, O.; Abe, Y.; Smith, D.R.; Fucharoen, S.; Svasti, S.; Umemura, T. Plasma microRNA-451 as a novel hemolytic marker for β0-thalassemia/HbE disease. Mol. Med. Rep. 2017, 15, 2495–2502. [Google Scholar] [CrossRef]
- Svasti, S.; Masaki, S.; Penglong, T.; Abe, Y.; Winichagoon, P.; Fucharoen, S.; Umemura, T. Expression of microRNA-451 in normal and thalassemic erythropoiesis. Ann. Hematol. 2010, 89, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Edelman, J.J.B.; Kao, S.C.-H.; Vallely, M.P.; van Zandwijk, N.; Reid, G. The impact of hemolysis on cell-free microRNA biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.S.; Soon, P.S.; Marsh, D.J. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS ONE 2016, 11, e0153200. [Google Scholar] [CrossRef]
- Simundic, A.-M.; Bölenius, K.; Cadamuro, J.; Church, S.; Cornes, M.P.; van Dongen-Lases, E.C.; Eker, P.; Erdeljanovic, T.; Grankvist, K.; Guimaraes, J.T.; et al. Joint EFLM-COLABIOCLI Recommendation for venous blood sampling: V 1.1, June 2018. Clin. Chem. Lab. Med. 2018, 56, 2015–2038. [Google Scholar] [CrossRef]
- Lippi, G.; Von Meyer, A.; Cadamuro, J.; Simundic, A.-M. Blood sample quality. Diagnosis 2019, 6, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.-L.; Zhang, Z.; Fan, W.-S.; Li, L.-A.; Ye, M.-X.; Zhang, Q.; Zhang, N.-N.; Li, Z.; Meng, Y.-G. Identification of MicroRNAs as potential biomarkers in ovarian endometriosis. Reprod. Sci. 2020, 27, 1715–1723. [Google Scholar] [CrossRef]
- He, Z.; Tian, M.; Fu, X. Reduced expression of miR-30c-5p promotes hepatocellular carcinoma progression by targeting RAB32. Mol. Ther. Nucleic Acids 2021, 26, 603–612. [Google Scholar] [CrossRef]
- Mehterov, N.; Vladimirov, B.; Sacconi, A.; Pulito, C.; Rucinski, M.; Blandino, G.; Sarafian, V. Salivary miR-30c-5p as Potential Biomarker for Detection of Oral Squamous Cell Carcinoma. Biomedicines 2021, 9, 1079. [Google Scholar] [CrossRef]
- Cao, J.-M.; Li, G.-Z.; Han, M.; Xu, H.-L.; Huang, K.-M. MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed. Pharmacother. 2017, 93, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Zhou, Y.; Xu, X.; Qi, X.; Liu, J.; Pu, Y.; Zhang, S.; Gao, X.; Luo, X.; Li, M.; et al. MiR-30c-5p loss-induced PELI1 accumulation regulates cell proliferation and migration via activating PI3K/AKT pathway in papillary thyroid carcinoma. J. Transl. Med. 2022, 20, 20. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Zhu, Y.; Sun, F.; Xu, D.; Wang, C. MicroRNA-30c-5p arrests bladder cancer G2/M phase and suppresses its progression by targeting PRC1-mediated blocking of CDK1/Cyclin B1 axis. Cell. Signal. 2023, 110, 110836. [Google Scholar] [CrossRef] [PubMed]
- Pollacco, J.; Sacco, K.; Portelli, M.; Schembri-Wismayer, P.; Calleja-Agius, J. Molecular links between endometriosis and cancer. Gynecol. Endocrinol. 2012, 28, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, J.B.; Kjær, S.K.; Mellemkjær, L.; Jensen, A. Endometriosis and risks for ovarian, endometrial and breast cancers: A nationwide cohort study. Gynecol. Oncol. 2016, 143, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, X.; Qiu, L.-W.; Peng, C.; Zhong, S.-P.; Ye, L. MicroRNA-30c inhibits metastasis of ovarian cancer by targeting metastasis-associated gene 1. J. Cancer Res. Ther. 2017, 13, 676–682. [Google Scholar] [CrossRef]
