Immunoassays: Analytical and Clinical Performance, Challenges, and Perspectives of SERS Detection in Comparison with Fluorescent Spectroscopic Detection
Abstract
1. Introduction
1.1. Fluorescence-Based Immunoassays Detection
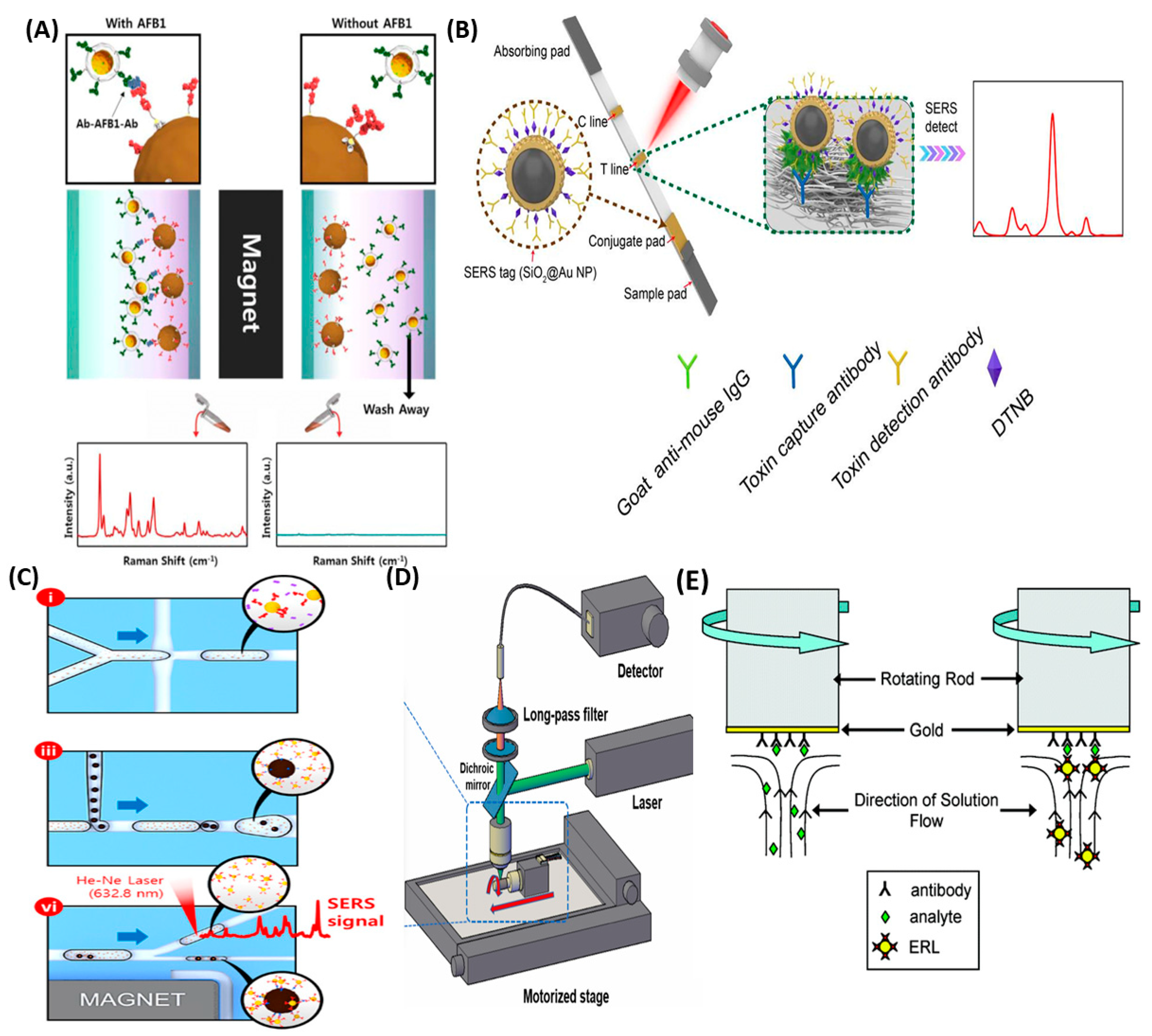
1.2. SERS-Based Immunoassays Detection
2. Analytical Reports on Immunoassays with the SERS and the Fluorescence Based Detection
2.1. Analytical Papers on the Fluorescent Immunoassays
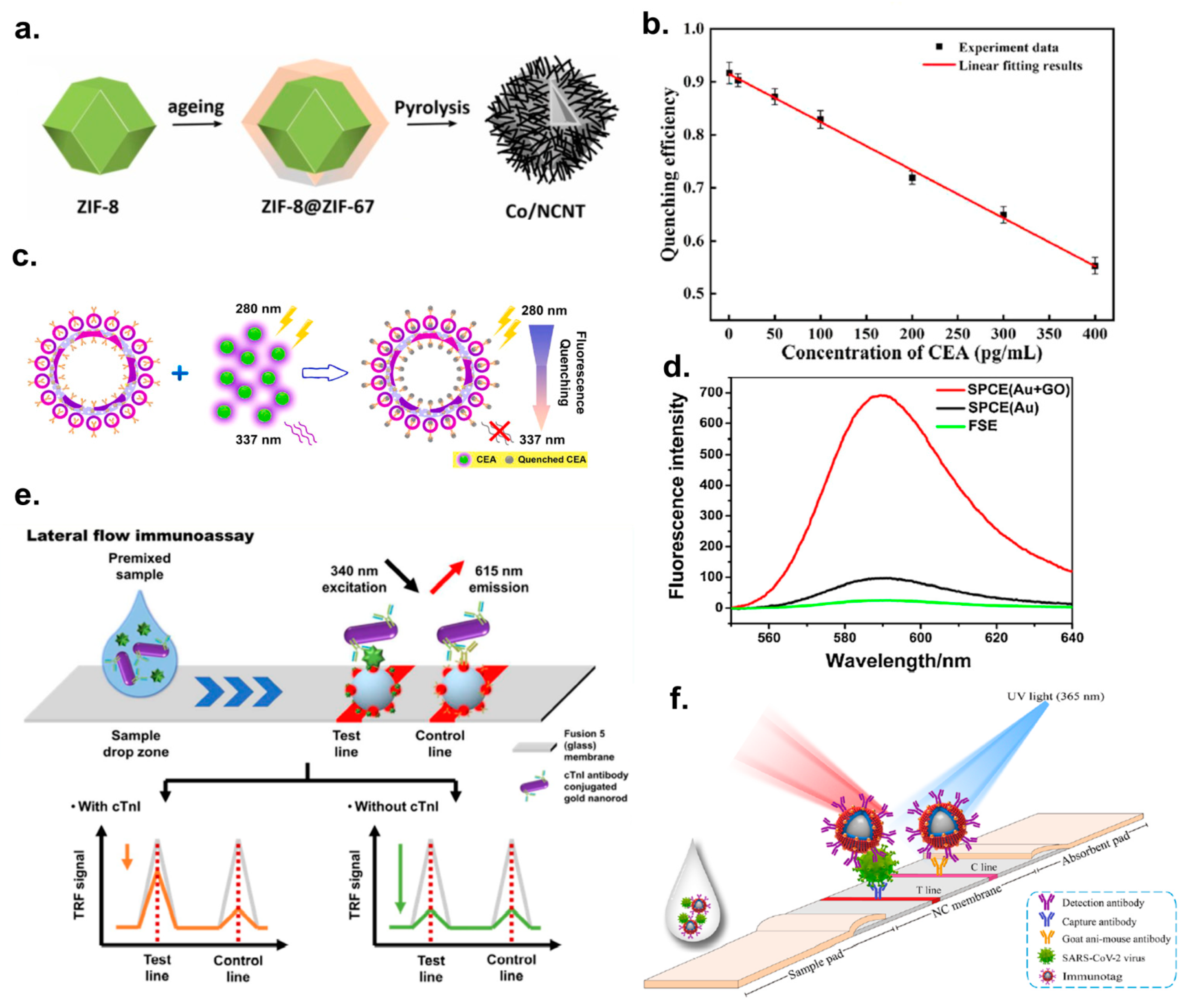
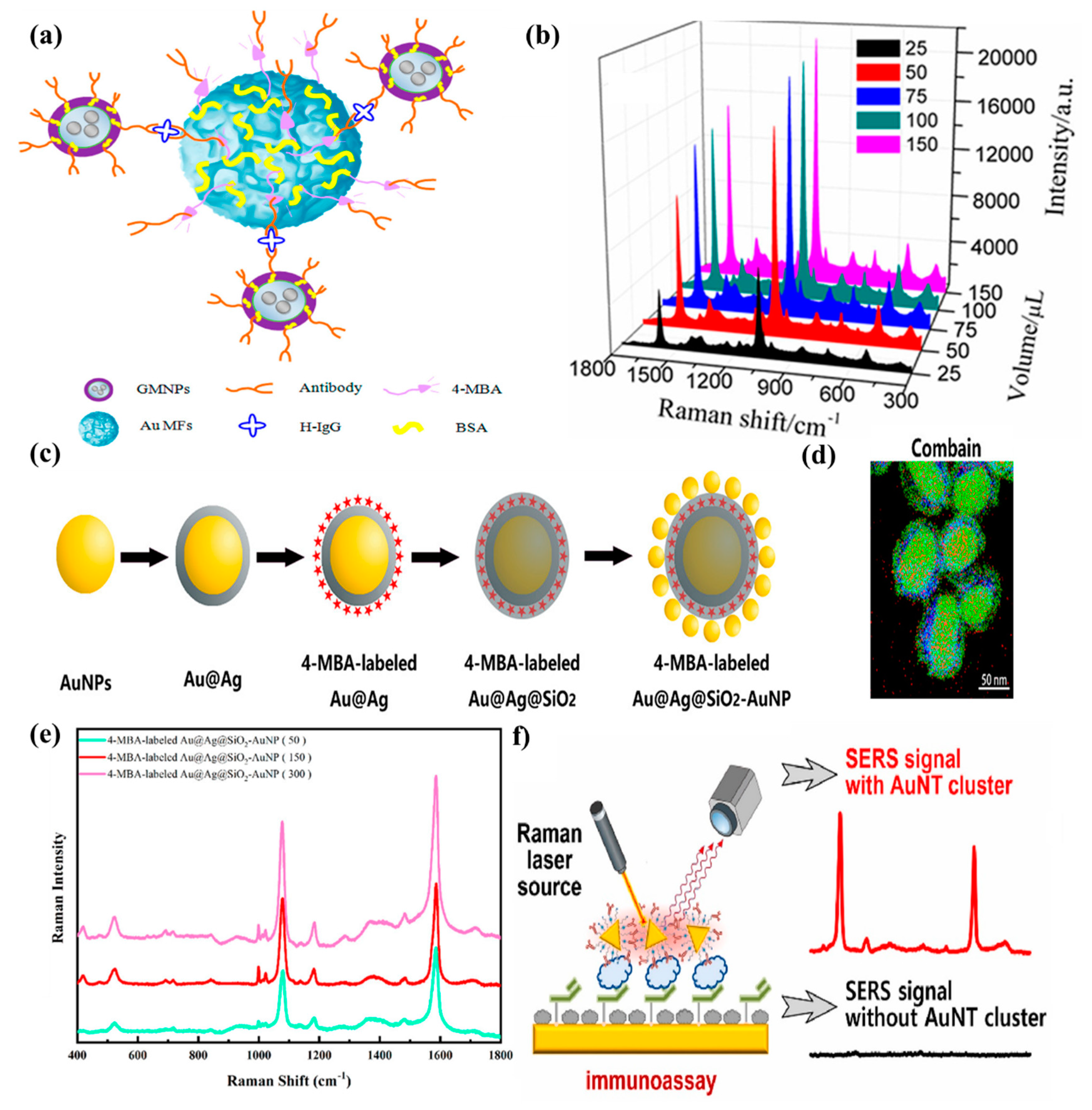
2.2. Analytical Papers on the SERS Immunoassays
2.3. Clinical Papers on the SERS- and Fluorescence-Based IA Detection
2.4. SERS vs. Fluorescence Immunoassays for Detection of Biomarkers
2.5. RSD Discussion
3. Challenges and Solutions for SERS and Fluorescent Detection
3.1. Common Challenges and Solutions for Both SERS and Fluorescence-Based Detection
3.2. Challenges and Solutions for Fluorescence Based Detection
3.3. Challenges and Solutions for SERS IA Detection
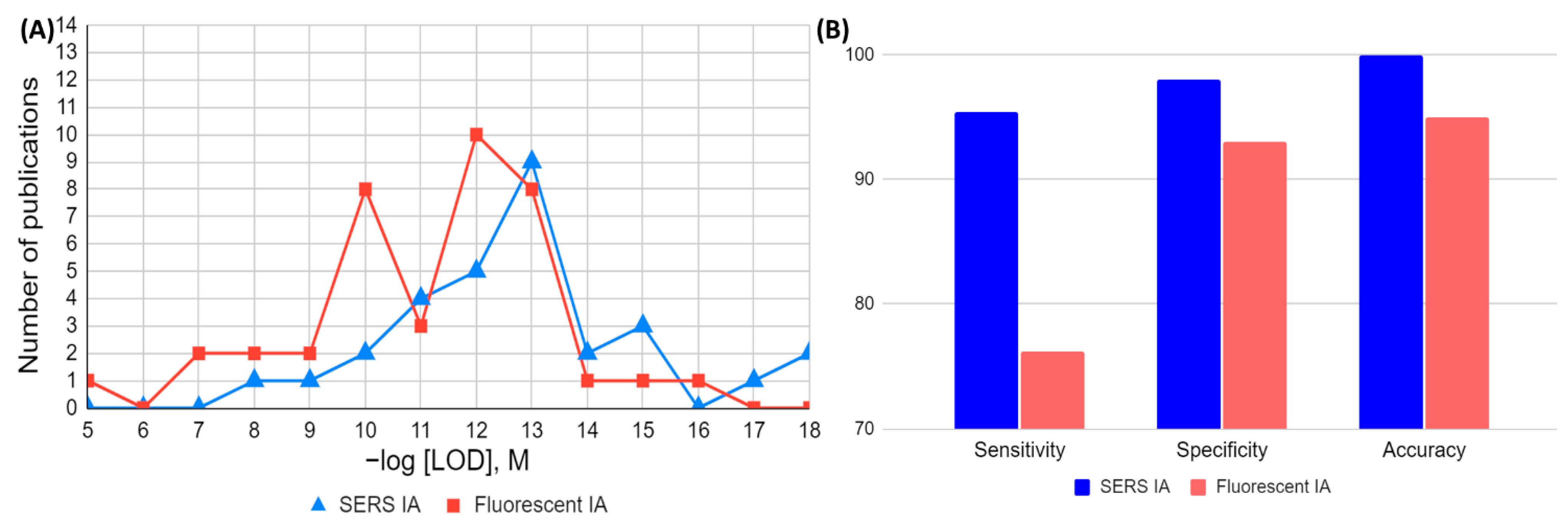
4. Graphical Comparison of FOMs in Analytical and Clinical Assays (SERS vs. Fluorescence)
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clarke, W.; Sokoll, L.J.; Rai, A.J. Chapter 12—Immunoassays. In Contemporary Practice in Clinical Chemistry, 4th ed.; Clarke, W., Marzinke, M.A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 201–214. [Google Scholar] [CrossRef]
- Bonwick, G.A.; Smith, C.J. Immunoassays: Their history, development and current place in food science and technology. Int. J. Food Sci. Technol. 2004, 39, 817–827. [Google Scholar] [CrossRef]
- Yalow, R.S.; Berson, S.A. Immunoassay of endogenous plasma insulin in man. J. Clin. Investig. 1960, 39, 1157–1175. [Google Scholar] [CrossRef] [PubMed]
- Hage, D.S. Immunoassays. Anal. Chem. 1999, 71, 294–304. [Google Scholar] [CrossRef]
- Itagaki, H. Chapter 3—Fluorescence Spectroscopy. In Experimental Methods in Polymer Science; Tanaka, T., Ed.; Academic Press: Boston, MA, USA, 2000; pp. 155–260. [Google Scholar] [CrossRef]
- Bose, A.; Thomas, I.; Abraham, E. Fluorescence spectroscopy and its applications: A Review. Int. J. Adv. Pharm. Res 2018, 8, 1–8. [Google Scholar]
- Zacharioudaki, D.-E.; Fitilis, I.; Kotti, M. Review of Fluorescence Spectroscopy in Environmental Quality Applications. Molecules 2022, 27, 4801. [Google Scholar] [CrossRef]
- Gan, S.D.; Patel, K.R. Enzyme Immunoassay and Enzyme-Linked Immunosorbent Assay. J. Investig. Dermatol. 2013, 133, e12. [Google Scholar] [CrossRef]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Van Weemen, B.K.; Schuurs, A. Immunoassay using antigen—Enzyme conjugates. FEBS Lett. 1971, 15, 232–236. [Google Scholar] [CrossRef]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef]
- Dong, J.; Ueda, H. ELISA-type assays of trace biomarkers using microfluidic methods. WIREs Nanomed. Nanobiotechnol. 2017, 9, e1457. [Google Scholar] [CrossRef] [PubMed]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Lateral flow assays: Principles, designs and labels. TrAC Trends Anal. Chem. 2016, 82, 286–306. [Google Scholar] [CrossRef]
- O’Farrell, B. Evolution in Lateral Flow–Based Immunoassay Systems. In Lateral Flow Immunoassay; Humana Press: Totowa, NJ, USA, 2009; pp. 1–33. [Google Scholar]
- Sharma, B.; Frontiera, R.R.; Henry, A.-I.; Ringe, E.; Van Duyne, R.P. SERS: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Feliu, N.; Hassan, M.; Garcia Rico, E.; Cui, D.; Parak, W.; Alvarez-Puebla, R. SERS Quantification and Characterization of Proteins and Other Biomolecules. Langmuir 2017, 33, 9711–9730. [Google Scholar] [CrossRef]
- Aitekenov, S.; Sultangaziyev, A.; Boranova, A.; Dyussupova, A.; Ilyas, A.; Gaipov, A.; Bukasov, R. SERS for Detection of Proteinuria: A Comparison of Gold, Silver, Al Tape, and Silicon Substrates for Identification of Elevated Protein Concentration in Urine. Sensors 2023, 23, 1605. [Google Scholar] [CrossRef]
- Schatz, G.C. SERS and the scientific career of Richard P. Van Duyne (1945–2019). J. Raman Spectrosc. 2021, 52, 268–278. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wu, L.; Zhu, D.; Cui, Y. SERS-Activated Platforms for Immunoassay: Probes, Encoding Methods, and Applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, A.; Dyussupova, A.; Sultangaziyev, A.; Shevchenko, Y.; Filchakova, O.; Bukasov, R. SERS immuno- and apta-assays in biosensing/bio-detection: Performance comparison, clinical applications, challenges. Talanta 2023, 265, 124818. [Google Scholar] [CrossRef] [PubMed]
- Grubisha, D.S.; Lipert, R.J.; Park, H.-Y.; Driskell, J.; Porter, M.D. Femtomolar Detection of Prostate-Specific Antigen: An Immunoassay Based on Surface-Enhanced Raman Scattering and Immunogold Labels. Anal. Chem. 2003, 75, 5936–5943. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Lipert, R.J.; Jain, M.; Kaur, S.; Chakraboty, S.; Torres, M.P.; Batra, S.K.; Brand, R.E.; Porter, M.D. Detection of the Potential Pancreatic Cancer Marker MUC4 in Serum Using Surface-Enhanced Raman Scattering. Anal. Chem. 2011, 83, 2554–2561. [Google Scholar] [CrossRef] [PubMed]
- Granger, J.H.; Granger, M.C.; Firpo, M.A.; Mulvihill, S.J.; Porter, M.D. Toward development of a surface-enhanced Raman scattering (SERS)-based cancer diagnostic immunoassay panel. Analyst 2013, 138, 410–416. [Google Scholar] [CrossRef]
- Crawford, A.C.; Laurentius, L.B.; Mulvihill, T.S.; Granger, J.H.; Spencer, J.S.; Chatterjee, D.; Hanson, K.E.; Porter, M.D. Detection of the tuberculosis antigenic marker mannose-capped lipoarabinomannan in pretreated serum by surface-enhanced Raman scattering. Analyst 2017, 142, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Yakes, B.J.; Lipert, R.J.; Bannantine, J.P.; Porter, M.D. Detection of Mycobacterium avium subsp. paratuberculosis by a sonicate immunoassay based on surface-enhanced Raman scattering. Clin. Vaccine Immunol. 2008, 15, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Granger, J.; Porter, M. SERS Detection of Clostridium botulinum Neurotoxin Serotypes A and B in Buffer and Serum: Towards the Development of a Biodefense Test Platform. Anal. Chim. Acta X 2018, 1, 100002. [Google Scholar] [CrossRef] [PubMed]
- Driskell, J.D.; Kwarta, K.M.; Lipert, R.J.; Porter, M.D.; Neill, J.D.; Ridpath, J.F. Low-Level Detection of Viral Pathogens by a Surface-Enhanced Raman Scattering Based Immunoassay. Anal. Chem. 2005, 77, 6147–6154. [Google Scholar] [CrossRef] [PubMed]
- Granger, J.H.; Schlotter, N.E.; Crawford, A.C.; Porter, M.D. Prospects for point-of-care pathogen diagnostics using surface-enhanced Raman scattering (SERS). Chem. Soc. Rev. 2016, 45, 3865–3882. [Google Scholar] [CrossRef] [PubMed]
- Crawford, A.C.; Skuratovsky, A.; Porter, M.D. Sampling Error: Impact on the Quantitative Analysis of Nanoparticle-Based Surface-Enhanced Raman Scattering Immunoassays. Anal. Chem. 2016, 88, 6515–6522. [Google Scholar] [CrossRef] [PubMed]
- Owens, N.A.; Pinter, A.; Porter, M.D. Surface-enhanced resonance Raman scattering for the sensitive detection of a tuberculosis biomarker in human serum. J. Raman Spectrosc. 2019, 50, 15–25. [Google Scholar] [CrossRef]
- Bukasov, R.; Sultangaziyev, A.; Kunushpayeva, Z.; Rapikov, A.; Dossym, D. Aluminum Foil vs. Gold Film: Cost-Effective Substrate in Sandwich SERS Immunoassays of Biomarkers Reveals Potential for Selectivity Improvement. Int. J. Mol. Sci. 2023, 24, 5578. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D.I. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Aitekenov, S.; Sultangaziyev, A.; Abdirova, P.; Yussupova, L.; Gaipov, A.; Utegulov, Z.; Bukasov, R. Raman, Infrared and Brillouin Spectroscopies of Biofluids for Medical Diagnostics and for Detection of Biomarkers. Crit. Rev. Anal. Chem. 2023, 53, 1561–1590. [Google Scholar] [CrossRef]
- Wang, X.; Yun, Y.; Sun, W.; Lu, Z.; Tao, X. A high-performance fluorescence immunoassay based on pyrophosphate-induced MOFs NH2-MIL-88B(Fe) hydrolysis for chloramphenicol detection. Sens. Actuators B Chem. 2022, 353, 131143. [Google Scholar] [CrossRef]
