New Insights into Pediatric Kidney Transplant Rejection Biomarkers: Tissue, Plasma and Urine MicroRNAs Compared to Protocol Biopsy Histology
Abstract
1. Introduction
2. Results
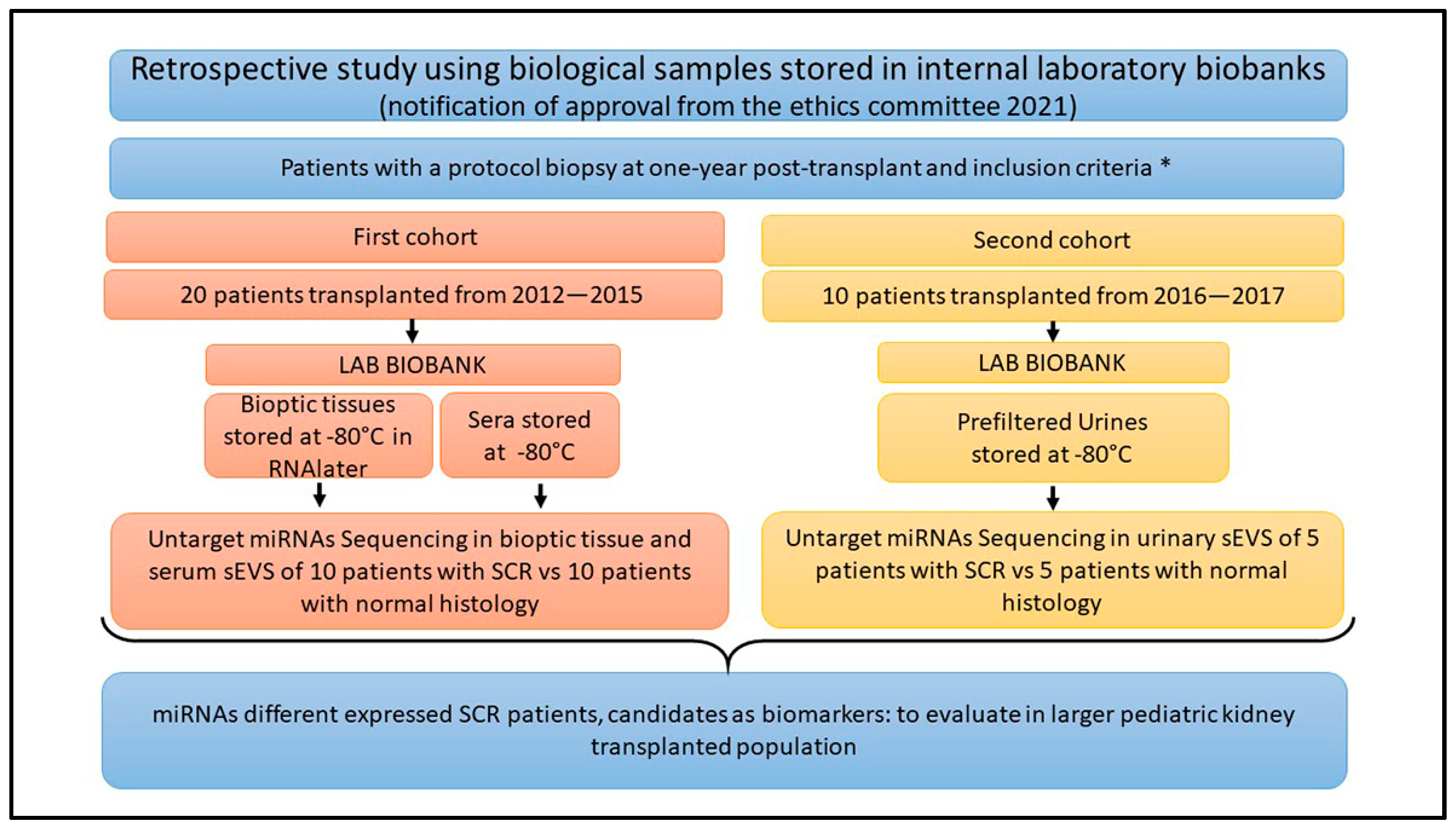
2.1. Population
2.2. EVS Extraction and Characterization
2.3. RNA Quality and Concentration
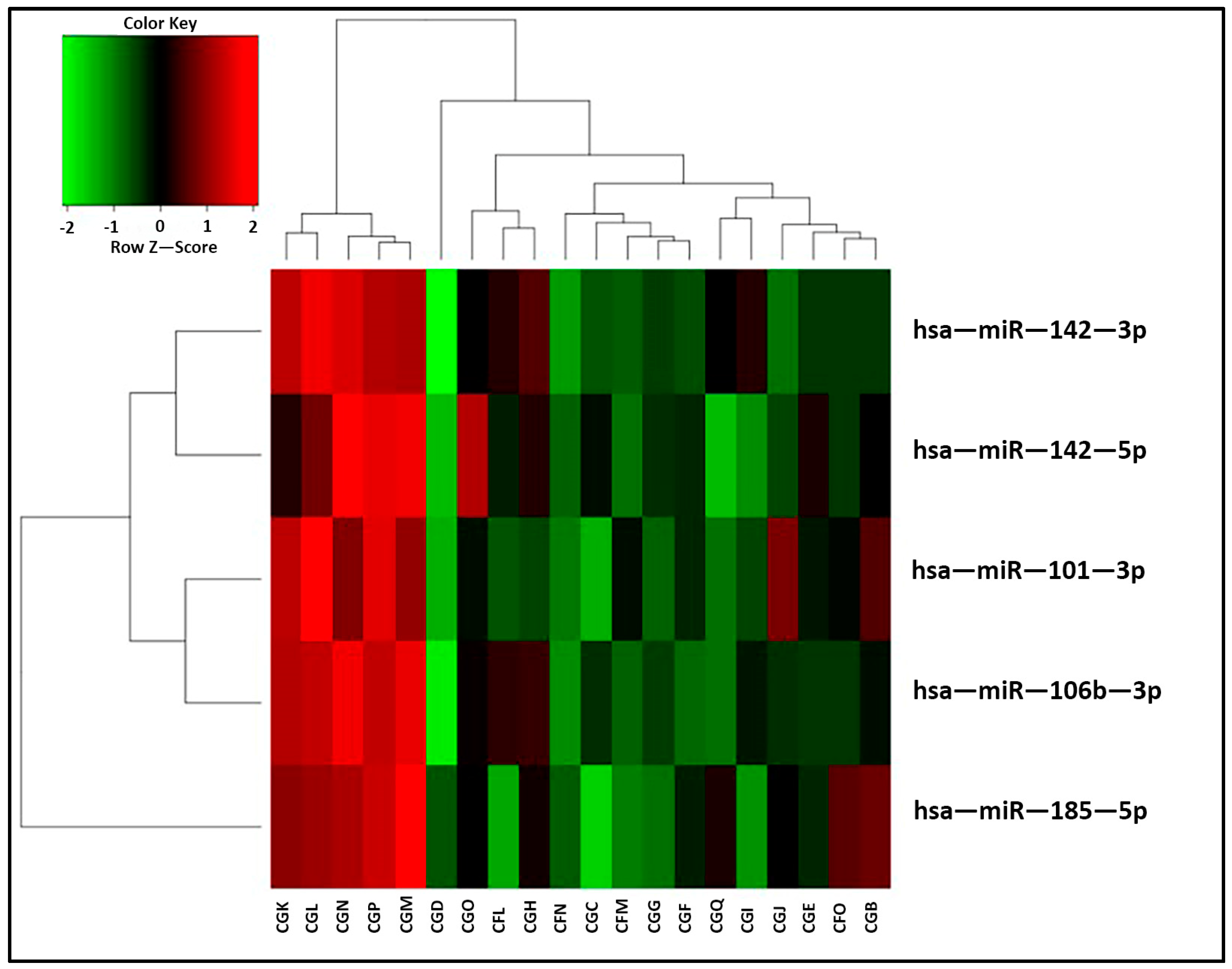
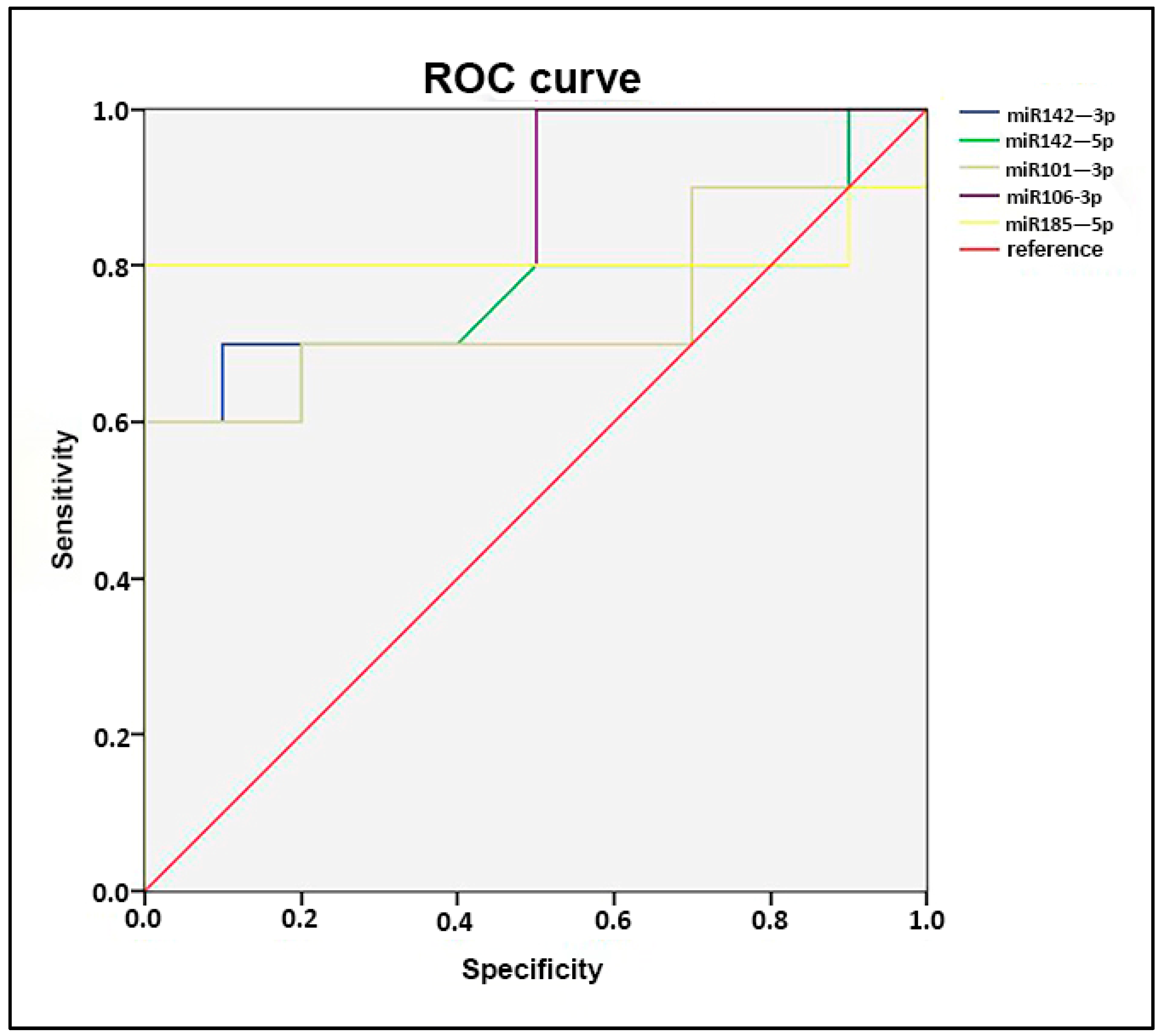
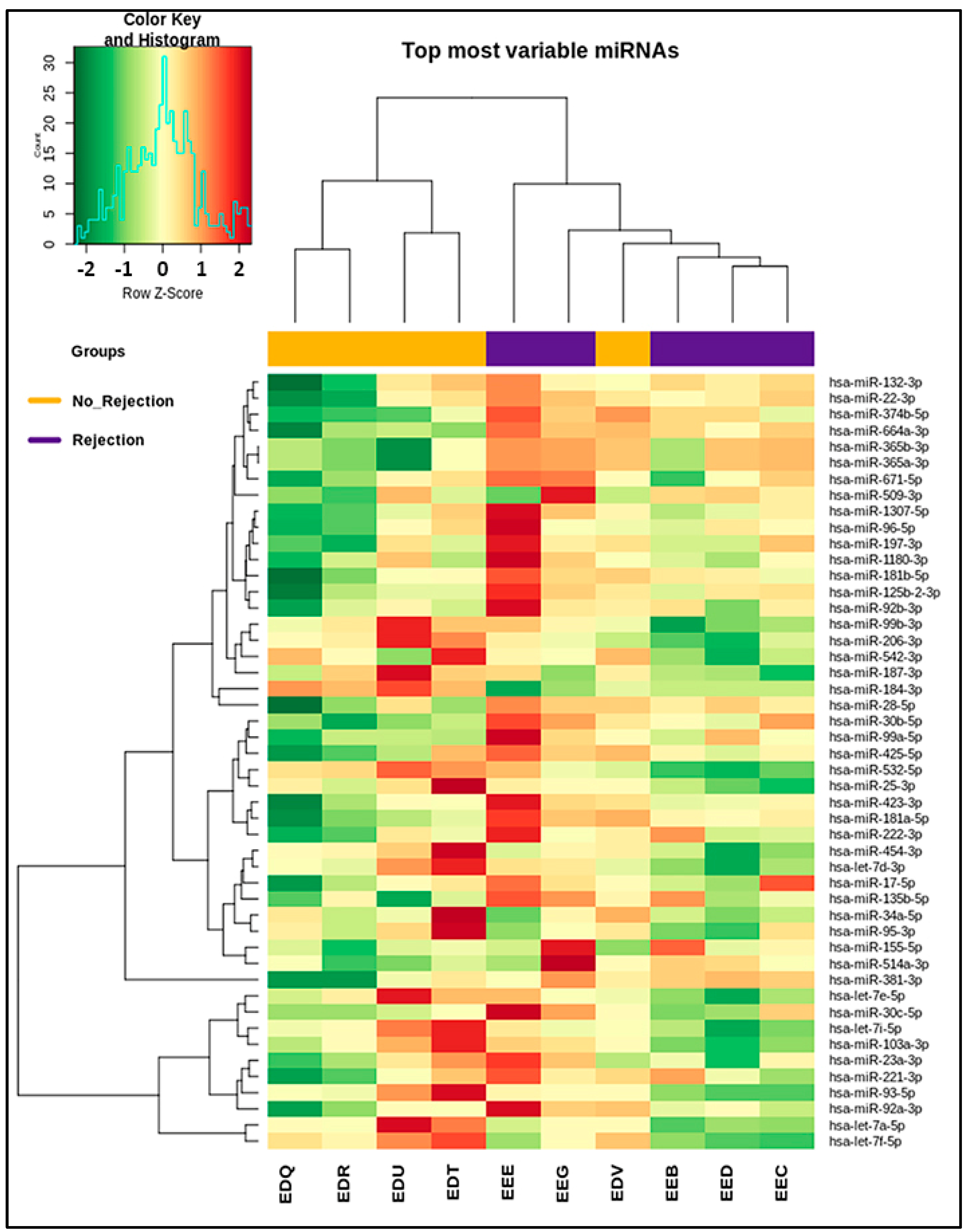
2.4. miRNA Profiling
3. Discussion
4. Materials and Methods
4.1. Patients’ Enrollment and Study Design
- Age < 18 years;
- First transplant;
- Single transplant (not two grafts, for example, kidney and liver);
- Not hyperimmune;
- No surgical complications;
- No delay in graft function;
- No clinical or subclinical rejection before one-year post-transplant;
- Stable graft at one-year post-transplant, with creatinine-based estimates of kidney transplant function or proteinuria.
4.2. Serum EV Isolation
4.3. Urinary EV Isolation
4.4. EV Characterization
4.5. Western Blot Analysis
4.6. Transmitted Electronic Microscopy (TEM)
4.7. Nanoparticle Tracking Analysis (NTA)
4.8. RNA Extraction and NGS Sequencing
4.9. Alignment and Comparison of Sequenced miRNAs
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McDonald, S.P.; Craig, J.C. Long-Term Survival of Children with End-Stage Renal Disease. N. Engl. J. Med. 2004, 350, 2654–2662. [Google Scholar] [CrossRef]
- Winterberg, P.D.; Garro, R. Long-Term Outcomes of Kidney Transplantation in Children. Pediatr. Clin. N. Am 2019, 66, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Gaston, R.S.; Cecka, J.M.; Kasiske, B.L.; Fieberg, A.M.; Leduc, R.; Cosio, F.C.; Gourishankar, S.; Grande, J.; Halloran, P.; Hunsicker, L.; et al. Evidence for Antibody-Mediated Injury as a Major Determinant of Late Kidney Allograft Failure. Transplantation 2010, 90, 68–74. [Google Scholar] [CrossRef]
- Deville, K.A.; Seifert, M.E. Biomarkers of Alloimmune Events in Pediatric Kidney Transplantation. Front. Pediatr. 2023, 10, 1087841. [Google Scholar] [CrossRef]