- Pei, B.; Li, T.; Qian, Q.; Fan, W.; He, X.; Zhu, Y.; Xu, L. Downregulation of microRNA-30c-5p was responsible for cell migration and tumor metastasis via COTL1-mediated microfilament arrangement in breast cancer. Gland. Surg. 2020, 9, 747–758. [Google Scholar] [CrossRef]
- Blagih, J.; Buck, M.D.; Vousden, K.H. p53, cancer and the immune response. J. Cell Sci. 2020, 133, jcs237453. [Google Scholar] [CrossRef]
- Goh, A.M.; Coffill, C.R.; Lane, D.P. The role of mutant p53 in human cancer. J. Pathol. 2011, 223, 116–126. [Google Scholar] [CrossRef]
- Lin, S.; Yu, L.; Song, X.; Bi, J.; Jiang, L.; Wang, Y.; He, M.; Xiao, Q.; Sun, M.; Olopade, O.I.; et al. Intrinsic adriamycin resistance in p53-mutated breast cancer is related to the miR-30c/FANCF/REV1-mediated DNA damage response. Cell Death Dis. 2019, 10, 666. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, J.; Fang, X.; Guo, G.; Dai, J.; Sheng, Z.; Li, D.; Chen, J.; Zhang, L.; Liu, C.; et al. Identification of microRNA hsa-miR-30c-5p as an inhibitory factor in the progression of hepatocellular carcinoma and investigation of its regulatory network via comprehensive analysis. Bioengineered 2021, 12, 7154–7166. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.-Q.; Zhang, T.; Xu, L.; Han, H.; Liu, S.-H. miR-30c-5p inhibits glioma proliferation and invasion via targeting Bcl2. Transl. Cancer Res. 2021, 10, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Xia, Y.; Wang, P.; Liu, B.; Chen, Y. Low expression of microRNA-30c promotes invasion by inducing epithelial mesenchymal transition in non-small cell lung cancer. Mol. Med. Rep. 2014, 10, 2575–2579. [Google Scholar] [CrossRef] [PubMed]
- Bashir, S.T.; Redden, C.R.; Raj, K.; Arcanjo, R.B.; Stasiak, S.; Li, Q.; Steelman, A.J.; Nowak, R.A. Endometriosis leads to central nervous system-wide glial activation in a mouse model of endometriosis. J. Neuroinflamm. 2023, 20, 59. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; Maddern, J.; Erickson, A.; Harrington, A.M.; Brierley, S.M. Peripheral and central neuroplasticity in a mouse model of endometriosis. J. Neurochem. 2023. [Google Scholar] [CrossRef] [PubMed]
- Coxon, L.; Wiech, K.; Vincent, K. Is There a Neuropathic-Like Component to Endometriosis-Associated Pain? Results From a Large Cohort Questionnaire Study. Front. Pain Res. 2021, 2, 743812. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.S.; Kotlyar, A.M.; A Flores, V. Endometriosis is a chronic systemic disease: Clinical challenges and novel innovations. Lancet 2021, 397, 839–852. [Google Scholar] [CrossRef]
- Ding, S.; Guo, X.; Zhu, L.; Wang, J.; Li, T.; Yu, Q.; Zhang, X. Macrophage-derived netrin-1 contributes to endometriosis- associated pain. Ann. Transl. Med. 2021, 9, 29. [Google Scholar] [CrossRef]
- Ding, S.; Yu, Q.; Wang, J.; Zhu, L.; Li, T.; Guo, X.; Zhang, X. Activation of ATF3/AP-1 signaling pathway is required for P2X3-induced endometriosis pain. Hum. Reprod. 2020, 35, 1130–1144. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.; Qiu, C.; Sun, Y.; Li, W.; Jiang, J.; Zhang, J.-M. Fractalkine/CX3CR1 Contributes to Fractalkine/CX3CR1 contributes to endometriosis-induced neuropathic pain and mechanical hypersensitivity in rats. Front. Cell. Neurosci. 2018, 12, 495. [Google Scholar] [CrossRef]
- Tang, Y.; Chen, Y.; Liu, R.; Li, W.; Hua, B.; Bao, Y. Wnt signaling pathways: A role in pain processing. NeuroMol. Med. 2022, 24, 233–249. [Google Scholar] [CrossRef]
