Unraveling the Molecular Basis of Color Variation in Dioscorea alata Tubers: Integrated Transcriptome and Metabolomics Analysis
Abstract
1. Introduction
2. Results
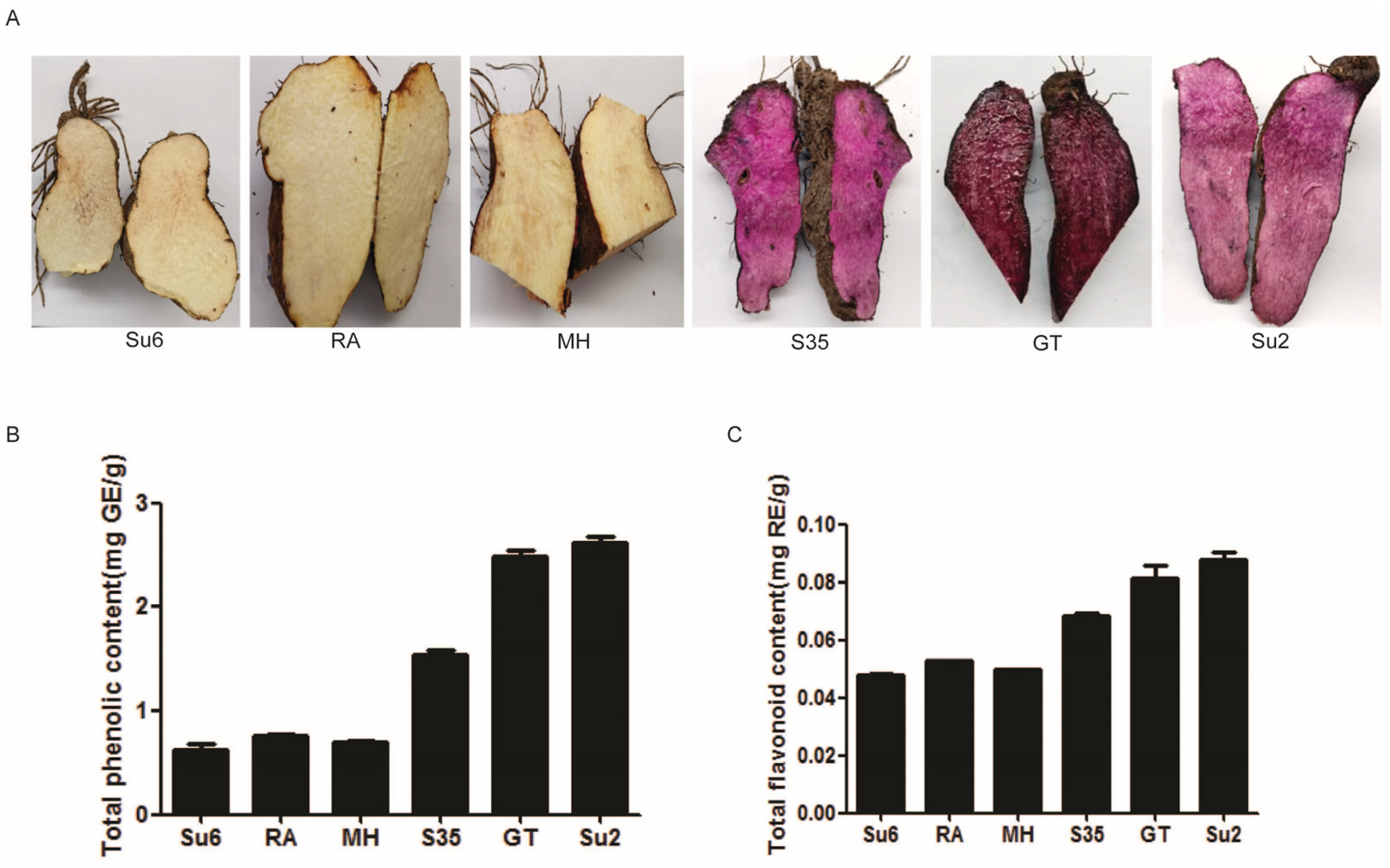
2.1. Phenolic Compounds and Antioxidant Activities of White and Purple Tubers of D. alata
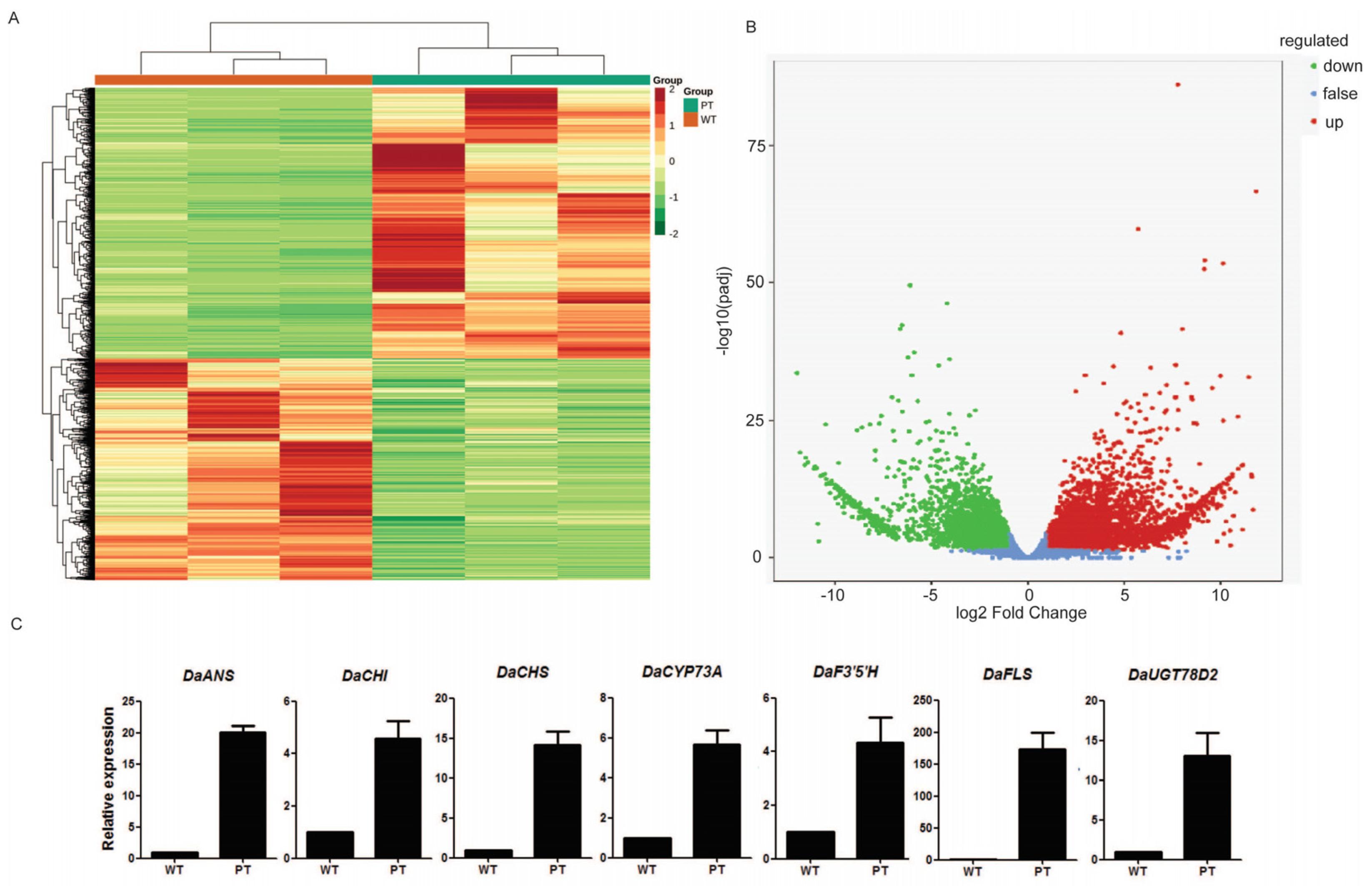
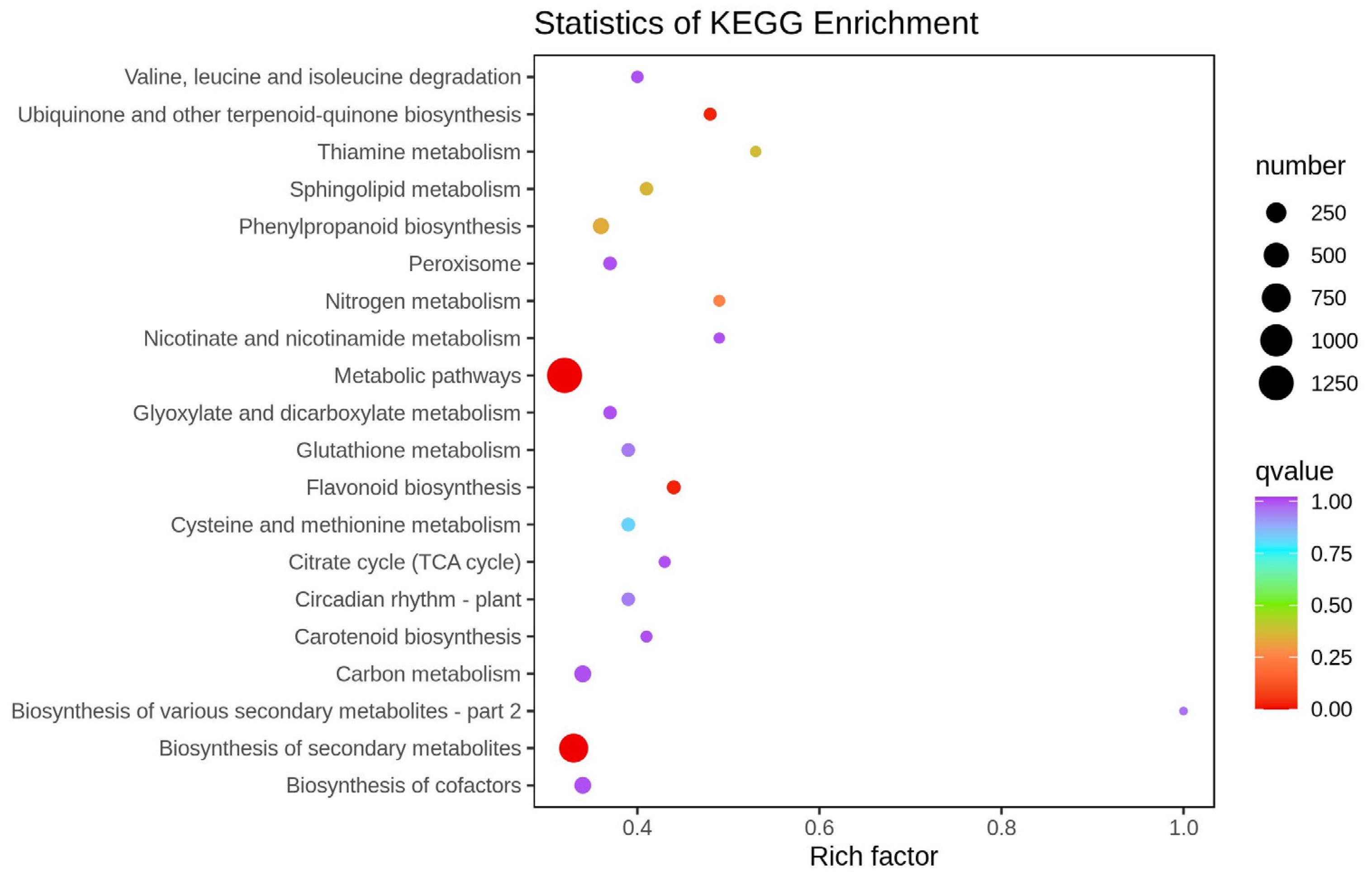
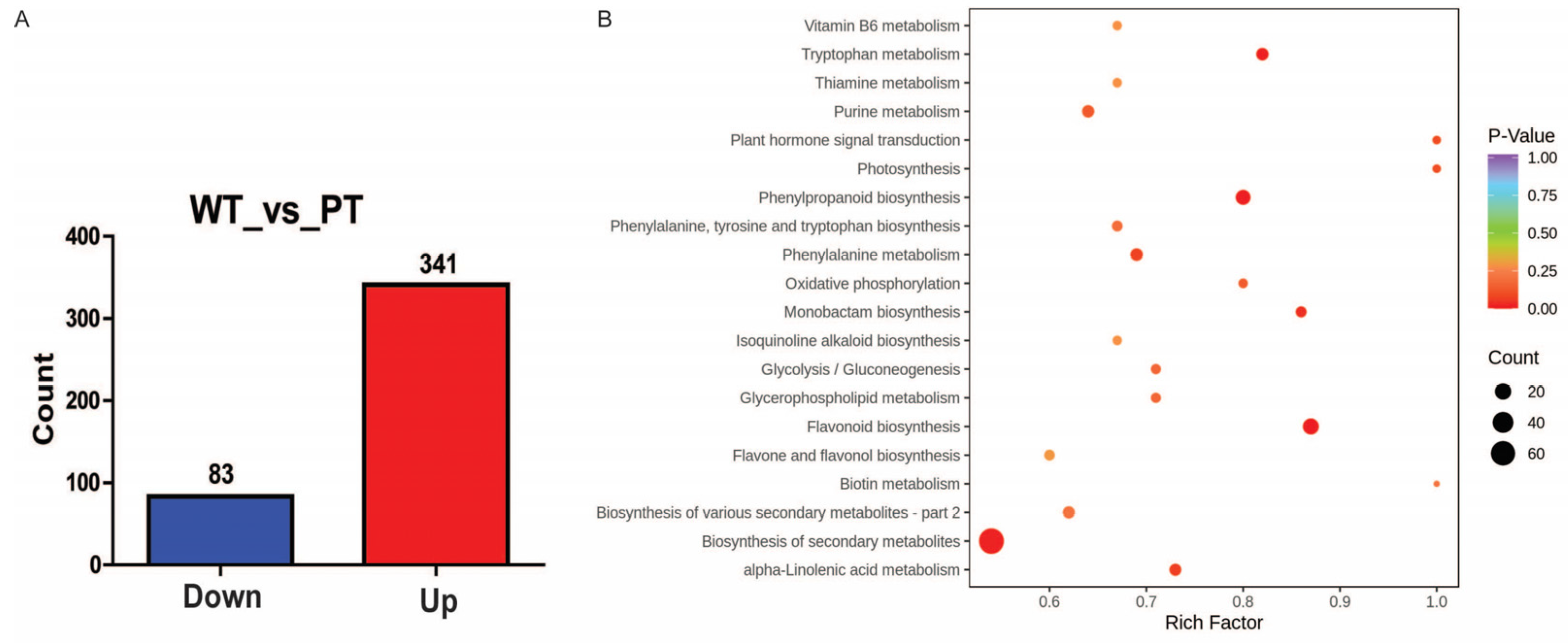
2.2. Transcriptome Analysis of White and Purple Tubers from Two Different D. alata Cultivars
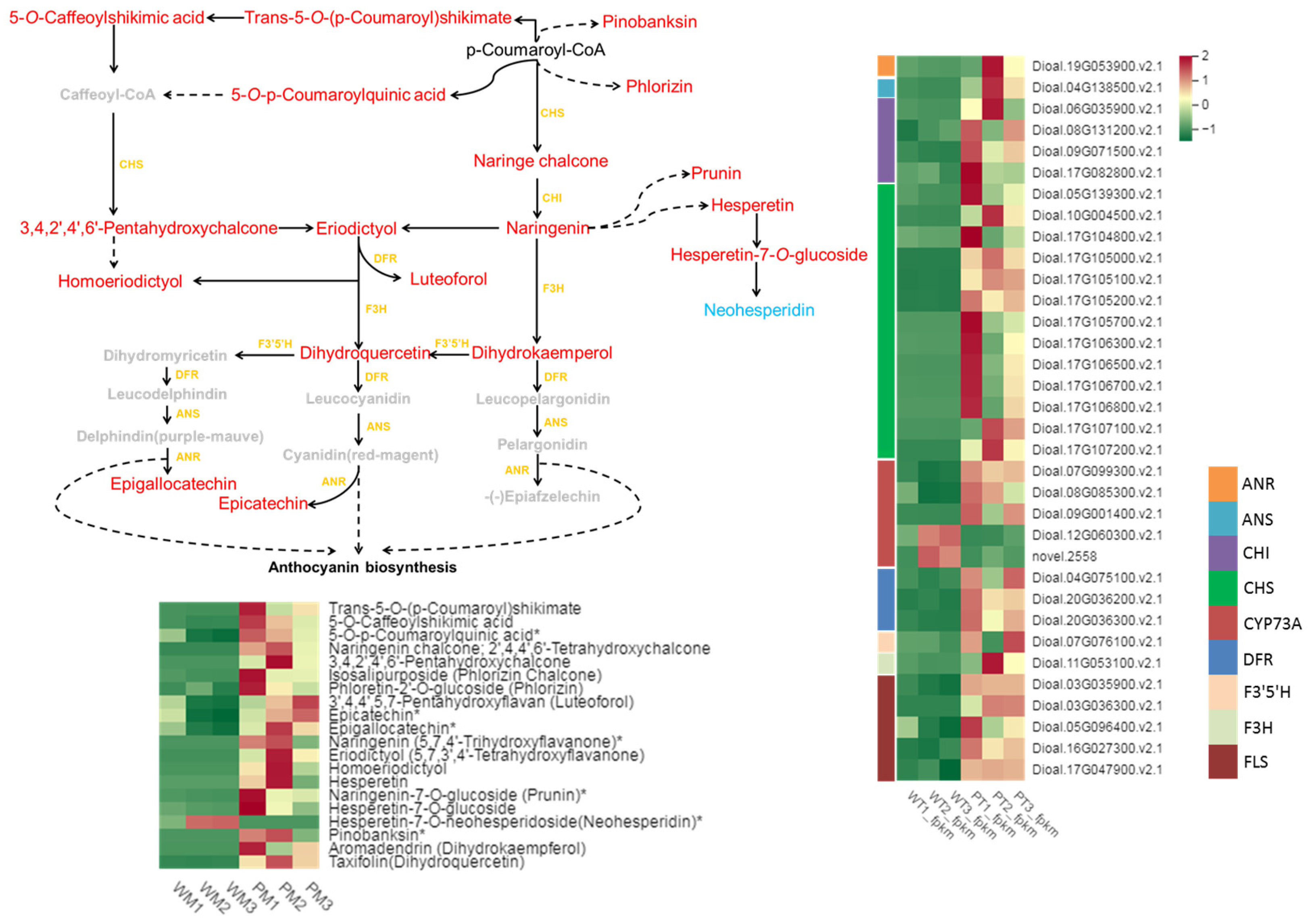
2.3. DEGs Related to Flavonoid Biosynthesis
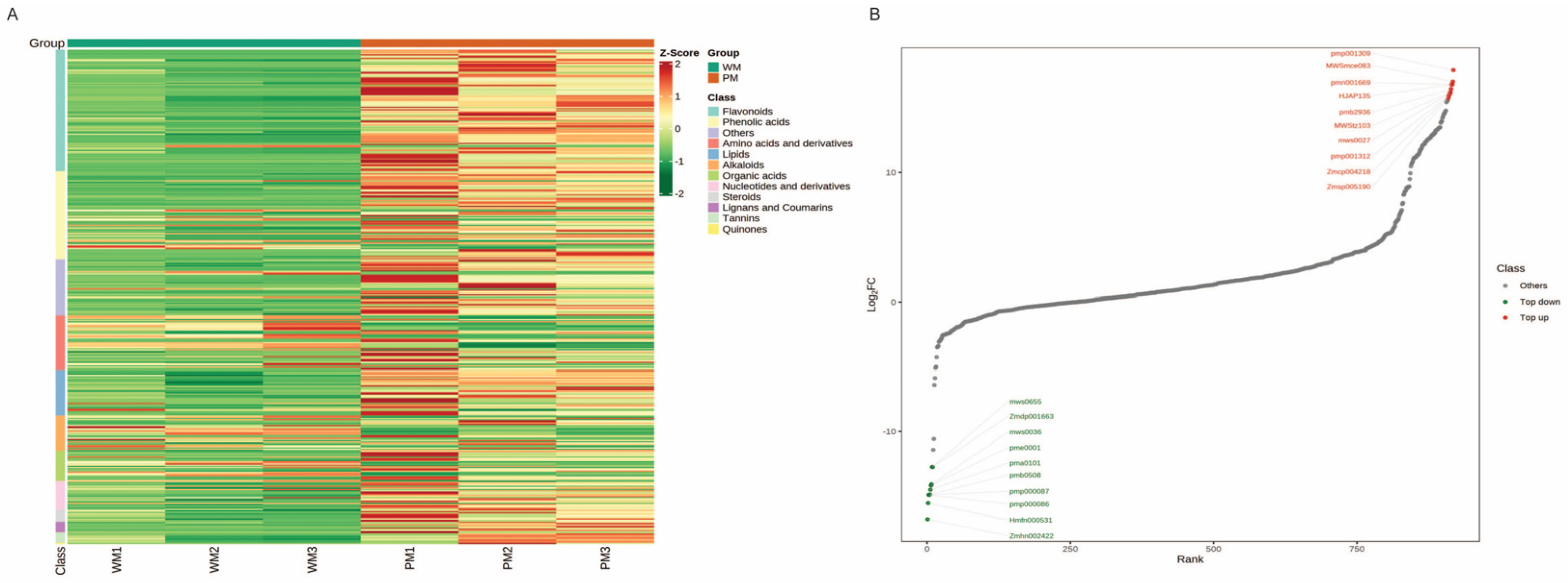
2.4. Metabolomics Analysis between the WT and PT
2.5. Flavonoid and Tannin Metabolites in WT and PT
2.6. Correlation Analysis between RNA-Seq and Metabolites Uncovers the Regulatory Pathway of Flavonoid Biosynthesis
3. Discussion
3.1. D. alata Tubers Exhibiting Different Colors Possess Different Polyphenol Contents and Antioxidant Activities
3.2. Analysis of DEGs and DEMs Refers to Flavonoid Pathway in WT and PT of D. alata
4. Materials and Methods
4.1. Plant Materials
4.2. Total Polyphenol Extraction and Contents and Antioxidant Activity Analysis
4.3. RNA Extraction, Library Preparation, and Sequencing
4.4. Transcriptome Data Analysis
4.5. Real-Time Quantitative Reverse-Transcription PCR Analysis
4.6. UPLC–MS/MS Conditions
4.7. Differential Metabolite Analysis
