Differential Expression of MicroRNA MiR-145 and MiR-155 Downstream Targets in Oral Cancers Exhibiting Limited Chemotherapy Resistance
Abstract
:1. Introduction
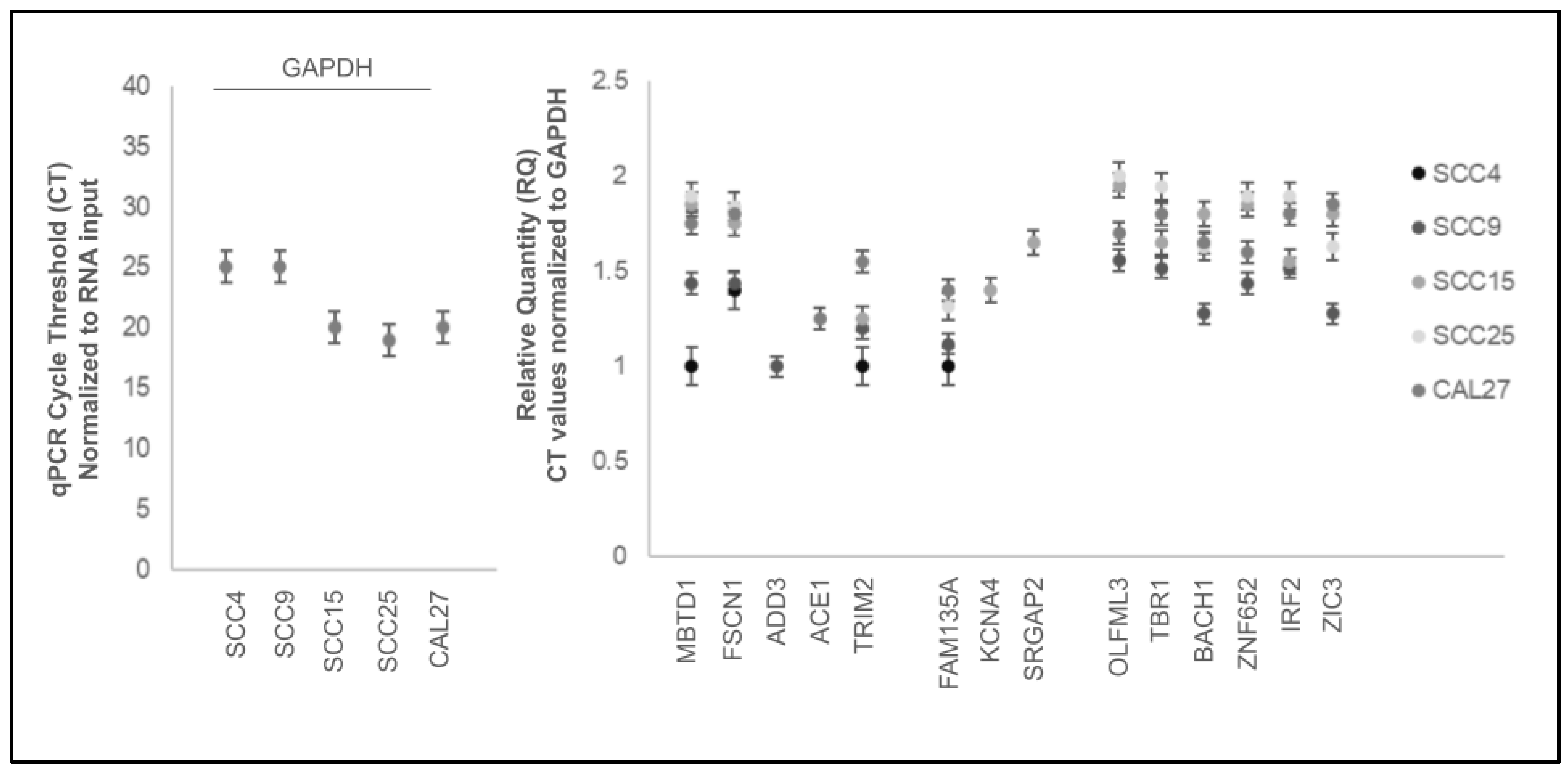
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Experimental Chemotherapy Agents
4.3. Proliferation Assays and Statistical Analyses
4.4. RNA Extraction
4.5. microRNA cDNA Synthesis
4.6. qPCR Screening
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424, Erratum in CA Cancer J. Clin. 2020, 70, 313. [Google Scholar] [CrossRef] [PubMed]
- Gómez, I.; Seoane, J.; Varela-Centelles, P.; Diz, P.; Takkouche, B. Is diagnostic delay related to advanced-stage oral cancer? A meta-analysis. Eur. J. Oral Sci. 2009, 117, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Varela-Centelles, P.; López-Cedrún, J.; Fernández-Sanromán, J.; Seoane-Romero, J.; de Melo, N.S.; Álvarez-Nóvoa, P.; Gómez, I.; Seoane, J. Key points and time intervals for early diagnosis in symptomatic oral cancer: A systematic review. Int. J. Oral Maxillofac. Surg. 2016, 46, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Varela-Centelles, P.; Seoane, J.; Lopez-Cedrun, J.; Fernandez-Sanroman, J.; García-Martin, J.; Takkouche, B.; Alvarez-Novoa, P.; Seoane-Romero, J. The length of patient and primary care time interval in the pathways to treatment in symptomatic oral cancer. A quantitative systematic review. Clin. Otolaryngol. 2018, 43, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Furness, S.; Glenny, A.-M.; Worthington, H.V.; Pavitt, S.; Oliver, R.; Clarkson, J.E.; Macluskey, M.; Chan, K.K.; Conway, D.I. Interventions for the treatment of oral cavity and oropharyngeal cancer: Chemotherapy. Cochrane Database Syst. Rev. 2011, 4, CD006386, Update in Cochrane Database Syst. Rev. 2021. [Google Scholar] [CrossRef]
- Sykes, E.A.; Weisbrod, N.; Rival, E.; Haque, A.; Fu, R.; Eskander, A. Methods, Detection Rates, and Survival Outcomes of Screening for Head and Neck Cancers: A Systematic Review. JAMA Otolaryngol. Neck Surg. 2023, 149, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Worthington, H.V.; Bulsara, V.M.; Glenny, A.-M.; Clarkson, J.E.; Conway, D.I.; Macluskey, M. Interventions for the treatment of oral cavity and oropharyngeal cancers: Surgical treatment. Emergencias 2023, 12, CD006205. [Google Scholar] [CrossRef]