- Peruzzi, L.; Deaglio, S. Rejection Markers in Kidney Transplantation: Do New Technologies Help Children? Pediatr. Nephrol. 2023, 38, 2939–2955. [Google Scholar] [CrossRef]
- Ibernon, M.; Moreso, F.; Serón, D. Subclinical Rejection in Renal Transplants Is Associated with Low Serum Mannose-Binding Lectin Levels. Kidney Int. Suppl. 2011, 1, 36–39. [Google Scholar] [CrossRef][Green Version]
- Mehta, R.B.; Tandukar, S.; Jorgensen, D.; Randhawa, P.; Sood, P.; Puttarajappa, C.; Zeevi, A.; Tevar, A.D.; Hariharan, S. Early Subclinical Tubulitis and Interstitial Inflammation in Kidney Transplantation Have Adverse Clinical Implications. Kidney Int. 2020, 98, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Nankivell, B.J.; Chapman, J.R. The Significance of Subclinical Rejection and the Value of Protocol Biopsies. Am. J. Transplant. 2006, 6, 2006–2012. [Google Scholar] [CrossRef]
- Bakdash, K.; Schramm, K.M.; Annam, A.; Brown, M.; Kondo, K.; Lindquist, J.D. Complications of Percutaneous Renal Biopsy. Semin. Intervent. Radiol. 2019, 36, 097–103. [Google Scholar] [CrossRef] [PubMed]
- Filippone, E.J.; Farber, J.L. The Problem of Subclinical Antibody-Mediated Rejection in Kidney Transplantation. Transplantation 2021, 105, 1176–1187. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Trionfini, P.; Benigni, A.; Remuzzi, G. MicroRNAs in Kidney Physiology and Disease. Nat. Rev. Nephrol. 2015, 11, 23–33. [Google Scholar] [CrossRef]
- Mahtal, N.; Lenoir, O.; Tinel, C.; Anglicheau, D.; Tharaux, P.L. MicroRNAs in Kidney Injury and Disease. Nat. Rev. Nephrol. 2022, 18, 643–662. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-W.; Lee, Y.H.; Tae, D.H.; Kim, Y.G.; Moon, J.-Y.; Jung, S.W.; Kim, J.S.; Hwang, H.S.; Jeong, K.-H.; Jeong, H.Y.; et al. Development and Validation of Urinary Exosomal MicroRNA Biomarkers for the Diagnosis of Acute Rejection in Kidney Transplant Recipients. Front. Immunol. 2023, 14, 1190576. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Suthanthiran, M.; Muthukumar, T. MicroRNAs and Transplantation. Clin. Lab. Med. 2019, 39, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Anglicheau, D.; Sharma, V.K.; Ding, R.; Hummel, A.; Snopkowski, C.; Dadhania, D.; Seshan, S.V.; Suthanthiran, M. MicroRNA Expression Profiles Predictive of Human Renal Allograft Status. Proc. Natl. Acad. Sci. USA 2009, 106, 5330–5335. [Google Scholar] [CrossRef] [PubMed]