4.8. Combined Transcriptome and Metabolome Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, Z.G.; Jiang, W.; Mantri, N.; Bao, X.Q.; Chen, S.L.; Tao, Z.M. Transciptome analysis reveals flavonoid biosynthesis regulation and simple sequence repeats in yam (Dioscorea alata L.) tubers. BMC Genom. 2015, 16, 346. [Google Scholar] [CrossRef] [PubMed]
- Sharif, B.M.; Burgarella, C.; Cormier, F.; Mournet, P.; Causse, S.; Van, K.N.; Kaoh, J.; Rajaonah, M.T.; Lakshan, S.R.; Waki, J. Genome-wide genotyping elucidates the geographical diversification and dispersal of the polyploid and clonally propagated yam (Dioscorea alata). Ann. Bot. 2020, 126, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Srivichai, S.; Hongsprabhas, P. Profiling anthocyanins in Thai purple yams (Dioscorea alata L.). Int. J. Food Sci. 2020, 2020, 1594291. [Google Scholar] [CrossRef] [PubMed]
- Wanasundera, J.P.; Ravindran, G. Nutritional assessment of yam (Dioscorea alata) tubers. Plant Foods Hum. Nutr. 1994, 46, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Champagne, A.; Hilbert, G.; Legendre, L.; Lebot, V. Diversity of anthocyanins and other phenolic compounds among tropical root crops from Vanuatu, South Pacific. J. Food Compos. Anal. 2011, 24, 315–325. [Google Scholar] [CrossRef]
- Wu, Z.G.; Jiang, W.; Nitin, M.; Bao, X.Q.; Chen, S.L.; Tao, Z.M. Characterizing diversity based on nutritional and bioactive compositions of yam germplasm (Dioscorea spp.) commonly cultivated in China. J. Food Drug Anal. 2016, 24, 367–375. [Google Scholar] [CrossRef]
- Effah-Manu, L.; Wireko-Manu, F.D.; Agbenorhevi, J.K.; Maziya-Dixon, B.; Oduro, I.N.; Baah-Ennum, T.Y. The effect of gender on end-user preferences for yam quality descriptors. Food Sci. Nutr. 2022, 10, 3890–3904. [Google Scholar] [CrossRef]
