ADAMTS Gene-Derived circRNA Molecules in Non-Small-Cell Lung Cancer: Expression Profiling, Clinical Correlations and Survival Analysis
Abstract
:1. Introduction
2. Results
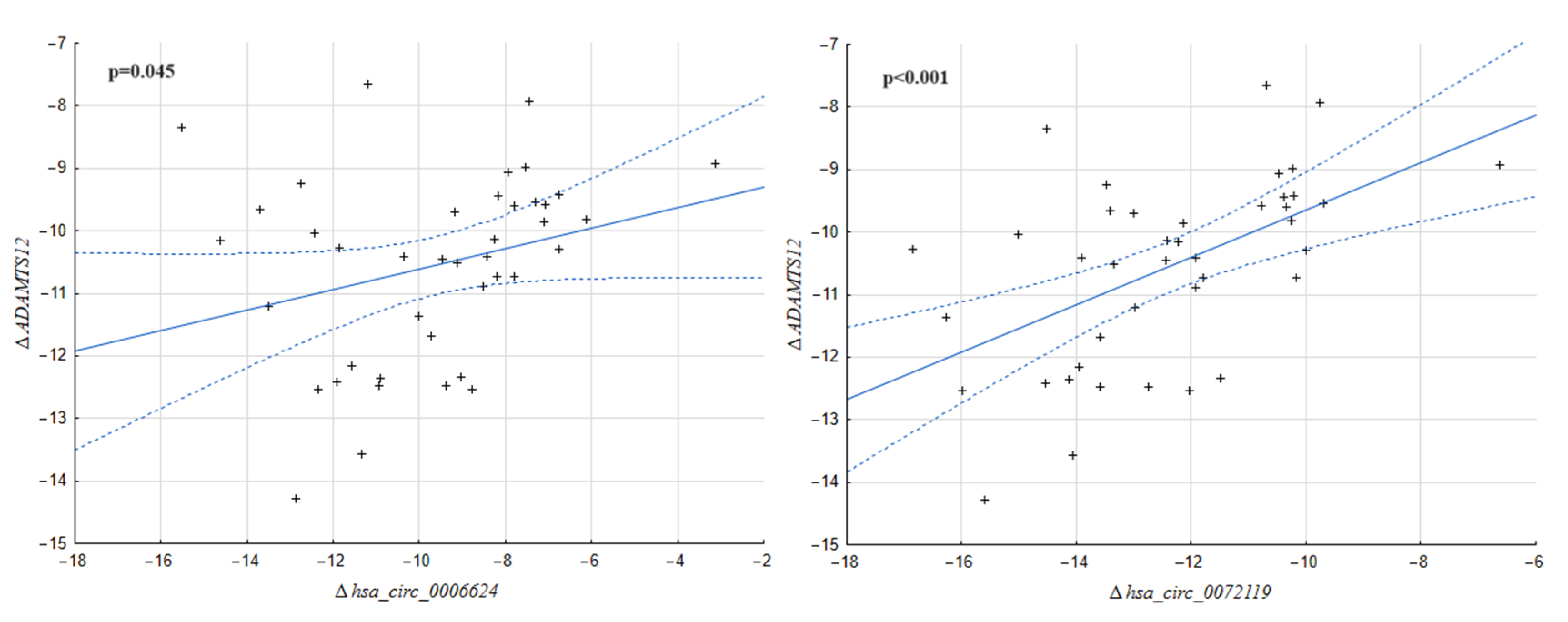
2.1. Correlation between the Expression of circRNA Molecules and Their Host Genes
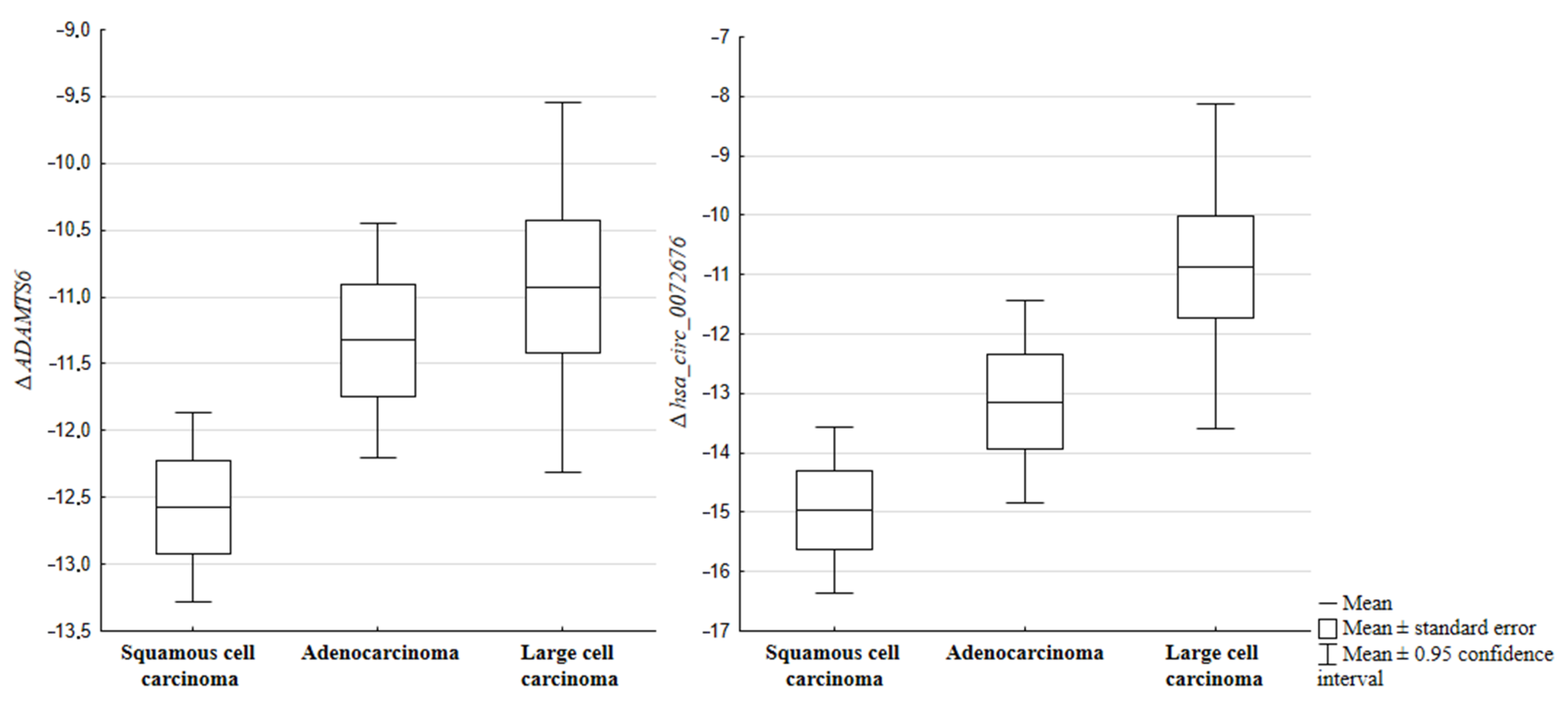
2.2. Expression of circRNAs and Their Host Genes in Relation to the Histopathological Subtype of NSCLC
2.3. Expression of circRNAs and Their Host Genes in Relation to Tumor Progression according to the TNM Classification
2.4. Expression of circRNAs and Their Host Genes in Relation to the Infiltration of Surrounding Tissues
2.5. Comparison of the Expression Levels of the Genes ADAMTS6, ADAMTS9 and ADAMTS12 between Tumor Tissue from NSCLC Patients with Histological Subtype Division and Normal Lung Tissue
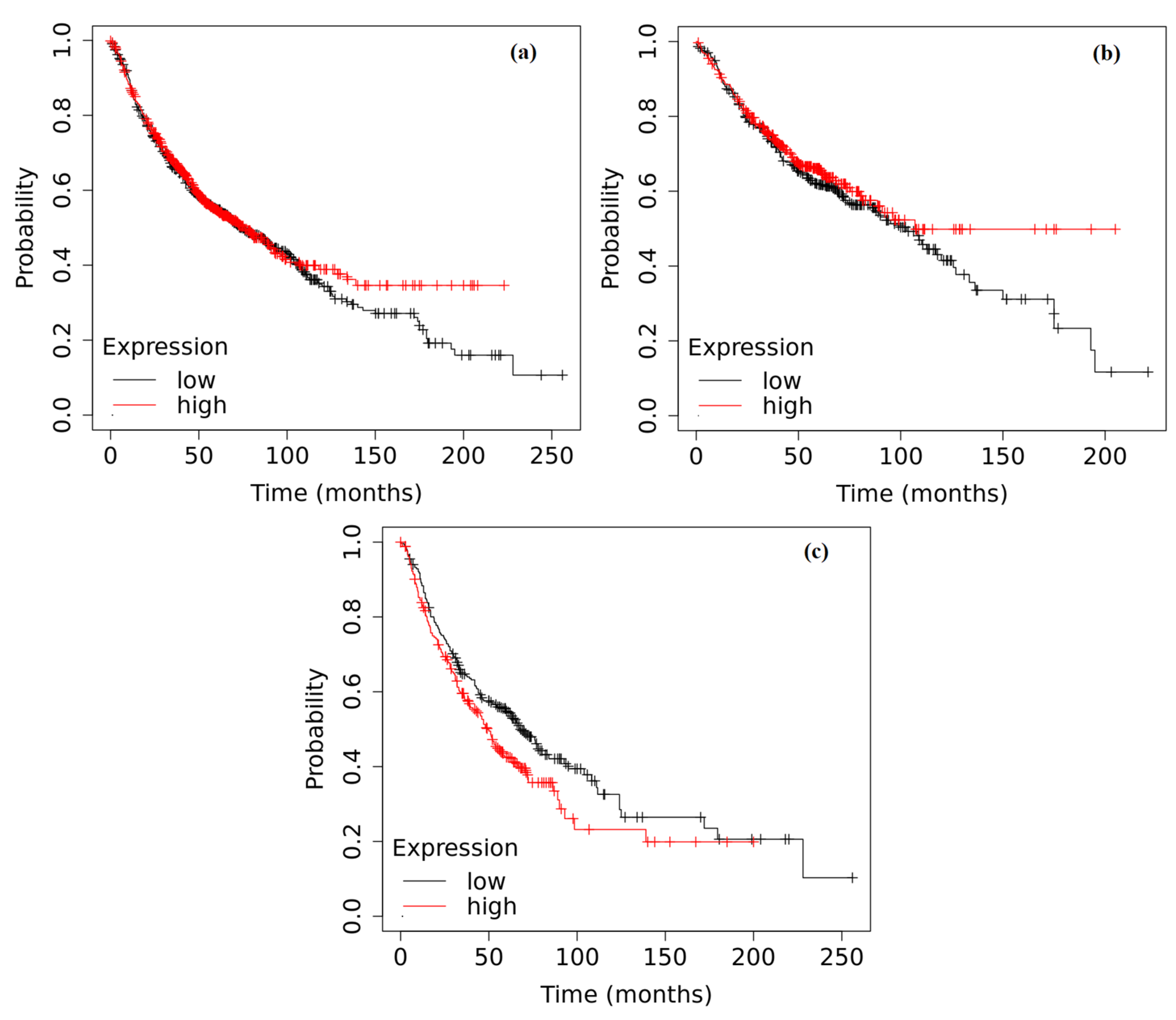