- Bungum, A.; Jensen, J.S.; Jakobsen, K.K.; Christensen, A.; Grønhøj, C.; von Buchwald, C. Impact of surgical resection margins less than 5 mm in oral cavity squamous cell carcinoma: A systematic review. Acta Oto-Laryngologica 2020, 140, 869–875. [Google Scholar] [CrossRef]
- De Koning, S.B.; Schaeffers, A.; Schats, W.; Brekel, M.v.D.; Ruers, T.; Karakullukcu, M. Assessment of the deep resection margin during oral cancer surgery: A systematic review. Eur. J. Surg. Oncol. 2021, 47, 2220–2232. [Google Scholar] [CrossRef]
- Glenny, A.M.; Furness, S.; Worthington, H.V.; Conway, D.I.; Oliver, R.; Clarkson, J.E.; Macluskey, M.; Pavitt, S.; Chan, K.K.; Brocklehurst, P.; et al. Interventions for the treatment of oral cavity and oropharyngeal cancer: Radiotherapy. Cochrane Database Syst. Rev. 2010, 12, CD006387. [Google Scholar] [CrossRef]
- Lau, A.; Li, K.-Y.; Yang, W.-F.; Su, Y.-X. Induction chemotherapy for squamous cell carcinomas of the oral cavity: A cumulative meta-analysis. Oral Oncol. 2016, 61, 104–114. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, Z.; Yuan, L.; Xu, S.; Yao, Z.; Qiao, L.; Li, K. Paclitaxel plus cisplatin vs. 5-fluorouracil plus cisplatin as first-line treatment for patients with advanced squamous cell esophageal cancer. Am. J. Cancer Res. 2016, 6, 2345–2350. [Google Scholar]
- Feng, Y.M.; Yang, D.-S.M.; Tang, H.-B.M.; Ding, Y.-S.M.; Li, X.-G.M. Efficacy and safety of cisplatin for the management of adult patients with oral cancer: A protocol for systematic review. Medicine 2019, 98, e18210. [Google Scholar] [CrossRef]
- Chan, K.K.; Glenny, A.-M.; Weldon, J.C.; Furness, S.; Worthington, H.V.; Wakeford, H. Interventions for the treatment of oral and oropharyngeal cancers: Targeted therapy and immunotherapy. Cochrane Database Syst. Rev. 2015, 2015, CD010341. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Foster, N.R.; Farrell, A.; Le-Lindqwister, N.A.; Mathew, J.; Costello, B.; Reynolds, J.; Meyers, J.P.; Jatoi, A. Oral cancer chemotherapy adherence and adherence assessment tools: A report from North Central Cancer Group Trial N0747 and a systematic review of the literature. J. Cancer Educ. 2013, 28, 770–776. [Google Scholar] [CrossRef]
- Correa, M.E.P.; Cheng, K.K.F.; Chiang, K.; Kandwal, A.; Loprinzi, C.L.; Mori, T.; Potting, C.; Rouleau, T.; Toro, J.J.; Ranna, V.; et al. Systematic review of oral cryotherapy for the management of oral mucositis in cancer patients and clinical practice guidelines. Support. Care Cancer 2020, 28, 2449–2456. [Google Scholar] [CrossRef]