- Dossa, K.; Morel, A.; Houngbo, M.E.; Mota, A.Z.; Malédon, E.; Irep, J.L.; Diman, J.L.; Mournet, P.; Causse, S.; Van, K.N. Genome-wide association studies reveal novel loci controlling tuber flesh color and oxidative browning in Dioscorea alata. J. Sci. Food Agric. 2023, in press. [Google Scholar] [CrossRef]
- Yin, C.; Tang, D.; Liu, X.; Li, Z.; Xiang, Y.; Gao, K.; Li, H.; Yuan, L.; Huang, B.; Li, J. Transcriptome analysis reveals important regulatory genes and pathways for tuber color variation in Pinellia ternata (Thunb.) Breit. Protoplasma 2023, 260, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [PubMed]
- Lo Piero, A.R. The state of the art in biosynthesis of anthocyanins and its regulation in pigmented Sweet Oranges [(Citrus sinensis) L. Osbeck]. J. Agric. Food Chem. 2015, 63, 4031–4041. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Yu, Q.; Lin, C.; Liu, H.; Dong, L.; Feng, X.; Liao, J. Analyzing Morphology, Metabolomics, and Transcriptomics Offers Invaluable Insights into the Mechanisms of Pigment Accumulation in the Diverse-Colored Labellum Tissues of Alpinia. Plants 2023, 12, 3766. [Google Scholar] [CrossRef]
- Gupta, M.; Ahmad, J.; Ahamad, J.; Kundu, S.; Goel, A.; Mishra, A. Flavonoids as promising anticancer therapeutics: Contemporary research, nanoantioxidant potential, and future scope. Phytother. Res. 2023, 37, 5159–5192. [Google Scholar] [CrossRef] [PubMed]