- Sun, Z.-H.; Zhang, X.-X.; Xu, D.; Liu, J.; Yu, R.-J.; Jing, C.; Han, H.-X.; Ma, W. Silver-amplified fluorescence immunoassay via aggregation-induced emission for detection of disease biomarker. Talanta 2021, 225, 121963. [Google Scholar] [CrossRef]
- Luo, L.; Jia, B.-Z.; Wei, X.-Q.; Xiao, Z.-L.; Wang, H.; Sun, Y.-M.; Shen, Y.-D.; Lei, H.-T.; Xu, Z.-L. Development of an inner filter effect-based fluorescence immunoassay for the detection of acrylamide using 9-xanthydrol derivatization. Sens. Actuators B Chem. 2021, 332, 129561. [Google Scholar] [CrossRef]
- Wang, Y.-W.; Chen, L.; Liang, M.; Xu, H.; Tang, S.; Yang, H.-H.; Song, H. Sensitive fluorescence immunoassay of alpha-fetoprotein through copper ions modulated growth of quantum dots in-situ. Sens. Actuators B Chem. 2017, 247, 408–413. [Google Scholar] [CrossRef]
- Chen, M.; Liu, Z.; Guan, Y.; Chen, Y.; Liu, W.; Liu, Y. Zeolitic imidazolate frameworks-derived hollow Co/N-doped CNTs as oxidase-mimic for colorimetric-fluorescence immunoassay of ochratoxin A. Sens. Actuators B Chem. 2022, 359, 131609. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, J.; Lu, Y.; Sun, J.; Yang, X. Fluorescence Immunoassay Based on the Phosphate-Triggered Fluorescence Turn-on Detection of Alkaline Phosphatase. Anal. Chem. 2018, 90, 3505–3511. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Li, H.; Sun, J.; Li, Y.; Mari, G.M.; Yu, X.; Yu, W.; Wen, K.; Shen, J.; Wang, Z. Magnetic assisted fluorescence immunoassay for sensitive chloramphenicol detection using carbon dots@CaCO3 nanocomposites. J. Hazard. Mater. 2021, 402, 123942. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.-X.; Cao, S.-H.; Wang, Z.-C.; Weng, Y.-H.; Huo, S.-X.; Zhai, Y.-Y.; Chen, M.; Pan, X.-H.; Li, Y.-Q. Graphene oxide-assisted surface plasmon-coupled emission for amplified fluorescence immunoassay. Sens. Actuators B Chem. 2017, 253, 804–808. [Google Scholar] [CrossRef]
- Li, Y.; Sun, J.; Wu, L.; Ji, J.; Sun, X.; Qian, Y. Surface-enhanced fluorescence immunosensor using Au nano-crosses for the detection of microcystin-LR. Biosens. Bioelectron. 2014, 62, 255–260. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, X.; Ren, X.; Li, W.; Sun, J.; Wang, X.; Huang, Y.; Guo, Y.; Zeng, H. Novel fluorescence immunoassay for the detection of zearalenone using HRP-mediated fluorescence quenching of gold-silver bimetallic nanoclusters. Food Chem. 2021, 355, 129633. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhu, Y.; Wang, J.; Gyimah, E.; Hu, X.; Zhang, Z. A novel fluorescence immunoassay based on AgNCs and ALP for ultrasensitive detection of sulfamethazine (SMZ) in environmental and biological samples. Talanta 2019, 199, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Tang, Y.; Zhang, K.; Tang, D. Wet NH3-Triggered NH2-MIL-125(Ti) Structural Switch for Visible Fluorescence Immunoassay Impregnated on Paper. Anal. Chem. 2018, 90, 14121–14125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Li, J.; Peng, C.; Zhu, S.; Sun, J.; Yang, X. Fluorescence Immunoassay Based on the Alkaline Phosphatase Triggered in Situ Fluorogenic Reaction of o-Phenylenediamine and Ascorbic Acid. Anal. Chem. 2019, 91, 2978–2984. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Huang, X.; Li, J.; Shan, S.; Lai, W.; Xiong, Y. A novel fluorescence immunoassay for the sensitive detection of Escherichia coli O157:H7 in milk based on catalase-mediated fluorescence quenching of CdTe quantum dots. Anal. Chim. Acta 2016, 947, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jin, R.; Kong, D.; Zhao, X.; Liu, F.; Yan, X.; Lin, Y.; Lu, G. Switchable fluorescence immunoassay using gold nanoclusters anchored cobalt oxyhydroxide composite for sensitive detection of imidacloprid. Sens. Actuators B Chem. 2019, 283, 207–214. [Google Scholar] [CrossRef]
- Luo, L.; Song, Y.; Zhu, C.; Fu, S.; Shi, Q.; Sun, Y.-M.; Jia, B.; Du, D.; Xu, Z.-L.; Lin, Y. Fluorescent silicon nanoparticles-based ratiometric fluorescence immunoassay for sensitive detection of ethyl carbamate in red wine. Sens. Actuators B Chem. 2018, 255, 2742–2749. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, J.; Wang, S.; Lu, S.; Sun, J.; Yang, X. Enzyme-induced in situ generation of polymer carbon dots for fluorescence immunoassay. Sens. Actuators B Chem. 2020, 306, 127583. [Google Scholar] [CrossRef]
- Han, H.; Wang, C.; Yang, X.; Zheng, S.; Cheng, X.; Liu, Z.; Zhao, B.; Xiao, R. Rapid field determination of SARS-CoV-2 by a colorimetric and fluorescent dual-functional lateral flow immunoassay biosensor. Sens. Actuators B Chem. 2022, 351, 130897. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wei, X.; Guo, X.; Wang, H.; Song, H.; Pan, C.; Xu, N. Immunoassay based on Au-Ag bimetallic nanoclusters for colorimetric/fluorescent double biosensing of dicofol. Biosens. Bioelectron. 2021, 194, 113611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Sheng, W.; Liu, Y.; Huang, N.; Zhang, W.; Wang, S. Multiplexed fluorescence immunoassay combined with magnetic separation using upconversion nanoparticles as multicolor labels for the simultaneous detection of tyramine and histamine in food samples. Anal. Chim. Acta 2020, 1130, 117–125. [Google Scholar] [CrossRef]
- Wang, Z.; Xing, K.; Ding, N.; Wang, S.; Zhang, G.; Lai, W. Lateral flow immunoassay based on dual spectral-overlapped fluorescence quenching of polydopamine nanospheres for sensitive detection of sulfamethazine. J. Hazard. Mater. 2022, 423, 127204. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Wang, Y.; Chen, Q.; Liu, X. Nanobody-alkaline phosphatase fusion-mediated phosphate-triggered fluorescence immunoassay for ochratoxin a detection. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 226, 117617. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Jiang, Z.; Jin, M.; Du, P.; Chen, G.; Cui, X.; Zhang, Y.; Qin, G.; Yan, F.; Abd El-Aty, A.M.; et al. Fluorescence immunoassay for multiplex detection of organophosphate pesticides in agro-products based on signal amplification of gold nanoparticles and oligonucleotides. Food Chem. 2020, 326, 126813. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wu, J.; Fang, X.; Kong, J. Washing-free centrifugal microchip fluorescence immunoassay for rapid and point-of-care detection of protein. Anal. Chim. Acta 2020, 1118, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wen, K.; Dong, B.; Zhang, J.; Bai, Y.; Liu, M.; Li, P.; Mujtaba, M.G.; Yu, X.; Yu, W.; et al. Novel inner filter effect-based fluorescence immunoassay with gold nanoclusters for bromadiolone detection in human serum. Sens. Actuators B Chem. 2019, 297, 126787. [Google Scholar] [CrossRef]
- Nishiyama, K.; Fukuyama, M.; Maeki, M.; Ishida, A.; Tani, H.; Hibara, A.; Tokeshi, M. One-step non-competitive fluorescence polarization immunoassay based on a Fab fragment for C-reactive protein quantification. Sens. Actuators B Chem. 2021, 326, 128982. [Google Scholar] [CrossRef]
- Shokri, E.; Hosseini, M.; Sadeghan, A.A.; Bahmani, A.; Nasiri, N.; Hosseinkhani, S. Virus-directed synthesis of emitting copper nanoclusters as an approach to simple tracer preparation for the detection of Citrus Tristeza Virus through the fluorescence anisotropy immunoassay. Sens. Actuators B Chem. 2020, 321, 128634. [Google Scholar] [CrossRef]
- Su, R.; Tang, X.; Feng, L.; Yao, G.-l.; Chen, J. Development of quantitative magnetic beads-based flow cytometry fluorescence immunoassay for aflatoxin B1. Microchem. J. 2020, 155, 104715. [Google Scholar] [CrossRef]
- Ao, L.; Liao, T.; Huang, L.; Lin, S.; Xu, K.; Ma, J.; Qiu, S.; Wang, X.; Zhang, Q. Sensitive and simultaneous detection of multi-index lung cancer biomarkers by an NIR-Ⅱ fluorescence lateral-flow immunoassay platform. Chem. Eng. J. 2022, 436, 135204. [Google Scholar] [CrossRef]