- Behera, M.; Owonikoko, T.K.; Kim, S.; Chen, Z.; Higgins, K.; Ramalingam, S.S.; Shin, D.M.; Khuri, F.R.; Beitler, J.J.; Saba, N.F. Concurrent therapy with taxane versus non-taxane containing regimens in locally advanced squamous cell carcinomas of the head and neck (SCCHN): A systematic review. Oral Oncol. 2014, 50, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Lala, M.; Chirovsky, D.; Cheng, J.D.; Mayawala, K. Clinical outcomes with therapies for previously treated recurrent/metastatic head-and-neck squamous cell carcinoma (R/M HNSCC): A systematic literature review. Oral Oncol. 2018, 84, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, Q.; Zhang, Y.; Wei, J.; Wang, B.; Zheng, Z.; Liu, S.; Liu, Z.; Meng, L.; Xin, Y.; et al. Efficacy and safety of systemic treatments for patients with recurrent/metastatic head and neck squamous cell carcinoma: A systematic review and network meta-analysis. Pharmacol. Res. 2021, 173, 105866. [Google Scholar] [CrossRef]
- Sha, J.; Bai, Y.; Ngo, H.X.; Okui, T.; Kanno, T. Overview of Evidence-Based Chemotherapy for Oral Cancer: Focus on Drug Resistance Related to the Epithelial-Mesenchymal Transition. Biomolecules 2021, 11, 893. [Google Scholar] [CrossRef]
- Atashi, F.; Vahed, N.; Emamverdizadeh, P.; Fattahi, S.; Paya, L. Drug resistance against 5-fluorouracil and cisplatin in the treatment of head and neck squamous cell carcinoma: A systematic review. J. Dent. Res. Dent. Clin. Dent. Prospects 2021, 15, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Khera, N.; Rajkumar, A.S.; Alkurdi, K.A.M.; Liu, Z.; Ma, H.; Waseem, A.; Teh, M.-T. Identification of multidrug chemoresistant genes in head and neck squamous cell carcinoma cells. Mol. Cancer 2023, 22, 146. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, S.; Yu, D. Metabolic Reprogramming of Chemoresistant Cancer Cells and the Potential Significance of Metabolic Regulation in the Reversal of Cancer Chemoresistance. Metabolites 2020, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.; Pruthi, N.; Seers, C.; Belobrov, S.; McCullough, M.; Celentano, A. Extracellular Vesicles in Oral Squamous Cell Carcinoma and Oral Potentially Malignant Disorders: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 1197. [Google Scholar] [CrossRef]