- Danger, R.; Paul, C.; Giral, M.; Lavault, A.; Foucher, Y.; Degauque, N.; Pallier, A.; Durand, M.; Castagnet, S.; Duong Van Huyen, J.-P.; et al. Expression of MiR-142-5p in Peripheral Blood Mononuclear Cells from Renal Transplant Patients with Chronic Antibody-Mediated Rejection. PLoS ONE 2013, 8, e60702. [Google Scholar] [CrossRef] [PubMed]
- Boštjančič, E.; Večerić-Haler, Ž.; Kojc, N. The Role of Immune-Related MiRNAs in the Pathology of Kidney Transplantation. Biomolecules 2021, 11, 1198. [Google Scholar] [CrossRef]
- Dharnidharka, V.R.; Fiorina, P.; Harmon, W.E. Kidney Transplantation in Children. N. Engl. J. Med. 2014, 371, 549–558. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exosome-Based Liquid Biopsies in Cancer: Opportunities and Challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef]
- Keller, S.; Ridinger, J.; Rupp, A.-K.; Janssen, J.W.; Altevogt, P. Body Fluid Derived Exosomes as a Novel Template for Clinical Diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef]
- Chancharoenthana, W.; Traitanon, O.; Leelahavanichkul, A.; Tasanarong, A. Molecular Immune Monitoring in Kidney Transplant Rejection: A State-of-the-Art Review. Front. Immunol. 2023, 14, 1206929. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Wu, L.; Boer, K.; Woud, W.W.; Udomkarnjananun, S.; Hesselink, D.A.; Baan, C.C. Urinary Extracellular Vesicles Are a Novel Tool to Monitor Allograft Function in Kidney Transplantation: A Systematic Review. Int. J. Mol. Sci. 2021, 22, 10499. [Google Scholar] [CrossRef] [PubMed]
- Merchant, M.L.; Rood, I.M.; Deegens, J.K.J.; Klein, J.B. Isolation and Characterization of Urinary Extracellular Vesicles: Implications for Biomarker Discovery. Nat. Rev. Nephrol. 