- Paulsmeyer, M.N.; Juvik, J.A. R3-MYB repressor Mybr97 is a candidate gene associated with the Anthocyanin3 locus and enhanced anthocyanin accumulation in maize. Theor. Appl. Genet. 2023, 136, 55. [Google Scholar] [CrossRef]
- Tsuda, T. Dietary anthocyanin-rich plants: Biochemical basis and recent progress in health benefits studies. Mol. Nutr. Food Res. 2012, 56, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Nakayama, T. Achievements and perspectives in biochemistry concerning anthocyanin modification for blue flower coloration. Plant Cell Physiol. 2015, 56, 28–40. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Czemmel, S.; Heppel, S.C.; Bogs, J. R2R3 MYB transcription factors: Key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 2012, 249 (Suppl. 2), S109–S118. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Yamagishi, M.; Toda, S.; Tasaki, K. The novel allele of the LhMYB12 gene is involved in splatter-type spot formation on the flower tepals of Asiatic hybrid lilies (Lilium spp.). New Phytol. 2014, 201, 1009–1020. [Google Scholar] [CrossRef]
- Broucke, E.; Dang, T.T.V. SnRK1 inhibits anthocyanin biosynthesis through both transcriptional regulation and direct phosphorylation and dissociation of the MYB/bHLH/TTG1 MBW complex. Plant J. 2023, 115, 1193–1213. [Google Scholar] [CrossRef]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Wu, M.; Si, M.; Li, X.; Song, L.; Liu, J.; Zhai, R.; Cong, L.; Yue, R.; Yang, C.; Ma, F. PbCOP1.1 Contributes to the Negative Regulation of Anthocyanin Biosynthesis in Pear. Plants 2019, 8, 39. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lui, A.C.W.; Lam, P.