- Othman, H.O.; Salehnia, F.; Hosseini, M.; Hassan, R.; Faizullah, A.; Ganjali, M.R. Fluorescence immunoassay based on nitrogen doped carbon dots for the detection of human nuclear matrix protein NMP22 as biomarker for early stage diagnosis of bladder cancer. Microchem. J. 2020, 157, 104966. [Google Scholar] [CrossRef]
- Guo, Y.; Zou, R.; Si, F.; Liang, W.; Zhang, T.; Chang, Y.; Qiao, X.; Zhao, J. A sensitive immunoassay based on fluorescence resonance energy transfer from up-converting nanoparticles and graphene oxide for one-step detection of imidacloprid. Food Chem. 2021, 335, 127609. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Choi, Y.H.; Shin, Y.-B.; Kim, H.-J.; Kim, M.-G. A fluorescence enhancement-based label-free homogeneous immunoassay of benzo[a]pyrene (BaP) in aqueous solutions. Chemosphere 2016, 150, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Kim, K.R.; Chun, H.J.; Jeong, K.Y.; Hong, D.-K.; Lee, K.-N.; Yoon, H.C. Time-resolved fluorescence resonance energy transfer-based lateral flow immunoassay using a raspberry-type europium particle and a single membrane for the detection of cardiac troponin I. Biosens. Bioelectron. 2020, 163, 112284. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, J.; Weng, G.-j.; Li, J.-j.; Zhao, J.-w. Gold nanoring core-shell satellites with abundant built-in hotspots and great analyte penetration: An immunoassay platform for the SERS/fluorescence-based detection of carcinoembryonic antigen. Chem. Eng. J. 2021, 409, 128173. [Google Scholar] [CrossRef]
- Xie, H.; Dong, J.; Duan, J.; Hou, J.; Ai, S.; Li, X. Magnetic nanoparticles-based immunoassay for aflatoxin B1 using porous g-C3N4 nanosheets as fluorescence probes. Sens. Actuators B Chem. 2019, 278, 147–152. [Google Scholar] [CrossRef]
- Zvereva, E.A.; Zherdev, A.V.; Formanovsky, A.A.; Abuknesha, R.A.; Eremin, S.A.; Dzantiev, B.B. Fluorescence polarization immunoassay of colchicine. J. Pharm. Biomed. Anal. 2018, 159, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Y.; Liu, G.; Zhang, C.; Wang, S. A time-resolved fluorescence immunoassay for the ultrasensitive determination of diethylstilbestrol based on the double-codified gold nanoparticles. Steroids 2014, 89, 41–46. [Google Scholar] [CrossRef]
- Dong, B.; Li, H.; Sun, J.; Mari, G.M.; Yu, X.; Ke, Y.; Li, J.; Wang, Z.; Yu, W.; Wen, K.; et al. Development of a fluorescence immunoassay for highly sensitive detection of amantadine using the nanoassembly of carbon dots and MnO2 nanosheets as the signal probe. Sens. Actuators B Chem. 2019, 286, 214–221. [Google Scholar] [CrossRef]
- Sun, M.; Du, L.; Gao, S.; Bao, Y.; Wang, S. Determination of 17β-oestradiol by fluorescence immunoassay with streptavidin-conjugated quantum dots as label. Steroids 2010, 75, 400–403. [Google Scholar] [CrossRef]
- Tian, Y.; Li, X.; Wang, F.; Gu, C.; Zhao, Z.; Si, H.; Jiang, T. SERS-based immunoassay and degradation of CA19-9 mediated by gold nanowires anchored magnetic–semiconductor nanocomposites. J. Hazard. Mater. 2021, 403, 124009. [Google Scholar] [CrossRef]
- Qu, Q.; Wang, J.; Zeng, C.; Wang, M.; Qi, W.; He, Z. AuNP array coated substrate for sensitive and homogeneous SERS-immunoassay detection of human immunoglobulin G. RSC Adv. 2021, 11, 22744–22750. [Google Scholar] [CrossRef]
- Baniukevic, J.; Hakki Boyaci, I.; Goktug Bozkurt, A.; Tamer, U.; Ramanavicius, A.; Ramanaviciene, A. Magnetic gold nanoparticles in SERS-based sandwich immunoassay for antigen detection by well oriented antibodies. Biosens. Bioelectron. 2013, 43, 281–288. [Google Scholar] [CrossRef]
- Xiao, M.; Xie, K.; Dong, X.; Wang, L.; Huang, C.; Xu, F.; Xiao, W.; Jin, M.; Huang, B.; Tang, Y. Ultrasensitive detection of avian influenza A (H7N9) virus using surface-enhanced Raman scattering-based lateral flow immunoassay strips. Anal. Chim. Acta 2019, 1053, 139–147. [Google Scholar] [CrossRef]
- Shi, Q.; Huang, J.; Sun, Y.; Yin, M.; Hu, M.; Hu, X.; Zhang, Z.; Zhang, G. Utilization of a lateral flow colloidal gold immunoassay strip based on surface-enhanced Raman spectroscopy for ultrasensitive detection of antibiotics in milk. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 197, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Achadu, O.J.; Abe, F.; Suzuki, T.; Park, E.Y. Molybdenum Trioxide Nanocubes Aligned on a Graphene Oxide Substrate for the Detection of Norovirus by Surface-Enhanced Raman Scattering. ACS Appl. Mater. Interfaces 2020, 12, 43522–43534. [Google Scholar] [CrossRef] [PubMed]
- Panikar, S.S.; Banu, N.; Haramati, J.; Gutierrez-Silerio, G.Y.; Bastidas-Ramirez, B.E.; Tellez-Bañuelos, M.C.; Camacho-Villegas, T.A.; de Toro-Arreola, S.; De la Rosa, E. Anti-fouling SERS-based immunosensor for point-of-care detection of the B7–H6 tumor biomarker in cervical cancer patient serum. Anal. Chim. Acta 2020, 1138, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, A.G.; Buyukgoz, G.G.; Soforoglu, M.; Tamer, U.; Suludere, Z.; Boyaci, I.H. Alkaline phosphatase labeled SERS active sandwich immunoassay for detection of Escherichia coli. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 194, 8–13. [Google Scholar] [CrossRef] [PubMed]
- She, P.; Chu, Y.; Liu, C.; Guo, X.; Zhao, K.; Li, J.; Du, H.; Zhang, X.; Wang, H.; Deng, A. A competitive immunoassay for ultrasensitive detection of Hg2+ in water, human serum and urine samples using immunochromatographic test based on surface-enhanced Raman scattering. Anal. Chim. Acta 2016, 906, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Ilhan, H.; Guven, B.; Dogan, U.; Torul, H.; Evran, S.; Çetin, D.; Suludere, Z.; Saglam, N.; Boyaci, İ.H.; Tamer, U. The coupling of immunomagnetic enrichment of bacteria with paper-based platform. Talanta 2019, 201, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liu, H.; Tian, Y.; Gu, C.; Zhao, Z.; Zeng, S.; Jiang, T. Recyclable SERS-Based Immunoassay Guided by Photocatalytic Performance of Fe3O4@TiO2@Au Nanocomposites. Biosensors 2020, 10, 25. [Google Scholar] [CrossRef]
- Wang, J.; Xu, C.; Lei, M.; Ma, Y.; Wang, X.; Wang, R.; Sun, J. Microcavity-based SERS chip for ultrasensitive immune detection of cardiac biomarkers. Microchem. J. 2021, 171, 106875. [Google Scholar] [CrossRef]
- Chang, H.; Kang, H.; Ko, E.; Jun, B.-H.; Lee, H.-Y.; Lee, Y.-S.; Jeong, D.H. PSA Detection with Femtomolar Sensitivity and a Broad Dynamic Range Using SERS Nanoprobes and an Area-Scanning Method. ACS Sens. 2016, 1, 645–649. [Google Scholar] [CrossRef]
- Kim, K.; Choi, N.; Jeon, J.H.; Rhie, G.-E.; Choo, J. SERS-Based Immunoassays for the Detection of Botulinum Toxins A and B Using Magnetic Beads. Sensors 2019, 19, 4081. [Google Scholar] [CrossRef]
- Kim, W.; Bang, A.; Kim, S.; Lee, G.-J.; Kim, Y.-H.; Choi, S. Adiponectin-targeted SERS immunoassay biosensing platform for early detection of gestational diabetes mellitus. Biosens. Bioelectron. 2022, 213, 114488. [Google Scholar] [CrossRef]
- Gao, R.; Lv, Z.; Mao, Y.; Yu, L.; Bi, X.; Xu, S.; Cui, J.; Wu, Y. SERS-Based Pump-Free Microfluidic Chip for Highly Sensitive Immunoassay of Prostate-Specific Antigen Biomarkers. ACS Sens. 2019, 4, 938–943. [Google Scholar] [CrossRef]
- Song, C.; Min, L.; Zhou, N.; Yang, Y.; Su, S.; Huang, W.; Wang, L. Synthesis of Novel Gold Mesoflowers as SERS Tags for Immunoassay with Improved Sensitivity. ACS Appl. Mater. Interfaces 2014, 6, 21842–21850. [Google Scholar] [CrossRef]
- Choi, N.; Lee, J.; Ko, J.; Jeon, J.H.; Rhie, G.-E.; deMello, A.J.; Choo, J. Integrated SERS-Based Microdroplet Platform for the Automated Immunoassay of F1 Antigens in Yersinia pestis. Anal. Chem. 2017, 89, 8413–8420. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, J.; Zhao, J.; Weng, G.-J.; Li, J.-J.; Zhao, J.-W. Growth of Spherical Gold Satellites on the Surface of Au@Ag@SiO2 Core–Shell Nanostructures Used for an Ultrasensitive SERS Immunoassay of Alpha-Fetoprotein. ACS Appl. Mater. Interfaces 2019, 11, 3617–3626. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, R.; Chen, H.; Weng, W.; Lin, Q.; Deng, D.; Li, Z.; Kong, J. Sensitive polydopamine bi-functionalized SERS immunoassay for microalbuminuria detection. Biosens. Bioelectron. 2019, 142, 111542. [Google Scholar] [CrossRef]