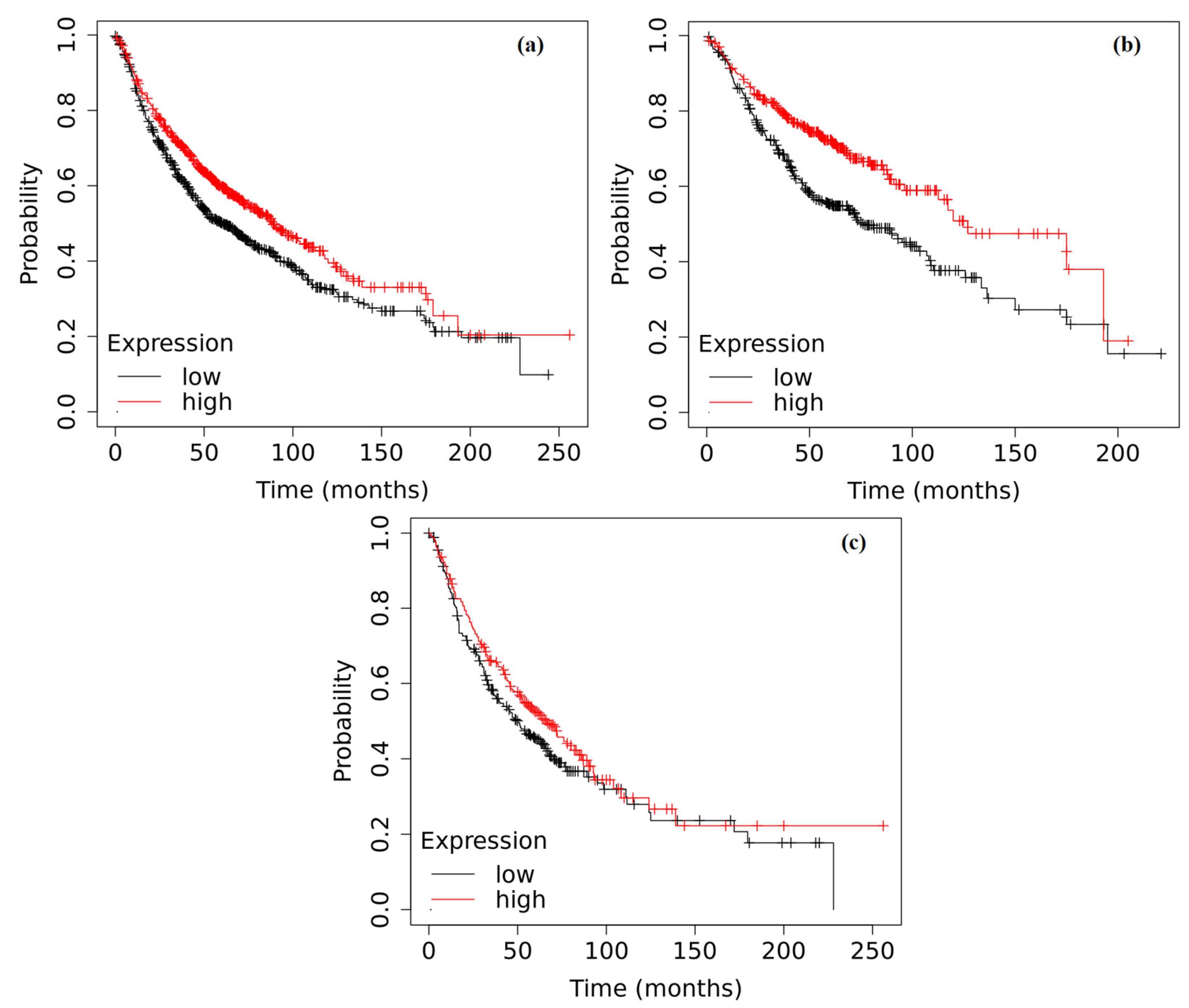
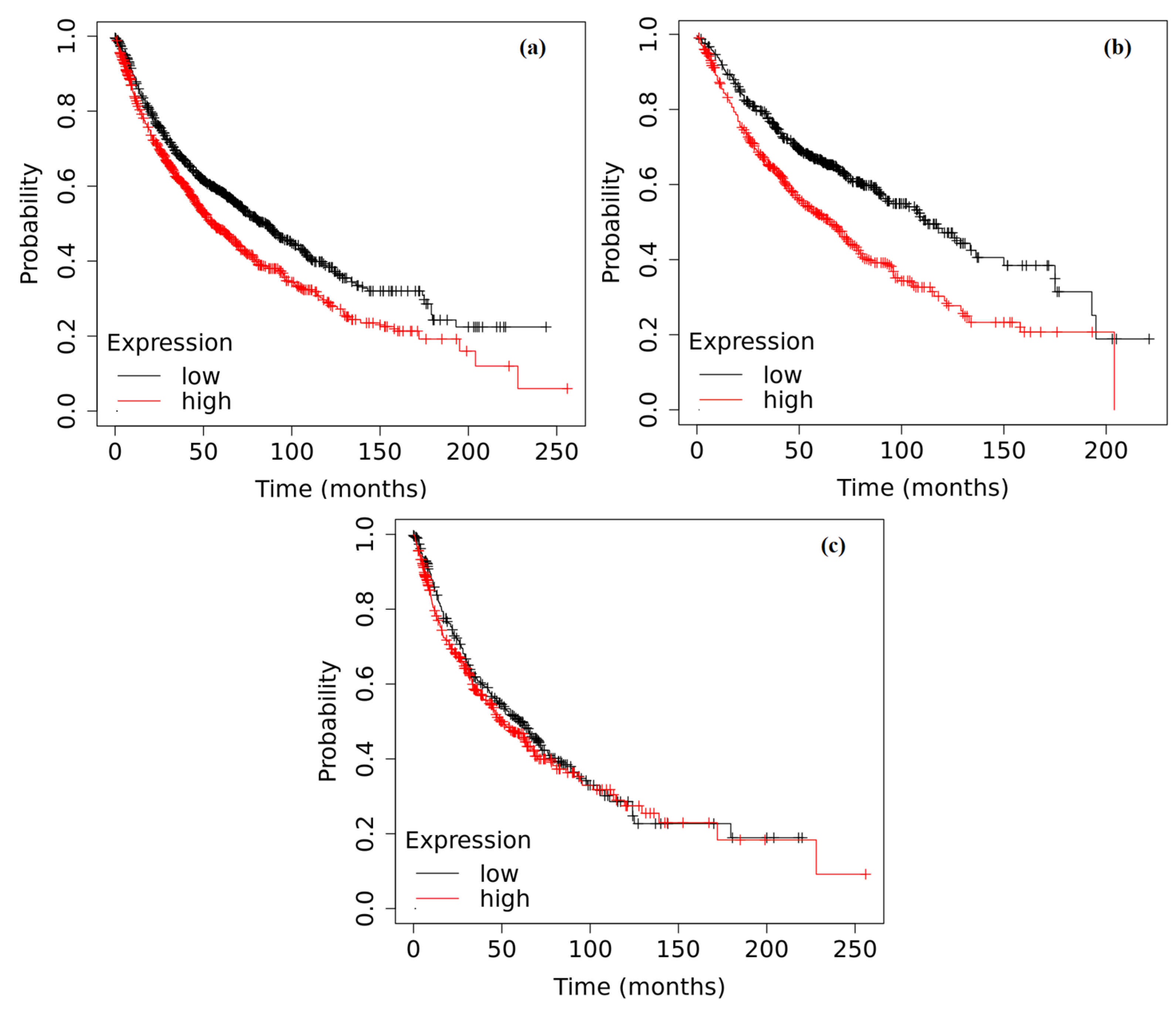
2.6. Survival Analysis Based on the Expression of ADAMTS6, ADAMTS9 and ADAMTS12 in the Tumor Tissue of Patients with NSCLC
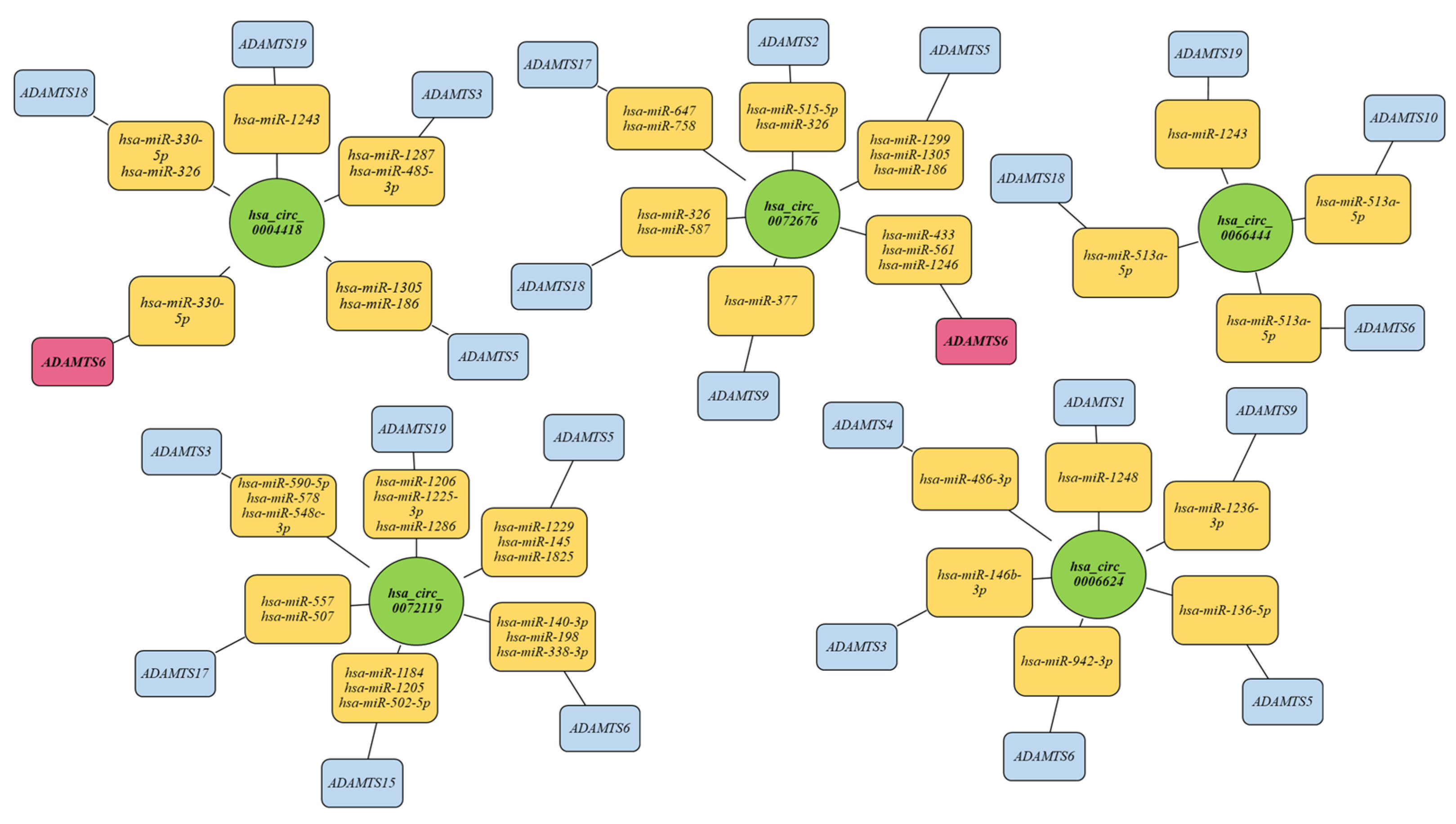
2.7. Analysis of Interactions between the Tested circRNA Molecules and miRNAs and mRNA of Genes Encoding ADAMTS Family Proteins, including Host Genes
3. Discussion
4. Materials and Methods
4.1. RNA Isolation
4.2. Reverse Transcription
4.3. qPCR
4.4. Degradation of Linear RNA
4.5. qPCR for circRNA
4.6. Analysis of the External Database
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef] [PubMed]
- Kiełbowski, K.; Ptaszyński, K.; Wójcik, J.; Wojtyś, M.E. The role of selected non-coding RNAs in the biology of non-small cell lung cancer. Adv. Med. Sci. 2023, 68, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Santer, L.; Bar, C.; Thum, T. Circular RNAs: A Novel Class of Functional RNA Molecules with a Therapeutic Perspective. Mol. Ther. 2019, 27, 1350–1363. [Google Scholar] [CrossRef]
- Patop, I.L.; Kadener, S. circRNAs in Cancer. Curr. Opin. Genet. Dev. 2018, 48, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.J.; Zhou, H.C.; Feng, Z.Y.; Xu, Z.H.; Tang, Y.; Li, P.Y.; Wu, M.H. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef]
- Kelwick, R.; Desanlis, I.; Wheeler, G.N.; Edwards, D.R. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015, 16, 113. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, J.T.; Yang, Z.L. The roles of ADAMTS in angiogenesis and cancer. Tumor Biol. 2015, 36, 4039–4051. [Google Scholar] [CrossRef]
- Meng, X.; Hu, D.; Zhang, P.; Chen, Q.; Chen, M. CircFunBase: A database for functional circular RNAs. Database 2019, 2019, baz003. [Google Scholar] [CrossRef]
- Dang, Y.; Ouyang, X.; Zhang, F.; Wang, K.; Lin, Y.; Sun, B.; Wang, Y.; Wang, L.; Huang, Q. Circular RNAs expression profiles in human gastric cancer. Sci. Rep. 2017, 7, 9060. [Google Scholar] [CrossRef]
- Rong, D.; Dong, C.; Fu, K.; Wang, H.; Tang, W.; Cao, H. Upregulation of circ_0066444 promotes the proliferation, invasion, and migration of gastric cancer cells. Onco Targets Ther. 2018, 11, 2753–2761. [Google Scholar] [CrossRef]
- Wu, P.; Mo, Y.Z.; Peng, M.; Tang, T.; Zhong, Y.; Deng, X.Y.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 2020, 19, 22. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Zhang, H.D.; Jiang, L.H.; Sun, D.W.; Hou, J.C.; Ji, Z.L. CircRNA: A novel type of biomarker for cancer. Breast Cancer 2018, 25, 1–7. [Google Scholar] [CrossRef]