-Y.; Liu, G.; Godwin, I.-D.; Clive, L. Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum. Plant Biotechnol. J. 2020, 18, 2170–2172. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, X.; Cui, L.; Li, Y.; Liang, Y.; Wang, S.; Chen, Y.; Zhou, L.; Zhang, Y.; Li, F. Transcriptome and metabolome analyses reveal anthocyanins pathways associated with fruit color changes in plum (Prunus salicina Lindl.). PeerJ 2022, 10, e14413. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Pan, J.; Yin, Z.G.; Feng, T.; Zhao, J.; Dong, X.; Zhou, Y. Integrated transcriptome and metabolome analyses revealed regulatory mechanisms of flavonoid biosynthesis in Radix Ardisia. PeerJ 2022, 10, e13670. [Google Scholar] [CrossRef] [PubMed]
- Mabhaudhi, T.; Chimonyo, V.G.P.; Hlahla, S.; Massawe, F.; Mayes, S.; Nhamo, L.; Modi, A.T. Prospects of orphan crops in climate change. Planta 2019, 250, 695–708. [Google Scholar] [CrossRef]
- Hunter, D.; Borelli, T.; Beltrame, D.M.O.; Oliveira, C.N.S.; Coradin, L.; Wasike, V.W.; Wasilwa, L.; Mwai, J.; Manjella, A.; Samarasinghe, G.W.L. The potential of neglected and underutilized species for improving diets and nutrition. Planta 2019, 250, 709–729. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, D.H.; Yang, J.H.; Lee, J.Y.; Lim, S.H. Increased flavonol levels in tobacco expressing AcFLS affect flower color and root growth. Int. J. Mol. Sci. 2020, 21, 1011. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.H.; Tsao, N.W.; Wang, S.Y.; Chu, F.H. Color variation in young and senescent leaves of Formosan sweet gum (Liquidambar formosana) by the gene regulation of anthocyanidin biosynthesis. Physiol. Plant 2021, 172, 1750–1763. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Shi, X.; Ren, X.; Qin, Z. Chemical composition and antioxidant activity of phenolic compounds from Dioscorea (Yam) leaves. Pak. J. Pharm. Sci. 2018, 31, 1031–1038. [Google Scholar]
- Wang, Y.; Meng, G.; Chen, S.; Chen, Y.; Jiang, J.; Wang, Y.P. Correlation analysis of phenolic contents and antioxidation in yellow- and black-seeded Brassica napus. Molecules 2018, 23, 1815. [Google Scholar] [CrossRef] [PubMed]
- Moriya, C.; Hosoya, T.; Agawa, S.; Sugiyama, Y.; Kozone, I.; Shin-Ya, K.; Terahara, N.; Kumazawa, S. New acylated anthocyanins from purple yam and their antioxidant activity. Biosci. Biotechnol. Biochem. 2015, 79, 1484–1492. [Google Scholar] [CrossRef] [PubMed]
- LaFountain, A.M.; Yuan, Y.W. Repressors of anthocyanin biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Shoyama, Y.; Nishioka, I.; Herath, W.; Uemoto, S.; Fujieda, K.; Okubo, H. Two acylated anthocyanins from Dioscorea alata. Phytochemistry 1990, 29, 2999–3001. [Google Scholar] [CrossRef]
- Barreca, D.; Gattuso, G.; Bellocco, E.; Calderaro, A.; Trombetta, D.; Smeriglio, A.; Laganà, G.; Daglia, M.; Meneghini, S.; Nabavi, S.M. Flavanones: Citrus phytochemical with health-promoting properties. Biofactors 2017, 43, 495–506. [Google Scholar] [CrossRef]
- Katsu, K.; Suzuki, R.; Tsuchiya, W.; Inagaki, N.; Yamazaki, T.; Hisano, T.; Yasui, Y.; Komori, T.; Koshio, M.; Kubota, S. A new buckwheat dihydroflavonol 4-reductase (DFR), with a unique substrate binding structure, has altered substrate specificity. BMC Plant Biol. 2017, 17, 239. [Google Scholar] [CrossRef]