2017, 13, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Ergunay, T.; Collino, F.; Bianchi, G.; Sedrakyan, S.; Perin, L.; Bussolati, B. Extracellular Vesicles in Kidney Development and Pediatric Kidney Diseases. Pediatr. Nephrol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Nassar, A.; Cashman, K.; Rao, S.; Dagher, M.; O’Brien, C.; Afif, J.; Cravedi, P.; Azzi, J.R. Liquid Biopsy for Non-Invasive Monitoring of Patients with Kidney Transplants. Front. Transplant. 2023, 2, 1148725. [Google Scholar] [CrossRef]
- Chen, Y.; Han, X.; Sun, Y.; He, X.; Xue, D. A Circulating Exosomal MicroRNA Panel as a Novel Biomarker for Monitoring Post-transplant Renal Graft Function. J. Cell Mol. Med. 2020, 24, 12154–12163. [Google Scholar] [CrossRef]
- Han, Q.-F.; Li, W.-J.; Hu, K.-S.; Gao, J.; Zhai, W.-L.; Yang, J.-H.; Zhang, S.-J. Exosome Biogenesis: Machinery, Regulation, and Therapeutic Implications in Cancer. Mol. Cancer 2022, 21, 207. [Google Scholar] [CrossRef]
- Mizenko, R.R.; Brostoff, T.; Rojalin, T.; Koster, H.J.; Swindell, H.S.; Leiserowitz, G.S.; Wang, A.; Carney, R.P. Tetraspanins Are Unevenly Distributed across Single Extracellular Vesicles and Bias Sensitivity to Multiplexed Cancer Biomarkers. J. Nanobiotechnol. 2021, 19, 250. [Google Scholar] [CrossRef]
- Karimi, N.; Dalirfardouei, R.; Dias, T.; Lötvall, J.; Lässer, C. Tetraspanins Distinguish Separate Extracellular Vesicle Subpopulations in Human Serum and Plasma—Contributions of Platelet Extracellular Vesicles in Plasma Samples. J. Extracell. Vesicles 2022, 11, e12213. [Google Scholar] [CrossRef]
- Koch, R.J.; Barrette, A.M.; Stern, A.D.; Hu, B.; Bouhaddou, M.; Azeloglu, E.U.; Iyengar, R.; Birtwistle, M.R. Validating Antibodies for Quantitative Western Blot Measurements with Microwestern Array. Sci. Rep. 2018, 8, 11329. [Google Scholar] [CrossRef]
- Momenbeitollahi, N.; Cloet, T.; Li, H. Pushing the Detection Limits: Strategies towards Highly Sensitive Optical-Based Protein Detection. Anal. Bioanal. Chem. 2021, 413, 5995–6011. [Google Scholar] [CrossRef]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA Integrity Number for Assigning Integrity Values to RNA Measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef]