- Zhou, R.Y.; Wu, Y.W.; Wang, W.X.; Su, W.J.; Liu, Y.C.; Wang, Y.M.; Fan, C.M.; Li, X.L.; Li, G.Y.; Li, Y.; et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018, 425, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Lei, M.; Zheng, G.T.; Ning, Q.Q.; Zheng, J.N.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 2020, 19, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, J.; Zhang, C.; Lin, C.; Zhang, J.; Zhang, W.; Zhang, W.; Lu, Y.; Zheng, L.; Li, X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J. Pathol. 2018, 246, 166–179. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, W.; Wang, Y.; Wang, H.; Liu, S. Extracellular vesicle-mediated crosstalk between pancreatic cancer and stromal cells in the tumor microenvironment. J. Nanobiotechnology 2022, 20, 208. [Google Scholar] [CrossRef]
- Richardson, N.C.; Kasamon, Y.; Pazdur, R.; Gormley, N. The saga of PI3K inhibitors in haematological malignancies: Survival is the ultimate safety endpoint. Lancet Oncol. 2022, 23, 563–566. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Rodriguez-Canales, J.; Parra-Cuentas, E.; Wistuba, I.I. Diagnosis and Molecular Classification of Lung Cancer. Cancer Treat. Res. 2016, 170, 25–46. [Google Scholar] [CrossRef]
- Cal, S.; López-Otín, C. ADAMTS proteases and cancer. Matrix Biol. 2015, 44–46, 77–85. [Google Scholar] [CrossRef]
- Koo, B.H.; Coe, D.M.; Dixon, L.J.; Somerville, R.P.; Nelson, C.M.; Wang, L.W.; Young, M.E.; Lindner, D.J.; Apte, S.S. ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am. J. Pathol. 2010, 176, 1494–1504. [Google Scholar] [CrossRef]
- El Hour, M.; Moncada-Pazos, A.; Blacher, S.; Masset, A.; Cal, S.; Berndt, S.; Detilleux, J.; Host, L.; Obaya, A.J.; Maillard, C.; et al. Higher sensitivity of Adamts12-deficient mice to tumor growth and angiogenesis. Oncogene 2010, 29, 3025–3032. [Google Scholar] [CrossRef]
- Du, W.; Wang, S.; Zhou, Q.; Li, X.; Chu, J.; Chang, Z.; Tao, Q.; Ng, E.K.; Fang, J.; Sung, J.J.; et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene 2013, 32, 3319–3328. [Google Scholar] [CrossRef] [PubMed]
- Fontanil, T.; Rúa, S.; Llamazares, M.; Moncada-Pazos, A.; Quirós, P.M.; García-Suárez, O.; Vega, J.A.; Sasaki, T.; Mohamedi, Y.; Esteban, M.M.; et al. Interaction between the ADAMTS-12 metalloprotease and fibulin-2 induces tumor-suppressive effects in breast cancer cells. Oncotarget 2014, 5, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, J.; Tian, Y.; Gao, Y.; Dong, X.; Chen, W.; Yuan, X.; Yin, W.; Xu, J.; Chen, K.; et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 2020, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 1 March 2023).