- Yang, K.; Hu, Y.; Dong, N. A novel biosensor based on competitive SERS immunoassay and magnetic separation for accurate and sensitive detection of chloramphenicol. Biosens. Bioelectron. 2016, 80, 373–377. [Google Scholar] [CrossRef]
- Jia, X.; Wang, K.; Li, X.; Liu, Z.; Liu, Y.; Xiao, R.; Wang, S. Highly sensitive detection of three protein toxins via SERS-lateral flow immunoassay based on SiO2@Au nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2022, 41, 102522. [Google Scholar] [CrossRef]
- Gao, R.; Chen, F.; Yang, D.; Zheng, L.; Jing, T.; Jia, H.; Chen, X.; Lu, Y.; Xu, S.; Zhang, D.; et al. Simultaneous SERS-based immunoassay of dual cardiac markers on pump-free hybrid microfluidic chip. Sens. Actuators B Chem. 2022, 369, 132378. [Google Scholar] [CrossRef]
- Kaladharan, K.; Chen, K.-H.; Chen, P.-H.; Goudar, V.S.; Ishdorj, T.-O.; Santra, T.S.; Tseng, F.-G. Dual-clamped one-pot SERS-based biosensors for rapid and sensitive detection of SARS-CoV-2 using portable Raman spectrometer. Sens. Actuators B Chem. 2023, 393, 134172. [Google Scholar] [CrossRef]
- Chen, R.; Wang, H.; Sun, C.; Zhao, Y.; He, Y.; Nisar, M.S.; Wei, W.; Kang, H.; Xie, X.; Du, C.; et al. Au@SiO2 SERS nanotags based lateral flow immunoassay for simultaneous detection of aflatoxin B1 and ochratoxin A. Talanta 2023, 258, 124401. [Google Scholar] [CrossRef]
- Mohammadi, M.; Antoine, D.; Vitt, M.; Dickie, J.M.; Sultana Jyoti, S.; Wall, J.G.; Johnson, P.A.; Wawrousek, K.E. A fast, ultrasensitive SERS immunoassay to detect SARS-CoV-2 in saliva. Anal. Chim. Acta 2022, 1229, 340290. [Google Scholar] [CrossRef]
- Zhu, G.; Hu, Y.; Gao, J.; Zhong, L. Highly sensitive detection of clenbuterol using competitive surface-enhanced Raman scattering immunoassay. Anal. Chim. Acta 2011, 697, 61–66. [Google Scholar] [CrossRef]
- Fu, X.; Wang, Y.; Liu, Y.; Liu, H.; Fu, L.; Wen, J.; Li, J.; Wei, P.; Chen, L. A graphene oxide/gold nanoparticle-based amplification method for SERS immunoassay of cardiac troponin I. Analyst 2019, 144, 1582–1589. [Google Scholar] [CrossRef]
- Tuckmantel Bido, A.; Brolo, A.G. Digital SERS Protocol Using Au Nanoparticle-Based Extrinsic Raman Labels for the Determination of SARS-CoV-2 Spike Protein in Saliva Samples. ACS Appl. Nano Mater. 2023, 6, 15426–15436. [Google Scholar] [CrossRef]
- Xie, T.; Xu, D.; Shang, Y.; Li, Y.; Gu, Y.; Yang, G.; Qu, L. Highly sensitive SERS detection of IL-6 in serum by Au@Fe3O4 nanoring-based sandwich immunoassay. Sens. Actuators B Chem. 2023, 375, 132897. [Google Scholar] [CrossRef]
- Yin, L.; You, T.; El-Seedi, H.R.; El-Garawani, I.M.; Guo, Z.; Zou, X.; Cai, J. Rapid and sensitive detection of zearalenone in corn using SERS-based lateral flow immunosensor. Food Chem. 2022, 396, 133707. [Google Scholar] [CrossRef]
- Cheng, Z.; Choi, N.; Wang, R.; Lee, S.; Moon, K.C.; Yoon, S.-Y.; Chen, L.; Choo, J. Simultaneous Detection of Dual Prostate Specific Antigens Using Surface-Enhanced Raman Scattering-Based Immunoassay for Accurate Diagnosis of Prostate Cancer. ACS Nano 2017, 11, 4926–4933. [Google Scholar] [CrossRef]
- Jiao, S.; Liu, J.; Sun, J.; Chang, Y.; Wang, S.; Dai, S.; Xu, R.; Dou, M.; Li, Q.; Wang, J.; et al. A highly sensitive and reproducible multiplex mycotoxin SERS array based on AuNPs-loaded inverse opal silica photonic crystal microsphere. Sens. Actuators B Chem. 2022, 355, 131245. [Google Scholar] [CrossRef]
- Cha, H.; Kim, H.; Joung, Y.; Kang, H.; Moon, J.; Jang, H.; Park, S.; Kwon, H.-J.; Lee, I.-C.; Kim, S.; et al. Surface-enhanced Raman scattering-based immunoassay for severe acute respiratory syndrome coronavirus 2. Biosens. Bioelectron. 2022, 202, 114008. [Google Scholar] [CrossRef]
- Ko, J.; Lee, C.; Choo, J. Highly sensitive SERS-based immunoassay of aflatoxin B1 using silica-encapsulated hollow gold nanoparticles. J. Hazard. Mater. 2015, 285, 11–17. [Google Scholar] [CrossRef]
- Yang, L.; Gao, M.X.; Zhan, L.; Gong, M.; Zhen, S.J.; Huang, C.Z. An enzyme-induced Au@ Ag core–shell nanoStructure used for an ultrasensitive surface-enhanced Raman scattering immunoassay of cancer biomarkers. Nanoscale 2017, 9, 2640–2645. [Google Scholar] [CrossRef]
- Yang, J.; Pan, M.; Liu, K.; Xie, X.; Wang, S.; Hong, L.; Wang, S. Core-shell AuNRs@Ag-enhanced and magnetic separation-assisted SERS immunosensing platform for amantadine detection in animal-derived foods. Sens. Actuators B Chem. 2021, 349, 130783. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, S.; Liu, Q.; Gao, B.; Zhao, X.; Sun, F. SERS-based lateral flow immunoassay strip for ultrasensitive and quantitative detection of acrosomal protein SP10. Microchem. J. 2022, 175, 107191. [Google Scholar] [CrossRef]
- Deng, D.; Yang, H.; Liu, C.; Zhao, K.; Li, J.; Deng, A. Ultrasensitive detection of diclofenac in water samples by a novel surface-enhanced Raman scattering (SERS)-based immunochromatographic assay using AgMBA@SiO2-Ab as immunoprobe. Sens. Actuators B Chem. 2019, 283, 563–570. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef]
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators B Chem. 2021, 329, 129196. [Google Scholar] [CrossRef] [PubMed]
- Banaei, N.; Moshfegh, J.; Kim, B. Surface enhanced Raman spectroscopy-based immunoassay detection of tumor-derived extracellular vesicles to differentiate pancreatic cancers from chronic pancreatitis. J. Raman Spectrosc. 2021, 52, 1810–1819. [Google Scholar] [CrossRef]
- Zamora-Mendoza, B.N.; Espinosa-Tanguma, R.; Ramírez-Elías, M.G.; Cabrera-Alonso, R.; Montero-Moran, G.; Portales-Pérez, D.; Rosales-Romo, J.A.; Gonzalez, J.F.; Gonzalez, C. Surface-enhanced raman spectroscopy: A non invasive alternative procedure for early detection in childhood asthma biomarkers in saliva. Photodiagn. Photodyn. Ther. 2019, 27, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Chen, S.; Duan, W.; Kong, X.; Wang, Y.; Zhou, L.; Li, P.; Zhang, C.; Du, L. Highly Sensitive Exosome Detection for Early Diagnosis of Pancreatic Cancer Using Immunoassay Based on Hierarchical Surface-Enhanced Raman Scattering Substrate. Small Methods 2022, 6, 2200154. [Google Scholar] [CrossRef]
- Lu, L.; Yu, J.; Liu, X.; Yang, X.; Zhou, Z.; Jin, Q.; Xiao, R.; Wang, C. Rapid, quantitative and ultra-sensitive detection of cancer biomarker by a SERRS-based lateral flow immunoassay using bovine serum albumin coated Au nanorods. RSC Adv. 2020, 10, 271–281. [Google Scholar] [CrossRef]
- Li, T.-D.; Zhang, R.; Chen, H.; Huang, Z.-P.; Ye, X.; Wang, H.; Deng, A.-M.; Kong, J.-L. An ultrasensitive polydopamine bi-functionalized SERS immunoassay for exosome-based diagnosis and classification of pancreatic cancer. Chem. Sci. 2018, 9, 5372–5382. [Google Scholar] [CrossRef]
- Jia, X.; Wang, C.; Rong, Z.; Li, J.; Wang, K.; Qie, Z.; Xiao, R.; Wang, S. Dual dye-loaded Au@Ag coupled to a lateral flow immunoassay for the accurate and sensitive detection of Mycoplasma pneumoniae infection. RSC Adv. 2018, 8, 21243–21251. [Google Scholar] [CrossRef]
- Lu, M.; Joung, Y.; Jeon, C.S.; Kim, S.; Yong, D.; Jang, H.; Pyun, S.H.; Kang, T.; Choo, J. Dual-mode SERS-based lateral flow assay strips for simultaneous diagnosis of SARS-CoV-2 and influenza a virus. Nano Converg. 2022, 9, 39. [Google Scholar] [CrossRef]
- Lee, S.H.; Hwang, J.; Kim, K.; Jeon, J.; Lee, S.; Ko, J.; Lee, J.; Kang, M.; Chung, D.R.; Choo, J. Quantitative Serodiagnosis of Scrub Typhus Using Surface-Enhanced Raman Scattering-Based Lateral Flow Assay Platforms. Anal. Chem. 2019, 91, 12275–12282. [Google Scholar] [CrossRef] [PubMed]