- Wang, S.; Duan, H.; Li, B.; Hong, W.; Li, X.; Wang, Y.; Guo, Z.C. BDNF and TrKB expression levels in patients with endometriosis and their associations with dysmenorrhoea. J. Ovarian Res. 2022, 15, 35. [Google Scholar] [CrossRef]
- Dwiningsih, S.R.; Meilani, C.; Hadi, S. Brain Derived Neurotrophic Factor as a Non-invasive Biomarker for Detection of Endometriosis. J. Reprod. Infertil. 2022, 23, 207–212. [Google Scholar] [CrossRef]
- Francés, R.; Mata-Garrido, J.; de la Fuente, R.; Carcelén, M.; Lafarga, M.; Berciano, M.T.; García, R.; Hurlé, M.A.; Tramullas, M. Identification of epigenetic interactions between MicroRNA-30c-5p and DNA methyltransferases in neuropathic pain. Int. J. Mol. Sci. 2022, 23, 13994. [Google Scholar] [CrossRef]
- Tramullas, M.; Francés, R.; de la Fuente, R.; Velategui, S.; Carcelén, M.; García, R.; Llorca, J.; Hurlé, M.A. MicroRNA-30c-5p modulates neuropathic pain in rodents. Sci. Transl. Med. 2018, 10, eaao6299. [Google Scholar] [CrossRef]
- Soni, U.K.; Chadchan, S.B.; Kumar, V.; Ubba, V.; Khan, M.T.A.; Vinod, B.S.V.; Konwar, R.; Bora, H.K.; Rath, S.K.; Sharma, S.; et al. A high level of TGF-B1 promotes endometriosis development via cell migration, adhesiveness, colonization, and invasiveness†. Biol. Reprod. 2019, 100, 917–938. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2019, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]



| miRNA qPCR Profiling n = 20 | miRNA Real-Time qPCR Validation n = 139 | |||
|---|---|---|---|---|
| Variable | Control Group n = 10 | Endometriosis Group n = 10 | Control Group n = 72 | Endometriosis Group n = 67 |
| Age ± SD (years) | 32 ± 4.6 | 34.9 ± 4.0 | 31.7 ± 5.2 | 37.5 ± 7.0 |
| Body Mass Index ± SD (kg/m2) | 22.5 ± 2.4 | 22.6 ± 3.2 | 24.1 ± 3.0 | 22.8 ± 3.1 |
| ASRM stage, n (%) | ||||
| I | NA | 0 (0) | NA | 1 (1.5) |
| II | NA | 4 (40) | NA | 6 (9.0) |
| III | NA | 4 (40) | NA | 19 (28.3) |
| IV | NA | 2 (20) | NA | 41 (61.2) |
| Phase of menstrual cycle, n (%) | ||||
| Proliferative | 10 (100) | 0 | 49 (68.1) | 0 |
| Secretory | 0 | 0 | 7 (9.7) | 0 |
| Unknown | 0 | 10 (100) | 16 (22.2) | 67 (100) |
| Pain symptoms, n (%) | ||||
| Any pain symptoms | 10 (100) | 2 (20) | 72 (100) | 6 (8.9) |
| Dyspareunia | 0 | 3 (30) | 0 | 30 (44.7) |
| Dysmenorrhea | 0 | 7 (70) | 0 | 51 (76.1) |
| Dyschezia | 0 | 3 (30) | 0 | 22 (32.8) |
| Dysuria | 0 | 1 (10) | 0 | 9 (13.4) |
| Chronic pelvic pain | 0 | 4 (40) | 0 | 42 (62.6) |
| Infertility, n (%) | ||||
| Primary | 0 | 3 (30) | 0 | 28 (41.7) |
| Secondary | 0 | 1 (10) | 0 | 1 (1.4) |
| Unknown | 0 | 5 (50) | 0 | 23 (34.3) |
| Proven fertility (live birth) | 10 (100) | 1 (10) | 72 (100) | 15 (22.3) |
| Tumoral makers ± SD | ||||
| CA-125 (U/mL) | NA | 32.4 ± 17.4 | NA | 185.3 ± 527.8 |
| CA-19.9 (U/mL) | NA | 28.1 ± 19.2 | NA | 94.99 ± 268.2 |
| HE4 (pmol/l) | NA | - | NA | 76.43 ± 31.05 |
| Comorbidities, n (%) | ||||
| Hypothyroidism | 0 | 0 | 2 (2.7) | 11 (16.4) |
| Asthma | 0 | 0 | 2 (2.7) | 7 (10.4) |
| Migraine | 0 | 0 | 1 (1.3) | 4 (5.9) |
| Allergic rhinoconjunctivitis | 0 | 0 | 1 (1.3) | 3 (4.4) |
| Vitiligo | 0 | 0 | 0 | 3 (4.4) |
| Hypertension | 0 | 0 | 0 | 3 (4.4) |
| Depression | 0 | 0 | 0 | 3 (4.4) |
| Fibromyalgia | 0 | 0 | 0 | 2 (2.9) |
| Raynaud Syndrome | 0 | 0 | 0 | 2 (2.9) |
| Other | 0 | 1 (10) | 4 (5.5) | 8 (11.9) |
| Differentially Expressed miRNAs | Assay Catalog No. | p-Value | Fold Change |
|---|---|---|---|