- Cuadrado-Payán, E.; Ramírez-Bajo, M.J.; Bañón-Maneus, E.; Rovira, J.; Diekmann, F.; Revuelta, I.; Cucchiari, D. Physiopathological Role of Extracellular Vesicles in Alloimmunity and Kidney Transplantation and Their Use as Biomarkers. Front. Immunol. 2023, 14, 1154650. [Google Scholar] [CrossRef]
- Soltaninejad, E.; Nicknam, M.H.; Nafar, M.; Ahmadpoor, P.; Pourrezagholi, F.; Sharbafi, M.H.; Hosseinzadeh, M.; Foroughi, F.; Yekaninejad, M.S.; Bahrami, T.; et al. Differential Expression of MicroRNAs in Renal Transplant Patients with Acute T-Cell Mediated Rejection. Transpl. Immunol. 2015, 33, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qian, W.; Quan, X.; Yang, H.; Zhao, G.; Wei, L. Differential MicroRNA Expressions in Human Peripheral Blood Mononuclear Cells Are Predictive of Renal Allograft Function. Transpl. Proc. 2019, 51, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Domenico, T.D.; Joelsons, G.; Montenegro, R.M.; Manfro, R.C. Upregulation of MicroRNA 142-3p in the Peripheral Blood and Urinary Cells of Kidney Transplant Recipients with Post-Transplant Graft Dysfunction. Braz. J. Med. Biol. Res. 2017, 50, e5533. [Google Scholar] [CrossRef]
- Aguado-Fraile, E.; Ramos, E.; Conde, E.; Rodríguez, M.; Martín-Gómez, L.; Lietor, A.; Candela, Á.; Ponte, B.; Liaño, F.; García-Bermejo, M.L. A Pilot Study Identifying a Set of MicroRNAs As Precise Diagnostic Biomarkers of Acute Kidney Injury. PLoS ONE 2015, 10, e0127175. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Hsu, C.-T.; Tsai, S.-F.; Chen, C.-H. Association between Circulating MicroRNAs (MiR-21-5p, MiR-20a-5p, MiR-29b-3p, MiR-126-3p and MiR-101-3p) and Chronic Allograft Dysfunction in Renal Transplant Recipients. Int. J. Mol. Sci. 2022, 23, 12253. [Google Scholar] [CrossRef]
- Ying, J.; Han, X.; Sang, X.; Cao, G. TGF-β/Smad and Wnt/Β-catenin Signaling Pathways Are Involved in Renal Fibrosis and Its Therapies. Clin. Transl. Med. 2020, 10, e127. [Google Scholar] [CrossRef]