- Squire, K.J.; Zhao, Y.; Tan, A.; Sivashanmugan, K.; Kraai, J.A.; Rorrer, G.L.; Wang, A.X. Photonic crystal-enhanced fluorescence imaging immunoassay for cardiovascular disease biomarker screening with machine learning analysis. Sens. Actuators B Chem. 2019, 290, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wu, Z.; Liu, B.; Wang, C.; Wang, Q.; Zhang, L.; Wang, Z.; Chen, C.; Fu, Y.; Li, C.; et al. A time-resolved fluorescence lateral flow immunoassay for rapid and quantitative serodiagnosis of Brucella infection in humans. J. Pharm. Biomed. Anal. 2021, 200, 114071. [Google Scholar] [CrossRef] [PubMed]
- Radon, T.P.; Massat, N.J.; Jones, R.; Alrawashdeh, W.; Dumartin, L.; Ennis, D.; Duffy, S.W.; Kocher, H.M.; Pereira, S.P.; Guarner, L.; et al. Identification of a three-biomarker panel in urine for early detection of pancreatic adenocarcinoma. Clin. Cancer Res. 2015, 21, 3512–3521. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Skotland, T.; Berge, V.; Sandvig, K.; Llorente, A. Exosomal proteins as prostate cancer biomarkers in urine: From mass spectrometry discovery to immunoassay-based validation. Eur. J. Pharm. Sci. 2017, 98, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Shimura, T.; Iwasaki, H.; Kitagawa, M.; Ebi, M.; Yamada, T.; Katano, T.; Nisie, H.; Okamoto, Y.; Ozeki, K.; Mizoshita, T.; et al. Urinary Cysteine-Rich Protein 61 and Trefoil Factor 3 as Diagnostic Biomarkers for Colorectal Cancer. Transl. Oncol. 2019, 12, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.; Prieto, C.; Folgueira, L. A prospective study to assess the diagnostic performance of the Sofia® Immunoassay for Influenza and RSV detection. J. Clin. Virol. 2016, 77, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Daag, J.V.; Ylade, M.; Adams, C.; Jadi, R.; Crisostomo, M.V.; Alpay, R.; Aportadera, E.T.C.; Yoon, I.-K.; White, L.; Deen, J.; et al. Evaluation of a new point-of-care test to determine prior dengue infection for potential use in pre-vaccination screening. Clin. Microbiol. Infect. 2021, 27, 904–908. [Google Scholar] [CrossRef]
- Smela, M.E.; Currier, S.S.; Bailey, E.A.; Essigmann, J.M. The chemistry and biology of aflatoxin B1: From mutational spectrometry to carcinogenesis. Carcinogenesis 2001, 22, 535–545. [Google Scholar] [CrossRef]
- Petzinger, E.; Ziegler, K. Ochratoxin A from a toxicological perspective. J. Vet. Pharmacol. Ther. 2000, 23, 91–98. [Google Scholar] [CrossRef]
- Galle, P.R.; Foerster, F.; Kudo, M.; Chan, S.L.; Llovet, J.M.; Qin, S.; Schelman, W.R.; Chintharlapalli, S.; Abada, P.B.; Sherman, M.; et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019, 39, 2214–2229. [Google Scholar] [CrossRef]
- Sharma, S.; Jackson, P.G.; Makan, J. Cardiac troponins. J. Clin. Pathol. 2004, 57, 1025. [Google Scholar] [CrossRef] [PubMed]
- Hall, P.; Selinger, B. A statistical justification to relating interlaboratory coefficients of variation with concentration levels. Anal. Chem. 1989, 61, 1465–1466. [Google Scholar] [CrossRef]
- Bernat, A.; Samiwala, M.; Albo, J.; Jiang, X.; Rao, Q. Challenges in SERS-based pesticide detection and plausible solutions. J. Agric. Food Chem. 2019, 67, 12341–12347. [Google Scholar] [CrossRef] [PubMed]
- Baschong, W.; Suetterlin, R.; Laeng, R.H. Control of Autofluorescence of Archival Formaldehyde-fixed, Paraffin-embedded Tissue in Confocal Laser Scanning Microscopy (CLSM). J. Histochem. Cytochem. 2001, 49, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Kunushpayeva, Z.; Rapikov, A.; Akhmetova, A.; Sultangaziyev, A.; Dossym, D.; Bukasov, R. Sandwich SERS immunoassay of human immunoglobulin on silicon wafer compared to traditional SERS substrate, gold film. Sens. Bio-Sens. Res. 2020, 29, 100355. [Google Scholar] [CrossRef]
- Blyth, R.I.R.; Mittendorfer, F.; Hafner, J.; Sardar, S.A.; Duschek, R.; Netzer, F.P.; Ramsey, M.G. An experimental and theoretical investigation of the thiophene/aluminum interface. J. Chem. Phys. 2001, 114, 935–942. [Google Scholar] [CrossRef]
- Sun, Q.; Reddy; Marquez, M.; Jena, P.; Gonzalez, C.; Wang, Q. Theoretical Study on Gold-Coated Iron Oxide Nanostructure: Magnetism and Bioselectivity for Amino Acids. J. Phys. Chem. C 2007, 111, 4159–4163. [Google Scholar] [CrossRef]
- Zhang, D.; Ansar, S.M.; Vangala, K.; Jiang, D. Protein adsorption drastically reduces surface-enhanced Raman signal of dye molecules. J. Raman Spectrosc. 2010, 41, 952–957. [Google Scholar] [CrossRef]
- Gnanasampanthan, T.; Beyer, C.D.; Yu, W.; Karthäuser, J.F.; Wanka, R.; Spöllmann, S.; Becker, H.-W.; Aldred, N.; Clare, A.S.; Rosenhahn, A. Effect of Multilayer Termination on Nonspecific Protein Adsorption and Antifouling Activity of Alginate-Based Layer-by-Layer Coatings. Langmuir 2021, 37, 5950–5963. [Google Scholar] [CrossRef]
- Sun, F.; Ella-Menye, J.-R.; Galvan, D.D.; Bai, T.; Hung, H.-C.; Chou, Y.-N.; Zhang, P.; Jiang, S.; Yu, Q. Stealth Surface Modification of Surface-Enhanced Raman Scattering Substrates for Sensitive and Accurate Detection in Protein Solutions. ACS Nano 2015, 9, 2668–2676. [Google Scholar] [CrossRef]
- Liu, L.; Li, D.; Deng, W. Stimuli-responsive microgels with fluorescent and SERS activities for water and temperature sensing. Biosens. Bioelectron. 2021, 180, 113138. [Google Scholar] [CrossRef]
- Alvarez-Puebla, R.A.; Liz-Marzán, L.M. SERS-Based Diagnosis and Biodetection. Small 2010, 6, 604–610. [Google Scholar] [CrossRef]
- Graham, D.; Faulds, K. Surface-enhanced Raman scattering as a detection technique for molecular diagnostics. Expert Rev. Mol. Diagn. 2009, 9, 537–539. [Google Scholar] [CrossRef][Green Version]
- Roth, S.; Hadass, O.; Cohen, M.; Verbarg, J.; Wilsey, J.; Danielli, A. Photobleaching: Improving the Sensitivity of Fluorescence-Based Immunoassays by Photobleaching the Autofluorescence of Magnetic Beads (Small 3/2019). Small 2019, 15, 1970016. [Google Scholar] [CrossRef]
- Esteve-Turrillas, F.A.; Abad-Fuentes, A. Applications of quantum dots as probes in immunosensing of small-sized analytes. Biosens. Bioelectron. 2013, 41, 12–29. [Google Scholar] [CrossRef]
- Sultangaziyev, A.; Bukasov, R. Review: Applications of surface-enhanced fluorescence (SEF) spectroscopy in bio-detection and biosensing. Sens. Bio-Sens. Res. 2020, 30, 100382. [Google Scholar] [CrossRef]
- Zhu, L.; Cui, X.; Wu, J.; Wang, Z.; Wang, P.; Hou, Y.; Yang, M. Fluorescence immunoassay based on carbon dots as labels for the detection of human immunoglobulin G. Anal. Methods 2014, 6, 4430–4436. [Google Scholar] [CrossRef]
- Ding, C.; Zhu, A.; Tian, Y. Functional Surface Engineering of C-Dots for Fluorescent Biosensing and in Vivo Bioimaging. Acc. Chem. Res. 2014, 47, 20–30. [Google Scholar] [CrossRef]
- Pirsaheb, M.; Mohammadi, S.; Salimi, A. Current advances of carbon dots based biosensors for tumor marker detection, cancer cells analysis and bioimaging. TrAC Trends Anal. Chem. 2019, 115, 83–99. [Google Scholar] [CrossRef]
- Sultangaziyev, A.; Akhmetova, A.; Kunushpayeva, Z.; Rapikov, A.; Filchakova, O.; Bukasov, R. Aluminum foil as a substrate for metal enhanced fluorescence of bacteria labelled with quantum dots, shows very large enhancement and high contrast. Sens. Bio-Sens. Res. 2020, 28, 100332. [Google Scholar] [CrossRef]
- Driskell, J.D.; Uhlenkamp, J.M.; Lipert, R.J.; Porter, M.D. Surface-Enhanced Raman Scattering Immunoassays Using a Rotated Capture Substrate. Anal. Chem. 2007, 79, 4141–4148. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Xu, M.; Xu, L.-J.; Wei, T.; Ma, X.; Zheng, X.-S.; Hu, R.; Ren, B. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef] [PubMed]
- Song, S.W.; Kim, D.; Kim, J.; You, J.; Kim, H.M. Flexible nanocellulose-based SERS substrates for fast analysis of hazardous materials by spiral scanning. J. Hazard. Mater. 2021, 414, 125160. [Google Scholar] [CrossRef] [PubMed]
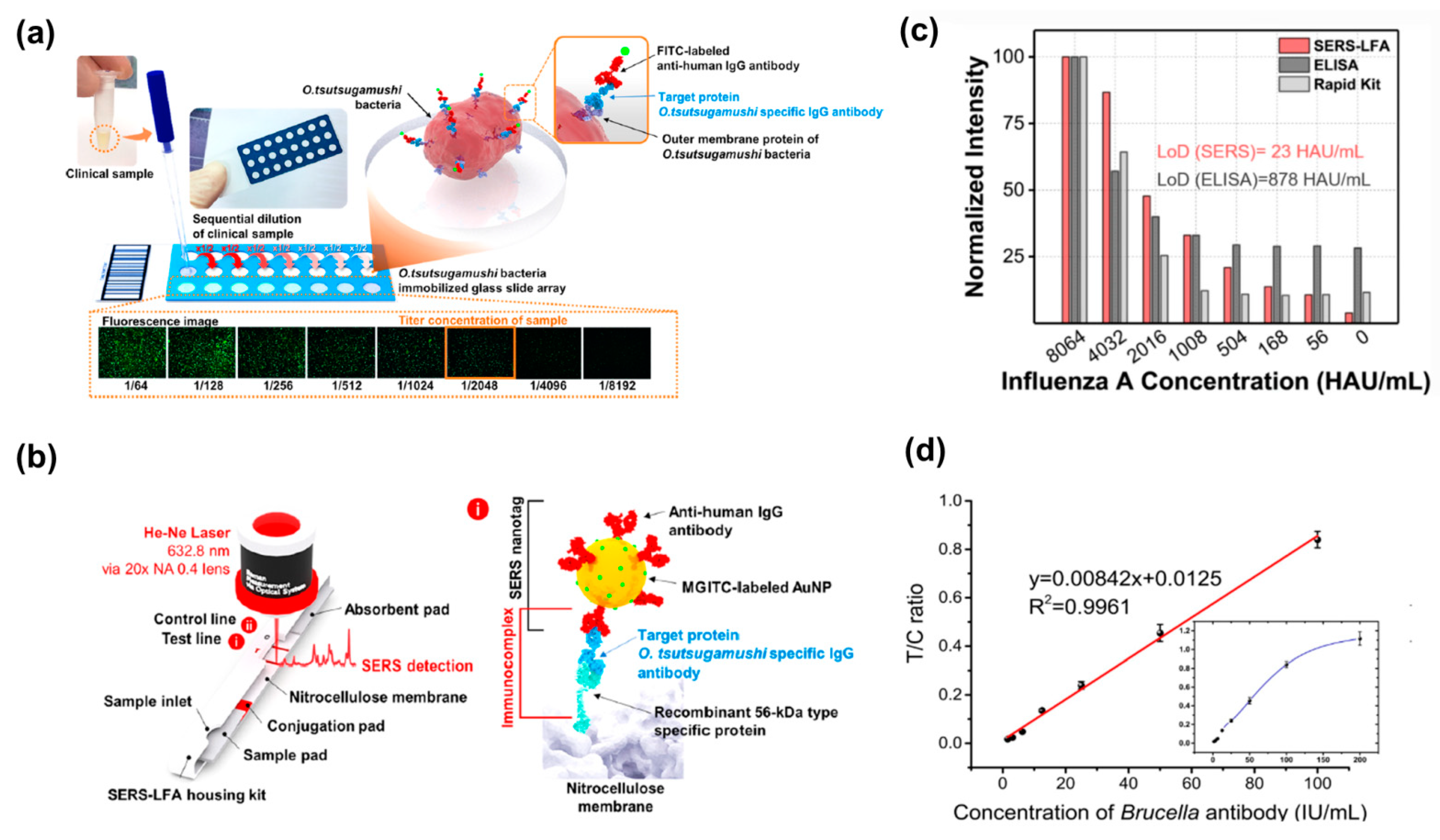

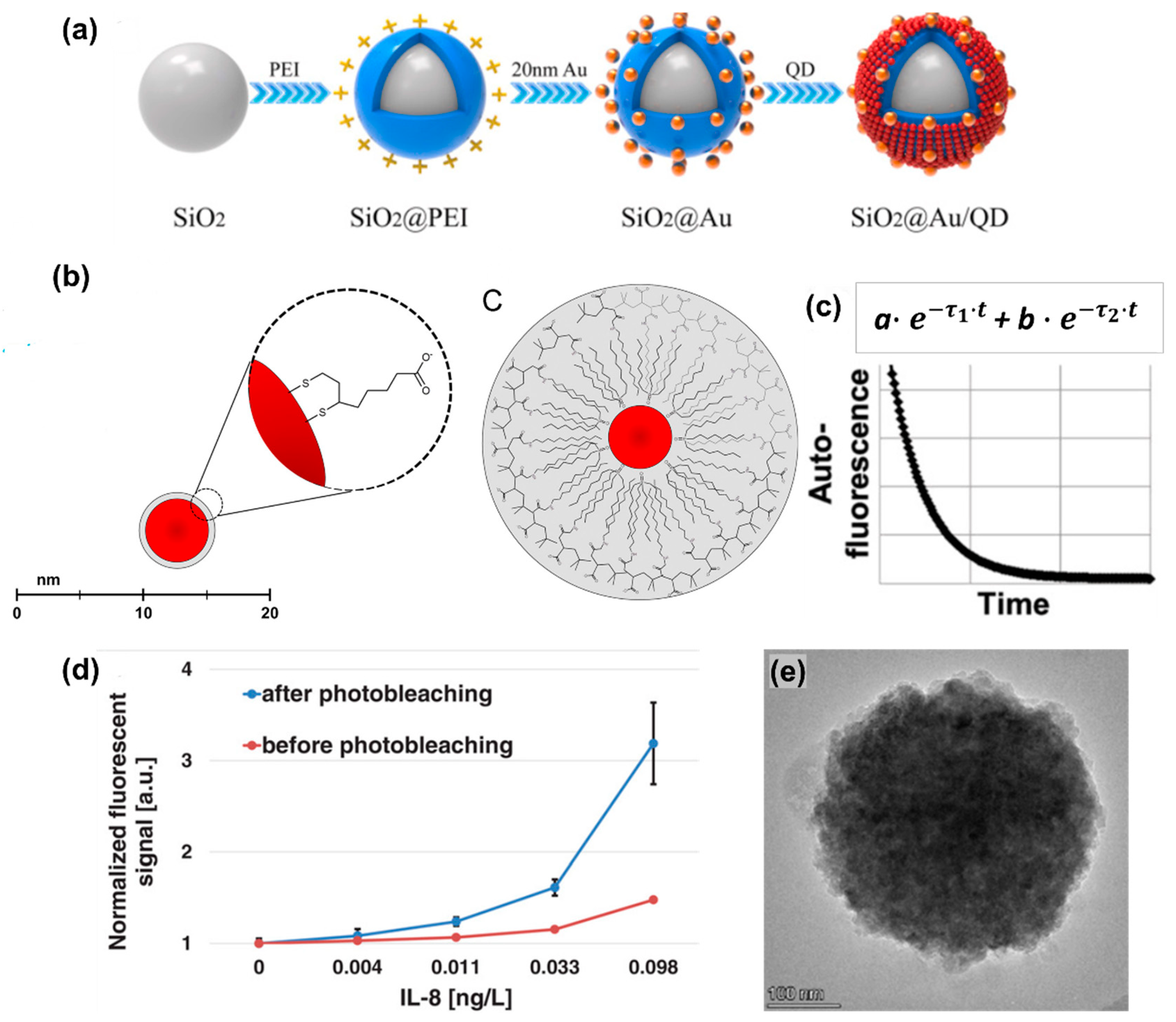
| Year, Author | Analytical Technique | Analyte | LOD (MW, kDa) | LOD, M | Other FoM (Linear Range (LR)/Dynamic Range (DR), Recovery, R2) |
|---|---|---|---|---|---|