- Jiang, J.; Shao, Y.; Li, A.; Lu, C.; Zhang, Y.; Wang, Y. Phenolic composition analysis and gene expression in developing seeds of yellow- and black-seeded Brassica napus. J. Integr. Plant Biol. 2013, 55, 537–551. [Google Scholar] [CrossRef]
- Faudale, M.; Viladomat, F.; Bastida, J.; Poli, F.; Codina, C. Antioxidant activity and phenolic composition of wild, edible, and medicinal fennel from different Mediterranean countries. J. Agric. Food Chem. 2008, 56, 1912–1920. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C.L.W.T. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Bredeson, J.V.; Lyons, J.B.; Oniyinde, I.O.; Okereke, N.R.; Kolade, O.; Nnabue, I.; Nwadili, C.O.; Hřibová, E.; Parker, M.; Nwogha, J. Chromosome evolution and the genetic basis of agronomically important traits in greater yam. Nat. Commun. 2022, 13, 2001. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R pipeline for comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, S.; Xie, P.; Wang, L.; Xue, J.Y.; Zhang, Y.; Lu, R.; Hang, Y.; Wang, Y.; Sun, X. Fitness benefits play a vital role in the retention of the Pi-ta susceptible alleles. Genetics 2022, 220, iyac019. [Google Scholar] [CrossRef]

| Sample | DPPH | ABTS |
|---|---|---|
| No. | (μmol Trolox/g DW ± SE) | (μmol Trolox/g DW ± SE) |
| Su6 | 5.60 ± 0.105 | 3.69 ±0.092 |
| RA | 8.75 ± 0.071 | 3.95 ± 0.108 |
| MH | 6.46 ± 0.007 | 3.53 ± 0.013 |
| S35 | 16.84 ± 0.353 | 4.01 ± 0.097 |