- Yuan, Q.; Xu, T.; Chen, Y.; Qu, W.; Sun, D.; Liu, X.; Sun, L. MiR-185-5p Ameliorates Endoplasmic Reticulum Stress and Renal Fibrosis by Downregulation of ATF6. Lab. Investig. 2020, 100, 1436–1446. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.-M.; He, L.-J.; Wang, P.-B.; Yu, Y.; Ye, Y.-P.; Liang, L. Antagonist Targeting MiR-106b-5p Attenuates Acute Renal Injury by Regulating Renal Function, Apoptosis and Autophagy via the Upregulation of TCF4. Int. J. Mol. Med. 2021, 48, 169. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Yang, X.; Han, Z.; Lu, P.; Wang, J.; Liu, X.; Wu, B.; Wang, Z.; Huang, Z.; Lu, Q.; et al. Serum MicroRNA-99a Helps Detect Acute Rejection in Renal Transplantation. Transpl. Proc. 2015, 47, 1683–1687. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, D.; Leierer, J.; Shapter, O.; Mohammed, S.; Gingell-Littlejohn, M.; Kingsmore, D.B.; Little, A.-M.; Kerschbaum, J.; Schneeberger, S.; Maglione, M.; et al. Identification of Molecular Markers of Delayed Graft Function Based on the Regulation of Biological Ageing. PLoS ONE 2016, 11, e0146378. [Google Scholar] [CrossRef]
- Millán, O.; Budde, K.; Sommerer, C.; Aliart, I.; Rissling, O.; Bardaji, B.; Matz, M.; Zeier, M.; Silva, I.; Guirado, L.; et al. Urinary MiR-155-5p and CXCL10 as Prognostic and Predictive Biomarkers of Rejection, Graft Outcome and Treatment Response in Kidney Transplantation. Br. J. Clin. Pharmacol. 2017, 83, 2636–2650. [Google Scholar] [CrossRef]
- Quintairos, L.; Colom, H.; Millán, O.; Fortuna, V.; Espinosa, C.; Guirado, L.; Budde, K.; Sommerer, C.; Lizana, A.; López-Púa, Y.; et al. Early Prognostic Performance of MiR155-5p Monitoring for the Risk of Rejection: Logistic Regression with a Population Pharmacokinetic Approach in Adult Kidney Transplant Patients. PLoS ONE 2021, 16, e0245880. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, L.; Ge, W.; Hui, Y.; Zhou, Z.; Hu, L.; Pan, H.; Huang, Y.; Shen, B. Multi-Omics Network Characterization Reveals Novel MicroRNA Biomarkers and Mechanisms for Diagnosis and Subtyping of Kidney Transplant Rejection. J. Transl. Med. 2021, 19, 346. [Google Scholar] [CrossRef]