| 2022, Wang [35] | FI based on PPi with MOFs | chloramphenicol (amide alcohols antibiotic) | 0.028 μg/L, 0.323 | 8.7 × 10−8 | LR 0.05–0.75 μg/L; 91.8–112.3%; -; 0.99 |
| 2021, Sun [36] | FI (silver-amplified) | α-fetoprotein (cancer biomarker) | 42 pg/mL, 66.478 | 6.3 × 10−13 | LR 0.1–5 μg/mL; -; 0.999 |
| 2021, Luo [37] | Fl | acrylamide food contamination) | 0.16 μg/L, 0.0718 | 2.4 × 10−9 | LR 0.21–6.48 μg/L; 82.3–105.6%; 0.988 |
| 2017, Wang [38] | Fl | α-fetoprotein (cancer biomarker) | 0.45 ng/mL, 66.478 | 6.8 × 10−12 | LR 1–80 ng/mL; -; 0.994 |
| 2022, Chen [39] | Dual-mode Fl (with colorimetry) | ochratoxin A (mycotoxin) | 0.17 ng/L, 0.404 | 4.2 × 10−13 | LR 0.001–10 μg/L; 95.0%− 103.8; 0.995 |
| 2018, Chen [40] | ELISA | α-fetoprotein (cancer biomarker) | 0.041 ng/mL, 66.478 | 6.2 × 10−13 | LR 0.2–1 ng/mL; -; 0.971 |
| 2018, Chen [40] | ELISA | α-fetoprotein (cancer biomarker) | 0.041 ng/mL, 66.478 | 6.2 × 10−13 | LR 1–4 ng/mL; -; 0.995 |
| 2021, Dong [41] | FI | chloramphenicol (amide alcohols antibiotic) | 0.03 μg/kg, 0.323 | 9.3 × 10−11 | LR 0–10 μg/kg; 83.7–105.0%; 0.993; assay time 30 min |
| 2017, Xie [42] | FI | IgG (against bacterial infections) | 0.006 ng/mL, 150 | 4.0 × 10−14 | LR 0.01–800 ng/mL; -; - |
| 2014, Li [43] | Surface-Enhanced Fluorescence (SEF) immunosensor | microcystin-LR (food contamination) | 0.007 ng/mL, 0.995 | 7.0 × 10−12 | LR 0.02–16 ng/mL 98.0–102.2%; 0.9981 |
| 2021, Liu [44] | Horseradish Peroxidase (HRP)-ELISA | zearalenone (mycotoxin) | 0.017 ng/mL, 0.318 | 5.3 × 10−11 | LR 0.02–0.625 ng/mL; 95.56–103.33%; 0.996; RSD = 1.0–8.39% |
| 2019, Zhu [45] | ELISA | sulfamethazine (against bacterial infections) | 0.05 μg/L, 0.278 | 1.8 × 10−7 | DR 0.1400–71.71 μg/L; 84.18–118.6%; - |
| 2016, Lv [46] | Paper-based analytical device (PAD) based FI | carcinoembryonic antigen (cancer marker) | 0.041 ng/mL, 180 | 2.3 × 10−13 | DR 0.1–200 ng/mL |
| 2019, Zhao [47] | Alkaline Phosphatase (ALP)-based FI | α-fetoprotein (cancer biomarker) | 0.21 ng/mL, 66.478 | 3.2 × 10−12 | LR 0.5–40 ng/mL; -; 0.997 |
| 2016, Chen [48] | Catalase (CAT) -based ELISA | E. coli (bacteria detection) | 5 × 102 CFU/mL | DR 5–5 × 106 CFU/mL; 65.88–105.6%; 0.9925 | |
| 2019, Li [49] | FIA | imidacloprid (pesticide residues screening) | 0.1 ng/mL, 0.256 | 3.9 × 10−10 | DR 0.1–50 ng/mL; 85.4–107.4%; 0.990 |
| 2018, Luo [50] | RF-ELISA | ethyl carbamate (food safety monitoring) | 2.6 μg/L, 0.891 | 2.9 × 10−5 | LR 3.9–105.0 μg/L; 86.0∼102.70%; -; CV = less than 11.6% |
| 2020, Liu [51] | ALP-based FI | alkaline phosphatase (liver damage or bone disorder marker) | 0.1 mU/mL, 86 | LR 0.1–6 mU/mL; 100.2–101.8%; 0.998; RSD = less than 5% | |
| 2020, Liu [51] | ALP-based FI | cardiac troponin I (cardiac marker) | 1 ng/mL, 24 | 4.2 × 10−11 | LR 1–30 ng/mL; -; 0.997 |
| 2020, Liu [51] | ALP-based FI | cardiac troponin I (cardiac marker) | 1 ng/mL, 24 | 4.2 × 10−11 | LR 30–250 ng/mL; -; 0.992 |
| 2022, Han [52] | LFIA | S1 protein of SARS-CoV-2 (COVID-19 marker) | 0.033 ng/mL, 76.9 | 4.3 × 10−13 | DR 0.05–1000 ng/mL; -; -; RSD = 6.89–7.16% |
| 2021, Pan [53] | Indirect competitive enzyme-linked immunosorbent assay (ic-ELISA) | Dicofol (food safety monitoring) | 0.62 ng/mL, 0.371 | 1.7 × 10−12 | LR 1.36–19.92 ng/mL; -; 0.985 |
| 2020, Zhang [54] | Upconversion Nanoparticles (UCNPs)-based FIA | tyramine (food safety monitoring) | 0.1 μg/L, 0.131 | 7.6 × 10−7 | LR 0.5–100 μg/L; -; 0.9988 |
| 2020, Zhang [54] | UCNPs-based FIA | histamine (food safety monitoring) | 0.01 μg/L, 0.111 | 9.0 × 10−8 | LR 0.1–100 μg/L; -; 0.9954 |
| 2022, Wang [55] | LFIA with Polydopamine Nanospheres (PDANs) | sulfamethazine (against bacterial infections) | 0.043 ng/mL, 0.278 | 1.5 × 10−13 | LR 0.05–10 ng/mL; 88.9–105.9%; 0.9910 |
| 2020, Wang [56] | PT-FIA | ochratoxin A (mycotoxin) | 0.12 ng/mL, 0.404 | 3.0 × 10−13 | LR 0.2–1.26 ng/mL; -;- |
| 2020, Zhang [57] | Multi-analyte FIA | triazophos (organophosphate pesticides monitoring) | 0.007 μg/L, 0.313 | 2.2 × 10−11 | LR 0.01–20 μg/L; 77.7–113.6%; 7.1–17.1% |
| 2020, Zhang [57] | Multi-analyte FIA | parathion (organophosphate pesticides monitoring) | 0.009 μg/L, 0.291 | 3.1 × 10−11 | LR 0.05–50 μg/L; -; - |
| 2020, Zhang [57] | Multi-analyte FIA | chlorpyrifos (organophosphate pesticides monitoring) | 0.087 μg/L, 0.351 | 2.5 × 10−10 | LR 0.5–1000 μg/L; -; - |
| 2020, Lin [58] | Microfluidic fluorescence immunoassay | procalcitonin (infectious disease biomarker) | 0.05 ng/mL, 13 | 3.9 × 10−12 | DR 0.10–70.00 ng/mL; -; 0.994; detection time 10 min |
| 2019, Li [59] | FI | bromadiolone (Rodenticides detection) | 0.047 ng/mL, 0.527 | 8.9 × 10−11 | LR 0.11–3.09 ng/mL; 77.9–85.6%; 0.987 |
| 2021, Nishiyama [60] | FPIA | c-reactive protein (marker of inflammation) | 1.58 μg/mL, 120 | 1.3 × 10−8 | DR 5–20 μg/mL; -; -; detection time 10 min |
| 2021, Shokri [61] | FAIA | CP25 protein (Citrus Tristeza virus detection) | 220 pg/mL, 0.620 | 3.6 × 10−10 | LR 0.4–25 ng/mL; -; 0.995 |
| 2020, Su [62] | FCMFI | aflatoxin B1 (mycotoxin) | 0.2034 μg/L, 0.312 | 6.5 × 10−10 | LR 0–3 μg/L; -; 0.9642; LOQ = 0.5659 μg/L |
| 2022, Ao [63] | LFIA | carcino-embryonic antigen (cancer marker) | 0.11 ng/mL, 180 | 6.1 × 10−13 | LR 0.11–100 ng/mL; -; - |
| 2022, Ao [63] | LFIA | cytokeratin 19 fragment (cancer biomarker) | 0.18 ng/mL, 40 | 4.5 × 10−12 | LR 0.18–100 ng/mL; -; - |
| 2022, Ao [63] | LFIA | neuron-specific enolase (cancer biomarker) | 0.28 ng/mL, 78 | 3.6 × 10−12 | LR 0.28–100 ng/mL; -; - |
| 2020, Othman [64] | NCD-based FI | nuclear matrix protein 22 (bladder cancer biomarker) | 0.047 ng/mL | LR 1.3–16.3 ng/mL; 96.50–103.61%; 0.99; RSD = 0.90–4.85% | |
| 2021, Guo [65] | FRET immunoassay | GO-imidaclothiz antigen (food safety monitoring) | 0.08 ng/mL, 0.262 | 3.1 × 10−10 | DR 0.08–50 ng/mL; -; 0.9934 |
| 2016, Li [66] | FRET immunoassay | BaP (environmental pollutant) | 0.06 ng/mL, 0.225 | 2.7 × 10−10 | LR 0.1–5 ng/mL; 80.5–87.0% and 92.9–92.1%; 0.9929 |
| 2020, Lee [67] | Single membrane-based LFIA | cardiac troponin I (cTnI) (cardiac marker) | 97 pg/mL, 24 | 4.0 × 10−12 | LR 0–1.16 ng/mL; -; 0.99; |
| 2021, Yang [68] | SERS/FI | carcinoembryonic antigen (cancer marker) | 0.1 pg/mL, 180 | 5.6 × 10−16 | LR 0.5–400 pg/mL; 105.50–106.73%; 0.9910 |
| 2019, Xie [69] | FI | aflatoxin B1 (mycotoxin) | 0.002 ng/mL, 0.312 | 6.4 × 10−12 | LR 0.01–0.5 ng/mL; 96.5–119.0% and 98.0–135.0%; 0.995; |
| 2018, Zvereva [70] | FPIA | Colchicine (inflammation marker?) | 1.8 ng/mL, 0.399 | 4.5 × 10−9 | DR 4.1–74.3 ng/mL; 79–108%; -; detection time 10 min |
| 2014, Wang [71] | TRFIA | diethylstilbestrol (environmental pollutant) | 0.4 fg/mL, 0.268 | 1.5 × 10−15 | LR 1.0 × 10−6–10 ng/mL; 88–105%; 0.9989 |
| 2019, Dong [72] | ELISA | Amantadine (food contamination) | 0.035 ng/mL, 0.151 | 2.3 × 10−10 | LR 0.048–1.1 ng/mL; 72.6%-90.4%; - |
| 2010, Sun [73] | FIA | 17β-oestradiol (environmental estrogen pollution) | 0.00542 ng/mL, 0.272 | 2.0 × 10−10 | LR 0.01–10,000 ng/mL; 86–113%; 0.9887 |
| 2021, Tian [74] | FPIA | α-fetoprotein (cancer biomarker) | 0.28 ng/mL, 66.478 | 4.2 × 10−12 | LR 0.5–500 ng/mL; 97.0–107.4%; 0.993; |
| carcinoembryonic antigen (cancer marker) | 0.36 ng/mL, 180 | 2.0 × 10−12 | LR 0.5–500 ng/mL; 93.0–104%; 0.996; | ||
| LOD Median, M | 1.5 × 10−11 | ||||
| Geometrical mean, M | 2.8 × 10−11 |
| Year, Authors | Analytical Technique (Objective, Excitation Laser, Assay Type) | Analyte | LOD (MW, kDa) | LOD (M) | Other FoM (Linear Range (LR)/Dynamic Range (DR)/Linear Dynamic Range (LDR), Recovery, RSD) |
|---|---|---|---|---|---|
| 2021, Qu [75] | SERS (-, 633 nm, Stagnant on solid substrate) | HIgG (human immunoglobulin G) | 0.1 μg/mL (150) | 6.7 × 10−10 | LR 0.1–200 μg/mL; 103.6–105.3%; 4.55–4.66% |
| 2013, Baniukevic [76] | SERS (20×, 785 nm, in solution) | gp51 (bovine leukemia virus antigen) | 0.95 μg/mL (51) | 1.9 × 10−8 | LR 0–0.06 mg/mL; 85.5–100%; 7.31–8.9%; LOQ 3.14 μg/mL; R2 = 0.9983 |