| GT | 25.85 ± 2.222 | 6.45 ± 0.156 |
| Su2 | 23.06 ± 0.662 | 6.58 ± 0.393 |
| Flavonoid | ABTS | DPPH | ||||
|---|---|---|---|---|---|---|
| WT | PT | WT | PT | WT | PT | |
| Phenolics | 0.885 * | 0.951 ** | 0.887 * | 0.945 ** | 0.782 | 0.980 ** |
| Flavonoids | - | - | 0.656 | 0.835 ** | 0.931 ** | 0.917 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Lu, R.-S.; Li, M.-H.; Lu, X.-Y.; Sun, X.-Q.; Zhang, Y.-M. Unraveling the Molecular Basis of Color Variation in Dioscorea alata Tubers: Integrated Transcriptome and Metabolomics Analysis. Int. J. Mol. Sci. 2024, 25, 2057. https://doi.org/10.3390/ijms25042057
Wang Y, Lu R-S, Li M-H, Lu X-Y, Sun X-Q, Zhang Y-M. Unraveling the Molecular Basis of Color Variation in Dioscorea alata Tubers: Integrated Transcriptome and Metabolomics Analysis. International Journal of Molecular Sciences. 2024; 25(4):2057. https://doi.org/10.3390/ijms25042057
Chicago/Turabian StyleWang, Yue, Rui-Sen Lu, Ming-Han Li, Xin-Yu Lu, Xiao-Qin Sun, and Yan-Mei Zhang. 2024. "Unraveling the Molecular Basis of Color Variation in Dioscorea alata Tubers: Integrated Transcriptome and Metabolomics Analysis" International Journal of Molecular Sciences 25, no. 4: 2057. https://doi.org/10.3390/ijms25042057
APA StyleWang, Y., Lu, R.-S., Li, M.-H., Lu, X.-Y., Sun, X.-Q., & Zhang, Y.-M. (2024). Unraveling the Molecular Basis of Color Variation in Dioscorea alata Tubers: Integrated Transcriptome and Metabolomics Analysis. International Journal of Molecular Sciences, 25(4), 2057. https://doi.org/10.3390/ijms25042057