- Roufosse, C.; Simmonds, N.; Clahsen-van Groningen, M.; Haas, M.; Henriksen, K.J.; Horsfield, C.; Loupy, A.; Mengel, M.; Perkowska-Ptasińska, A.; Rabant, M.; et al. A 2018 Reference Guide to the Banff Classification of Renal Allograft Pathology. Transplantation 2018, 102, 1795–1814. [Google Scholar] [CrossRef]
- Collino, F.; Lopes, J.A.; Corrêa, S.C.; Abdelhay, E.S.; Takiya, C.M.; Wendt, C.H.; Miranda, K.R.; Vieyra, A.; Lindoso, R.S. Adipose-Derived Mesenchymal Stromal Cells Under Hypoxia: Changes in Extracellular Vesicles Secretion and Improvement of Renal Recovery after Ischemic Injury. Cell. Physiol. Biochem. 2019, 52, 1463–1483. [Google Scholar] [CrossRef] [PubMed]

| miRNA | p-Value |
|---|---|
| hsa-miR-101-3p | 0.0429 |
| hsa-miR-185-5p | 0.0428 |
| hsa-miR-106b-3p | 0.0315 |
| hsa-miR-142-3p | 0.0125 |
| hsa-miR-142-5p | 0.0059 |
| miRNA | logFC | p-Value |
|---|---|---|
| hsa-miR-184-3p | −1.641 | 0.000 |
| hsa-miR-99a-5p | 1.112 | 0.000 |
| hsa-miR-93-5p | −0.709 | 0.000 |
| hsa-let-7f-5p | −0.719 | 0.001 |
| hsa-miR-155-5p | 1.325 | 0.001 |
| hsa-miR-514a-3p | 1.443 | 0.001 |
| hsa-miR-532-5p | −0.688 | 0.002 |
| hsa-miR-125b-2-3p | 1.057 | 0.002 |
| hsa-miR-1307-5p | 0.612 | 0.003 |
| hsa-miR-30b-5p | 0.700 | 0.004 |
| hsa-miR-22-3p | 0.859 | 0.006 |
| hsa-let-7i-5p | −0.483 | 0.007 |
| hsa-miR-509-3p | 1.165 | 0.007 |
| hsa-miR-187-3p | −0.914 | 0.008 |
| hsa-miR-423-3p | 0.789 | 0.008 |
| hsa-miR-132-3p | 0.768 | 0.008 |
| hsa-miR-181a-5p | 0.886 | 0.009 |
| hsa-miR-222-3p | 0.792 | 0.010 |
| hsa-let-7a-5p | −0.558 | 0.010 |
| hsa-miR-664a-3p | 0.944 | 0.011 |
| hsa-miR-96-5p | 0.572 | 0.011 |
| hsa-miR-181b-5p | 0.915 | 0.012 |
| hsa-miR-103a-3p | −0.374 | 0.015 |
| hsa-miR-92b-3p | 0.894 | 0.016 |
| hsa-miR-542-3p | −0.819 | 0.017 |
| hsa-miR-30c-5p | 0.487 | 0.018 |
| hsa-miR-671-5p | 0.628 | 0.022 |
| hsa-miR-135b-5p | 0.759 | 0.023 |
| hsa-miR-206-3p | −0.663 | 0.023 |
| hsa-miR-99b-3p | −0.742 | 0.024 |
| hsa-miR-197-3p | 0.625 | 0.025 |
| hsa-let-7d-3p | −0.445 | 0.025 |
| hsa-miR-23a-3p | 0.402 | 0.025 |
| hsa-miR-381-3p | 1.103 | 0.026 |
| hsa-miR-92a-3p | 0.560 | 0.027 |
| hsa-let-7e-5p | −0.533 | 0.032 |
| hsa-miR-365a-3p | 0.799 | 0.032 |