| 2019, Xiao [77] | SERS (-, 785 nm, LFIA) | H7N9 (avian influenza virus) | 0.0018 HAU | LR 0.0025–0.5 HAU; -; -; detection time 20 min | |
| 2018, Shi [78] | SERS (50×, 632.8 nm, LFIA) | NEO (neomycin) (broad spectrum antibiotic) | 0.216 pg/mL (0.615) | 3.5 × 10−13 | -; 89.7–105.6%; 2.4–5.3% (n = 3); IC50 0.04 ng/mL |
| 2020, Achadu [79] | SERS (sandwich-type immunoassay) (100⨯, 785 nm, Stagnant on solid substrate) | norovirus (NoV) (cause of gastritis and colitis) | 5.2 fg/mL | LR 10 fg/mL–100 ng/mL; -;7.95–8.67% | |
| 2020, Panikar [80] | SERS (50×, 785 nm, Stagnant on solid substrate) | B7-H6 (tumor biomarker) | 10.8 fg/mL | 1.0 × 10−13 M | NS 10−10–10−14 M; -; 4.8% |
| 2018, Bozkurt [81] | SERS (-, 785 nm, in solution) | E. coli (Bacteria detection) | 10 cfu/mL | LR 1.7 × 101–1.7 × 106 cfu/mL; -; -; R2 = 0.992 | |
| 2016, She [82] | SERS (-, 785 nm, Stagnant on solid substrate) | Hg2+ (hazardous pollutants in the environment) | 0.45 pg/mL (0.201) | 2.2 × 10−12 | DR 10−3–100 ng/mL; 88.3–107.3%; 1.5–9.5% (n = 3); IC50 0.12 ng/mL; |
| 2019, Ilhan [83] | SERS (10×, 785 nm, LFIA) | E. coli (Bacteria detection) | 0.52 cfu/mL | LR 101–107 cfu/mL; -; -; R2 = 0.984; LOQ = 1.57 cfu/mL | |
| 2020, Du [84] | SERS (-, 785 nm, Stagnant on solid substrate) | PSA (prostate cancer biomarker) | 1.871 pg/mL (27.76) | 6.7 × 10−14 | LR 10−4–10−12 g/mL; -; -; R2 = 0.987 |
| 2021, Wang [85] | SERS (-, 785 nm, Stagnant on solid substrate) | cardiac troponin I (cTnI) (cardiac marker) | 3.16 pg/mL (24) | 1.3 × 10−13 | LR 0.01–100 ng/mL; 94.9–121.6%; -; CV below 15% |
| 2021, Wang [85] | SERS (-, 785 nm, Stagnant on solid substrate) | creatine kinase isoenzyme MB (CK-MB) (cardiac marker) | 4.27 pg/mL (87) | 4.9 × 10−14 | LR 0.01–100 ng/mL; 94.9–121.6%; -; CV below 15% |
| 2016, Chang [86] | SERS (-, 532 nm, Stagnant on solid substrate) | PSA (prostate cancer biomarker) | 3.4 fM (27.76) | 3.4 × 10−15 | DR 0.001–1000 ng/mL; -; - |
| 2019, Kim [87] | SERS (20×, 633 nm, in solution) | botulinum neurotoxins (BoNTs) Type A (antitoxin) | 5.7 ng/mL (150) | 3.8 × 10−11 | DR 0 ng/mL–1.0 μg/mL; -; -; assay time less than 2 h |
| 2019, Kim [87] | SERS (20×, 633 nm, in solution) | botulinum neurotoxins (BoNTs) Type B (antitoxin) | 1.3 ng/mL (159) | 8.5 × 10−12 | DR 0 ng/mL–1.0 μg/mL; -; -; assay time less than 2 h |
| 2022, Kim [88] | SERS (-, 638 nm; 725 nm, Stagnant on solid substrate) | adiponectin (Gestational diabetes mellitus (GDM) biomarker) | 3.0 × 10–16 g/mL (30) | 1.0 × 10−17 | NS 10–15–10–6 g/mL; -; -; R2 = 0.994; |
| 2019, Gao [89] | SERS (10×, 632.8 nm, in solution) | PSA (prostate cancer biomarker) | 0.01 ng/mL (27.76) | 3.6 × 10−13 | LR 0.01–100 ng/mL -; - |
| 2014, Song [90] | SERS (-, 785 nm, Stagnant on solid substrate) | HIgG (defending against viruses or bacteria) | 1 fg/mL (150) | 6.7 × 10−18 | LR 1 fg/mL-1ng/mL; -; - |
| 2017, Choi [91] | SERS (20×, 632.8 nm, on substrate, in solution) | F1 antigen for Yersinia pestis (biological weapon) | 59.6 pg/mL (15.5) | 3.9 × 10−12 | -; -; -; R2 = 0.959; assay time 10 min |
| 2019, Yang [92] | SERS (10×, 633 nm, Stagnant on solid substrate) | AFP (liver, ovaries, testicles cancer biomarker) | 0.3 fg/mL (67.5) | 4.4 × 10−18 | LR 1 fg/mL–1 ng/mL; 94.36–102.12%; 8.21% |
| 2021, Tian [74] | SERS (-, 785 nm, in solution) | CA 19-9 (pancreatic cancer biomarker) | 5.65 × 10−4 IU/mL | LR 0.001–1000 IU/mL; -; 5.65% | |
| 2019, Huang [93] | SERS (-, 785 nm, Stagnant on solid substrate) | albumin (diabetic nephropathy and cardiovascular disease marker) | 0.2 mg/L (66.5) | 3.0 × 10−9 | LR 10–300 mg/L; -; - |
| 2016, Yang [94] | SERS (-, 632.8 nm, in solution) | chloramphenicol (CAP) (amide alcohols antibiotic) | 1.0 pg/mL (0.323) | 3.1 × 10−12 | DR 1–1 × 104 pg/mL; 81.42–96.92%; 9.7–14.4% |
| 2022, Jia [95] | SERS (-, 785 nm, LFIA) | ricin (biological warfare agent) | 0.1 ng/mL (65) | 1.5 × 10−12 | LDR 0.05–103 ng/mL; -; <2.5%; detection time 15 min |
| 2022, Jia [95] | SERS (-, 785 nm, LFIA) | SEB (biological warfare agent) | 0.05 ng/mL (28) | 1.8 × 10−12 | LDR 0.05–103 ng/mL; -; <4.75%; detection time 15 min |
| 2022, Jia [95] | SERS (-, 785 nm, LFIA) | BoNT/A (biological warfare agent) | 1 ng/mL (150) | 6.7 × 10−12 | LDR 0.05–103 ng/mL; -; <3.2%; detection time 15 min |
| 2022, Gao [96] | SERS (50×, 632.8 nm, in solution) | creatine kinase MB isoenzyme (CK-MB) (cardiac marker) | 7.92 pg/mL (87) | 9.1 × 10−14 | -; -; - |
| 2022, Gao [96] | SERS (50×, 632.8 nm, in solution) | cardiac troponin (cTnI) (cardiac marker) | 2.94 pg/mL (24) | 1.2 × 10−13 | -; -; - |
| 2023, Kaladharan [97] | SERS (-, 785 nm, Stagnant on solid substrate) | SARS-CoV-2 spike protein (COVID-19 marker) | 50 pg/mL | -; -; -; reaction time 20 min | |
| 2023, Kaladharan [97] | SERS (-, 785 nm, Stagnant on solid substrate) | SARS-CoV-2 virus-like-particle (VLP) (COVID-19 marker) | 50 pg/mL (in PBS); 400 pg/mL (untreated saliva) | LR 1000 ng/mL–100 pg/mL; -; -; reaction time 20 min | |
| 2023, Chen [98] | SERS (-, 785 nm, LFIA) | aflatoxin B1 (AFB1) (mycotoxin) | 0.24 pg/mL (0.312) | 7.7 × 10−13 | LDR 250 fg/mL–25 ng/mL; 91.0% ± 6.3–104.8% ± 5.6%; - |
| 2023, Chen [98] | SERS (-, 785 nm, LFIA) | ochratoxin A (OTA) (mycotoxin) | 0.37 pg/mL (0.404) | 9.2 × 10−13 | LDR 250 fg/mL–25 ng/mL; 87.0% ± 4.2–112.0% ± 3.3%; - |
| 2022, Mohammadi [99] | SERS (-, 785 nm, in solution) | SARS-CoV-2 (spike protein) (COVID-19 marker) | 4.7 fg/mL | -; -; -; R2 = 0.9932 | |
| 2013, Granger [24] | SERS (-, 632.8 nm, Stagnant on solid substrate) | MMP-7 (cancer marker) | 2.28 pg/mL | 7.9 × 10−14 | -; -; -; R2 = 0.989 |
| 2013, Granger [24] | SERS (-, 632.8 nm, Stagnant on solid substrate) | CA 19-9 (pancreatic cancer biomarker) | 34.5 pg/mL | 1.6 × 10−13 | -; -; -; R2 = 0.991 |
| 2011, Zhu [100] | SERS (-, 632.8 nm, Stagnant on solid substrate) | Clenbuterol (drug for the pulmonary disease) | 0.1 pg/mL (0.277) | 3.6 × 10−13 | DR 0.1–100 pg/mL; 96.9–116.5%; 9.1% |
| 2019, Fu [101] | SERS (20×, 633 nm, in solution) | Cardiac troponin I (cTnI) (cardiac marker) | 5 pg/mL | 2.1 × 10−13 (24) | LR 0.01–1000 ng/mL; -; - |
| 2023, Tuckmantel Bido [102] | SERS (50×, 632.8 nm, Stagnant on solid substrate) | SARS-CoV-2 spike protein (COVID-19 marker) | 34.9 pM | 3.5 × 10−11 | -; -; -; LOQ = 105.7 pM |
| 2023, Xie [103] | SERS (-, -, in solution) | IL-6 (ovarian cancer biomarker) | 0.028 pg/mL (21) | 1.3 × 10−15 | LR 0.1–1000 pg/mL; 80–117%; 5.8–10.9% |
| 2022, Yin [104] | SERS (-, 532, 638 and 785 nm, LFIA) | Zearalenone (ZEN) (mycotoxin) | 3.6 μg/kg (0.318) | -; 86.06–111.23%; 8.45∼11.37%; coincidence rate 86.06–111.23%; | |
| 2017, Cheng [105] | SERS (20×, 633 nm, in solution) | free PSA (f-PSA) (prostate cancer biomarker) | 0.012 ng/mL (27.76) | 4.3 × 10−13 | LR 4.0–10.0 ng/mL; -; -; assay time < 1 h |
| 2017, Cheng [105] | SERS (20×, 633 nm, in solution) | complexed PSA (c-PSA) (prostate cancer biomarker) | 0.15 ng/mL (27.76) | 5.4 × 10−12 | LR 4.0–10.0 ng/mL; -; -; assay time < 1 h |
| 2022, Jiao [106] | SERS (-, 785 nm, Stagnant on solid substrate) | ochratoxin A (OTA) (mycotoxin) | 2.46 pg/mL (0.404) | 6.1 × 10−12 | LR 0.001–10 ng/mL; 77.68–104.69%; -; assay time 90 min |
| 2022, Jiao [106] | SERS (-, 785 nm, Stagnant on solid substrate) | fumonisin B1 (FB1) (mycotoxin) | 0.20 pg/mL (0.722) | 2.8 × 10−13 | LR 0.001–10 ng/mL; 76.39–117.73%; -; assay time 90 min |
| 2022, Jiao [106] | SERS (-, 785 nm, Stagnant on solid substrate) | deoxynivalenol (DON) (mycotoxin) | 68.98 pg/mL (0.296) | 3.3 × 10−11 | LR 0.1–1000 ng/mL; 70.95–113.16%; -; assay time 90 min |
| 2022, Cha [107] | SERS (20×, 633 nm, in solution) | SARS-CoV-2 (COVID-19 marker) | 2.56 fg/mL | -; -; -; detection time < 30 min | |
| 2015, Ko [108] | SERS (-, 633 nm, in solution) | aflatoxin B1 (AFB1) (mycotoxin) | 0.1 ng/mL (0.312) | 3.2 × 10−10 | -; -; -; analysis time < 30 min |
| 2017, Yang [109] | SERS (-, 532 nm, Stagnant on solid substrate) | α-fetoprotein (AFP) (liver, ovaries, testicles cancer biomarker) | 0.081 pg/mL (67.5) | 1.2 × 10−15 | LR 0.5–100 pg/mL; -; 3.4–7.4% |