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Győrffy, B. Transcriptome-level discovery of survival-associated biomarkers and therapy targets in non-small-cell lung cancer. Br. J. Pharmacol. 2023, 181, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2019, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Lin, Y.-C.-D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA–target interactions. Nucleic Acids Res. 2021, 50, D222–D230. [Google Scholar] [CrossRef]


| Clinical Parameter | Number of Patients | |||
|---|---|---|---|---|
| Smoking tobacco products | Yes—40 | No—21 | ||
| Pack–year | Mean—39.69 (SD—18.33) | |||
| Histological subtype | Sqamous cell carcinoma 31 | Adenocarcinoma 20 | Large-cell carcinoma 5 | Other 6 |
| Primary tumor size (T in TNM classification) | T1—12 | T2—23 | T3—16 | T4—10 |
| Lymph node involvement (N in TNM classification) | N0—16 | N1—27 | N2—16 | N3—2 |
| Presence of distant metastasis (M in TNM classification) | Mx—25 | M0—30 | M1—6 | |
| Another cancer | Yes—12 | No—59 | ||
| Primer Name | Sequence |
|---|---|
| hsa-ADAMTS6 0002 forward | 5′-CTGTGACAGTCCAGCGTAAGT-3′ |
| hsa-ADAMTS6 0002 reverse | 5′-TGACACAGCGGTTGCTTTTG-3′ |
| hsa-ADAMTS6 0003 forward | 5′-GTGACAGTCCAGCACCTTCAG-3′ |
| hsa-ADAMTS6 0003 reverse | 5′-ACGCTTGGGAGGCTCATTAT-3′ |
| hsa-ADAMTS9 0002 forward | 5′-GACGCTGCATGGAGTACTGG-3′ |
| hsa-ADAMTS9 0002 reverse | 5′-GACATACAACAACACGCCGC-3′ |
| hsa-ADAMTS12 0001 forward | 5′-CTCAGTGGCACGGTTCTACA-3′ |
| hsa-ADAMTS12 0001 reverse | 5′-TCTACTCGGACTGGACCCAC-3′ |
| hsa-ADAMTS12 0004 forward | 5′-CCTCCTTTCCTGCAACAGAGA-3′ |
| hsa-ADAMTS12 0004 reverse | 5′-GCGGCCCTTCTTTATGCAATG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietrzak, J.; Świechowski, R.; Wosiak, A.; Wcisło, S.; Balcerczak, E. ADAMTS Gene-Derived circRNA Molecules in Non-Small-Cell Lung Cancer: Expression Profiling, Clinical Correlations and Survival Analysis. Int. J. Mol. Sci. 2024, 25, 1897. https://doi.org/10.3390/ijms25031897
Pietrzak J, Świechowski R, Wosiak A, Wcisło S, Balcerczak E. ADAMTS Gene-Derived circRNA Molecules in Non-Small-Cell Lung Cancer: Expression Profiling, Clinical Correlations and Survival Analysis. International Journal of Molecular Sciences. 2024; 25(3):1897. https://doi.org/10.3390/ijms25031897
Chicago/Turabian StylePietrzak, Jacek, Rafał Świechowski, Agnieszka Wosiak, Szymon Wcisło, and Ewa Balcerczak. 2024. "ADAMTS Gene-Derived circRNA Molecules in Non-Small-Cell Lung Cancer: Expression Profiling, Clinical Correlations and Survival Analysis" International Journal of Molecular Sciences 25, no. 3: 1897. https://doi.org/10.3390/ijms25031897
APA StylePietrzak, J., Świechowski, R., Wosiak, A., Wcisło, S., & Balcerczak, E. (2024). ADAMTS Gene-Derived circRNA Molecules in Non-Small-Cell Lung Cancer: Expression Profiling, Clinical Correlations and Survival Analysis. International Journal of Molecular Sciences, 25(3), 1897. https://doi.org/10.3390/ijms25031897