| hsa-miR-365b-3p | 0.799 | 0.032 |
| hsa-miR-28-5p | 0.883 | 0.032 |
| hsa-miR-1180-3p | 0.678 | 0.034 |
| hsa-miR-374b-5p | 0.735 | 0.035 |
| hsa-miR-221-3p | 0.629 | 0.037 |
| hsa-miR-34a-5p | −0.671 | 0.038 |
| hsa-miR-95-3p | −0.734 | 0.041 |
| hsa-miR-425-5p | 0.606 | 0.043 |
| hsa-miR-454-3p | −0.462 | 0.045 |
| hsa-miR-17-5p | 0.486 | 0.049 |
| hsa-miR-25-3p | −0.450 | 0.049 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carraro, A.; De Gaspari, P.; Antoniello, B.; Marzenta, D.; Vianello, E.; Bussolati, B.; Tritta, S.; Collino, F.; Bertoldi, L.; Benvenuto, G.; et al. New Insights into Pediatric Kidney Transplant Rejection Biomarkers: Tissue, Plasma and Urine MicroRNAs Compared to Protocol Biopsy Histology. Int. J. Mol. Sci. 2024, 25, 1911. https://doi.org/10.3390/ijms25031911
Carraro A, De Gaspari P, Antoniello B, Marzenta D, Vianello E, Bussolati B, Tritta S, Collino F, Bertoldi L, Benvenuto G, et al. New Insights into Pediatric Kidney Transplant Rejection Biomarkers: Tissue, Plasma and Urine MicroRNAs Compared to Protocol Biopsy Histology. International Journal of Molecular Sciences. 2024; 25(3):1911. https://doi.org/10.3390/ijms25031911
Chicago/Turabian StyleCarraro, Andrea, Piera De Gaspari, Benedetta Antoniello, Diana Marzenta, Emanuele Vianello, Benedetta Bussolati, Stefania Tritta, Federica Collino, Loris Bertoldi, Giuseppe Benvenuto, and et al. 2024. "New Insights into Pediatric Kidney Transplant Rejection Biomarkers: Tissue, Plasma and Urine MicroRNAs Compared to Protocol Biopsy Histology" International Journal of Molecular Sciences 25, no. 3: 1911. https://doi.org/10.3390/ijms25031911
APA StyleCarraro, A., De Gaspari, P., Antoniello, B., Marzenta, D., Vianello, E., Bussolati, B., Tritta, S., Collino, F., Bertoldi, L., Benvenuto, G., Vedovelli, L., Benetti, E., & Negrisolo, S. (2024). New Insights into Pediatric Kidney Transplant Rejection Biomarkers: Tissue, Plasma and Urine MicroRNAs Compared to Protocol Biopsy Histology. International Journal of Molecular Sciences, 25(3), 1911. https://doi.org/10.3390/ijms25031911