| 2021, Yang [110] | SERS (50×, 785 nm, in solution) | amantadine (AMD) (broad-spectrum antiviral drug) | 0.0038 μg/L (0.151) | 2.5 × 10−11 | LR 0.01–50 μg/L; 82.0–106.0%; 4.7–9.6%; detection time 30 min |
| 2022, Liu [111] | SERS (-, 785 nm, LFIA) | acrosomal protein SP10 (sperm concentration detection biomarker) | 25.12 fg/mL | LDR 100 fg/mL–10 ng/mL; 92.5–110.9%; 6% | |
| 2019, Deng [112] | SERS (-, 785 nm, stagnant on solid substrate) | pharmaceutical diclofenac (DCF) (non-steroidal anti-inflammatory drug) | 0.07 pg/mL (0.334) | 2.1 × 10−13 | -; 92.5–106.0%; 4.5–5.7%; IC50 = 9 pg/mL; detection time 15 min; |
| median (30), M | 4.32 × 10−13 | ||||
| average (30), M | 5.85 × 10−10 | ||||
| geometric mean (30), M | 5.38 × 10−13 |
| Type of Assay (Total Number of Works, Number of Works Involved in Calculations) | Median LOD, M | Geometric Mean of LOD, M |
|---|---|---|
| Stagnant on solid substrate (18, 16) | 1.2 × 10−13 | 8.5 × 10−14 |
| LFIA (7, 3) | 1.2 × 10−12 | 1.3 × 10−12 |
| In solution (15, 11) | 3.1 × 10−12 | 2.6 × 10−12 |
| Year, Authors | Analytical Technique | Analyte | Sensitivity | Specificity | Accuracy | Number of Samples | Other FoM |
|---|---|---|---|---|---|---|---|
| 2021, Liu [114] | SERS LFIA | IgG; IgM (defending against viruses or bacteria) | 100% | 100% | 68 (19+; 49−) | AUC: IgM + IgG (1–0.997); IgM (0.997–0.941) and IgG alone (0.986–0.977) | |
| 2021, Banaei [115] | SERS | extracellular vesicles (EVs) (delivery of biomolecules to recipient cells) | 95% | 96% | - | 15 (10+; 5−) | |
| 2019, Zamora-Mendoza [116] | SERS | proteins (IL-8, IL-10, sCD163) (immunomodulatory cytokine) | 85 | 82 | 84 | 44 (26+; 18−) | |
| 2022, Li [117] | SERS | LRG1-positive exosomes (LRG1-Exos) and GPC1-positive exosomes (GPC1-Exos) | 91.4% | 86.7% | - | 50 (15 normal, 35 tumor) | AUC of the 2-molecule panel 0.95; LOD 15 particles/µL |
| 2020, Lu [118] | SERS LFIA | AFP (ovaries, testicles, liver cancer biomarker) | - | 100 | 19 (+ − not given) | LOD 9.2 pg/mL; range 10 pg/mL-500 ng/mL | |
| 2018, Li [119] | SERS IA | pancreatic cancer derived exosomes | 95.7 | - | - | 103 (71+, 32−) | LOD 9 × 10−19 M |
| 2018, Jia [120] | SERS LFIA | human IgM (pathogens neutralization) | - | - | 100 | 20 (all MP-specific IgM positive) | LOD 0.1 ng/mL; detection rate = 100% |
| 2022, Lu [121] | SERS LFIA | Influenza (virus) | 100 | 100 | 100 | 39(28 SARS-CoV-2 +; 6 influenza +; 5−) | LOD = 23 HAU/mL |
| SARS-CoV-2 (COVID-19 marker) | LOD = 5.2 PFU/mL | ||||||
| 2019, Lee [122] | SERS LFIA | O. tsutsugamushi IgG (defending against viruses or bacteria) | 100 | 100 | 100 | 40 (16+; 24−) | - |
| median, M (Number of publications) | 95.35 (6) | 98 (6) | 100 (6) | ||||
| 2019, Squire [123] | FIA | NT-proBNP (diagnostic screening) | 65 | 93 | 78 | 40 | range 0–100 pg/mL; R2 = 0.86 |
| 2021, Lu [124] | TF-LFIA | brucella antibody (infectious disease detection) | 98.57 | 100 | 99.63 | LOD = 0.3 IU/mL; LLOQ = 1.6 IU/mL; R2 = 0.9961 | |
| 2019, Lee [122] | IFA | O. tsutsugamushi IgG (scrub typhus biomarker) | 96 | 94 | 95 | 40 (16+; 24−) | |
| 2015, Radon [125] | GeLC-MS/MS analysis, immunoassay—ELISA | LYVE-1, REG1A, and TFF1 (prostate cancer) | 76.9% | 86.8% | - | 279 (192+, 87−) | |
| 2017, Wang [126] | LC-MS, immunoassay—ELISA | Flotilin-2, PARK7 (Prostate cancer) | 68% | 93% | 42 (26+, 16−) | ||
| 2019, Shimur [127] | Immunoassay—ELISA | uCyr61, uTFF3, (Colorectal cancer) | 75.5% | 69.8% | 258 (148+, 110−) | ||
| 2016, Gomez [128] | FIA | Influenza A (respiratory virus) | 75.3% | 98.6% | 1057 | ||
| 2016, Gomez [128] | FIA | Influenza B (respiratory virus) | 50.0% | 92.4% | 1057 | ||
| 2016, Gomez [128] | FIA | RSV (respiratory virus) | 92.1% | 91.8% | 261 | ||
| 2021, Daag [129] | Immunoglobulin fluorescence immunoassay | Dengue serostatus (dengue fever detection) | 95% | 98% | 1000 | ||
| median, M (number of publications) | 76.2 (8) | 93 (8) | 95 (3) |
| Analyte | SERS | Fluorescence | Year, Author | ||
|---|---|---|---|---|---|
| Median LOD, M (Number of Works) | Geometric Mean LOD, M | Median LOD, M | Geometric Mean LOD, M | ||
| Aflatoxin B1 (AFB1) | 1.6 × 10−10 (2) | 1.6 × 10−11 (2) | 3.3 × 10−10 (2) | 6.5 × 10−11 (2) | 2023, Chen [98]; 2015, Ko [108]; 2020, Su [62]; 2019, Xie [69] |
| Ochratoxin A (OTA) | 3.5 × 10−12 (2) | 2.4 × 10−12 (2) | 3.6 × 10−13 (2) | 3.5 × 10−13 (2) | 2023, Chen [98]; 2022, Jiao [106]; 2022, Chen [39]; 2020, Wang [56] |
| Alpha-fetoprotein (AFP) | 6.0 × 10−16 (2) | 7.3 × 10−17 (2) | 3.2 × 10−12 (5) | 2.0 × 10−12 (5) | 2019, Yang [92]; 2017, Yang [109]; 2021, Sun [36]; 2017, Wang [38]; 2018, Chen [40]; 2019, Zhao [47]; 2021, Tian [74] |
| Cardiac troponin I (cTnI) | 1.3 × 10−13 (3) | 1.6 × 10−13 (3) | 2.3 × 10−11 (2) | 1.3 × 10−11 (2) | 2021, Wang [85]; 2022, Gao [96]; 2019, Fu [101]; 2020, Liu [51]; 2020, Lee [67] |
| Median Value, M | 1.82 × 10−12 (9) | 1.28 × 10−12 (9) | 1.31 × 10−11 (11) | 7.50 × 10−12 (11) | |
| Method (Number of Works Involved in Calculations) | Average RSD, % | Median LOD, M |
|---|---|---|
| SERS-based IA (13) | 6.2 ± 1.9 | 1.5 × 10−12 |
| FIA (5) | 5.4 ± 2.4 | 2.2 × 10−11 |
| Challenges | Solutions |
|---|---|
| The challenges and toxicity of the synthesis of QD probes for fluorescent immunoassays with the use of heavy metals such as cadmium and lead [41]. | The use of carbon dots (CDs) with low costs, simplicity of preparation, good photostability, and lower toxicity [41]. |
| Comparatively low sensitivity of the fluorescence immunoassays [52]. | Use of alternative signal markers, such as fluorescent microspheres, carbon-based nanoparticles (NPs), and SERS nanomaterials [52]. |
| Autofluorescence of the quantum dots from different substances [52]. | Photobleaching of the quantum dots to reduce autofluorescence [146]. |
| Challenges | Solutions |
|---|---|
| Significant costs of gold film-covered substrates. Relatively high non-specific protein binding onto gold film and possible contamination of gold film with S-containing compounds | Use the Al and Si cost-effective substrates [32,137]. |
| Relatively long assay time for stationary assays and sometimes relatively slow Raman measurements/readout | Use a wide-field Raman spectrometer; perform the analysis using the rotating substrate, microdroplet channels, strips and other alternative substrates, which significantly decrease the assay time [91,95,153,154]. |
| Detection environment, including solution, temperature, and pH, may introduce an interfering species [154]. | Use isolated systems like microdroplet channels and control the detection environment [91]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terzapulo, X.; Kassenova, A.; Bukasov, R. Immunoassays: Analytical and Clinical Performance, Challenges, and Perspectives of SERS Detection in Comparison with Fluorescent Spectroscopic Detection. Int. J. Mol. Sci. 2024, 25, 2080. https://doi.org/10.3390/ijms25042080
Terzapulo X, Kassenova A, Bukasov R. Immunoassays: Analytical and Clinical Performance, Challenges, and Perspectives of SERS Detection in Comparison with Fluorescent Spectroscopic Detection. International Journal of Molecular Sciences. 2024; 25(4):2080. https://doi.org/10.3390/ijms25042080
Chicago/Turabian StyleTerzapulo, Xeniya, Aiym Kassenova, and Rostislav Bukasov. 2024. "Immunoassays: Analytical and Clinical Performance, Challenges, and Perspectives of SERS Detection in Comparison with Fluorescent Spectroscopic Detection" International Journal of Molecular Sciences 25, no. 4: 2080. https://doi.org/10.3390/ijms25042080
APA StyleTerzapulo, X., Kassenova, A., & Bukasov, R. (2024). Immunoassays: Analytical and Clinical Performance, Challenges, and Perspectives of SERS Detection in Comparison with Fluorescent Spectroscopic Detection. International Journal of Molecular Sciences, 25(4), 2080. https://doi.org/10.3390/ijms25042080