| [miR-210-3p] * | YP00204333 | 0.005 | −1.73 |
| [miR-185-5p] * | YP00206037 | 0.006 | −1.44 |
| [miR-25-3p] * | YP00204361 | 0.007 | −1.44 |
| miR-660-5p * | YP00205911 | 0.007 | −1.44 |
| [miR-142-3p] | YP00204291 | 0.008 | 1.55 |
| [miR-92a-3p] * | YP00204258 | 0.010 | −1.37 |
| miR-28-5p | YP00204322 | 0.010 | 1.85 |
| let-7a-5p | YP00205727 | 0.010 | 1.35 |
| miR-532-5p * | YP00204003 | 0.014 | −1.43 |
| miR-16-2-3p * | YP00204309 | 0.014 | −1.43 |
| [let-7c-5p] | YP00204767 | 0.015 | 1.41 |
| miR-16-5p * | YO00205702 | 0.017 | −1.44 |
| miR-766-3p | YP00204499 | 0.019 | 1.73 |
| miR-451a * | YP02119305 | 0.020 | −1.70 |
| miR-7-1-3p | YP00205888 | 0.023 | 2.24 |
| miR-19b-3p * | YP00204450 | 0.023 | −1.29 |
| miR-132-3p | YP00206035 | 0.027 | 1.57 |
| miR-192-5p * | YP00204099 | 0.029 | −1.43 |
| miR-215-5p * | YP00204598 | 0.030 | −1.51 |
| [miR-30c-5p] | YP00204783 | 0.031 | 1.33 |
| let-7g-5p | YP00204565 | 0.031 | 1.13 |
| miR-338-3p | YP00204719 | 0.034 | 1.58 |
| miR-19a-3p * | YP00205862 | 0.034 | −1.25 |
| miR-140-3p * | YP00204304 | 0.037 | −1.26 |
| miR-30b-5p | YP00204765 | 0.039 | 1.29 |
| miR-141-3p * | YP00204504 | 0.040 | −1.73 |
| let-7d-5p | YP00204124 | 0.046 | 1.36 |
| [miR-342-3p] * | YP00205625 | 0.046 | −1.49 |
| miR-150-5p * | YP00204660 | 0.048 | −1.69 |
| Reference miRNA candidates | Assay catalog no. | Stability value | |
| miR-30e-5p | YP00204714 | 0.003 | |
| miR-652-3p | YP00204387 | 0.003 | |
| miR-29b-3p | YP00204679 | 0.004 | |
| miR-21-5p | YP00204230 | 0.004 | |
| miR-15b-5p | YP00204243 | 0.004 | |
| miR-22-3p | YP00204606 | 0.004 | |
| miR-107 | YP00204468 | 0.004 | |
| Technical controls (spike-ins) | Assay catalog no. | p-value | |
| RNA isolation control | |||
| UniSp2 | YP00203950 | 0.796 | |
| UniSp4 | YP00203953 | 0.971 | |
| UniSp5 | YP00203955 | 0.218 | |
| cDNA synthesis control | |||
| UniSp6 | YP00203954 | 0.684 | |
| Interplate calibrator | |||
| UniSp3 | YP02119288 | 0.143 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chico-Sordo, L.; Ruiz-Martínez, T.; Toribio, M.; González-Martín, R.; Spagnolo, E.; Domínguez, F.; Hernández, A.; García-Velasco, J.A. Identification of miR-30c-5p microRNA in Serum as a Candidate Biomarker to Diagnose Endometriosis. Int. J. Mol. Sci. 2024, 25, 1853. https://doi.org/10.3390/ijms25031853
Chico-Sordo L, Ruiz-Martínez T, Toribio M, González-Martín R, Spagnolo E, Domínguez F, Hernández A, García-Velasco JA. Identification of miR-30c-5p microRNA in Serum as a Candidate Biomarker to Diagnose Endometriosis. International Journal of Molecular Sciences. 2024; 25(3):1853. https://doi.org/10.3390/ijms25031853
Chicago/Turabian StyleChico-Sordo, Lucía, Tamara Ruiz-Martínez, Mónica Toribio, Roberto González-Martín, Emanuela Spagnolo, Francisco Domínguez, Alicia Hernández, and Juan A. García-Velasco. 2024. "Identification of miR-30c-5p microRNA in Serum as a Candidate Biomarker to Diagnose Endometriosis" International Journal of Molecular Sciences 25, no. 3: 1853. https://doi.org/10.3390/ijms25031853
APA StyleChico-Sordo, L., Ruiz-Martínez, T., Toribio, M., González-Martín, R., Spagnolo, E., Domínguez, F., Hernández, A., & García-Velasco, J. A. (2024). Identification of miR-30c-5p microRNA in Serum as a Candidate Biomarker to Diagnose Endometriosis. International Journal of Molecular Sciences, 25(3), 1853. https://doi.org/10.3390/ijms25031853