- Karkhane, M.; Lashgarian, H.E.; Hormozi, M.; Fallahi, S.; Cheraghipour, K.; Marzban, A. Oncogenesis and Tumor Inhibition by MicroRNAs and its Potential Therapeutic Applications: A Systematic Review. MicroRNA 2020, 9, 198–215. [Google Scholar] [CrossRef]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, C.; Pang, Q.; Liu, H.C. MicroRNA-217: A regulator of human cancer. Biomed. Pharmacother. 2021, 133, 110943. [Google Scholar] [CrossRef]
- Xiao, W.; Zhong, Y.; Wu, L.; Yang, D.; Ye, S.; Zhang, M. Prognostic value of microRNAs in lung cancer: A systematic review and meta-analysis. Mol. Clin. Oncol. 2019, 10, 67–77. [Google Scholar] [CrossRef]
- Padroni, L.; De Marco, L.; Fiano, V.; Milani, L.; Marmiroli, G.; Giraudo, M.T.; Macciotta, A.; Ricceri, F.; Sacerdote, C. Identifying MicroRNAs Suitable for Detection of Breast Cancer: A Systematic Review of Discovery Phases Studies on MicroRNA Expression Profiles. Int. J. Mol. Sci. 2023, 24, 15114. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, I.L.; Penna, K.G.B.D.; Carneiro, M.A.d.S.; Libera, L.S.D.; Ramos, J.E.P.; Saddi, V.A. Tissue micro-RNAs associated with colorectal cancer prognosis: A systematic review. Mol. Biol. Rep. 2021, 48, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Al Rawi, N.; Elmabrouk, N.; Abu Kou, R.; Mkadmi, S.; Rizvi, Z.; Hamdoon, Z. The role of differentially expressed salivary microRNA in oral squamous cell carcinoma. A systematic review. Arch. Oral Biol. 2021, 125, 105108. [Google Scholar] [CrossRef]
- Setti, G.; Pezzi, M.E.; Viani, M.V.; Pertinhez, T.A.; Cassi, D.; Magnoni, C.; Bellini, P.; Musolino, A.; Vescovi, P.; Meleti, M. Salivary MicroRNA for Diagnosis of Cancer and Systemic Diseases: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 907. [Google Scholar] [CrossRef]
- Dinesh, Y.; Ramani, P.; Yuwanati, M.; Ramalingam, K.; Gheena, S. MicroRNA Profiling in Circulating Exosomes in Oral Squamous Cell Carcinoma: A Systematic Review. Cureus 2023, 15, e43235. [Google Scholar] [CrossRef]
- Palaia, G.; Pippi, R.; Rocchetti, F.; Caputo, M.; Macali, F.; Mohsen, A.; Del Vecchio, A.; Tenore, G.; Romeo, U. Liquid biopsy in the assessment of microRNAs in oral squamous cell carcinoma: A systematic review. J. Clin. Exp. Dent. 2022, 14, e875–e884. [Google Scholar] [CrossRef]
- Dioguardi, M.; Spirito, F.; Sovereto, D.; Alovisi, M.; Troiano, G.; Aiuto, R.; Garcovich, D.; Crincoli, V.; Laino, L.; Cazzolla, A.P.; et al. MicroRNA-21 Expression as a Prognostic Biomarker in Oral Cancer: Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2022, 19, 3396. [Google Scholar] [CrossRef] [PubMed]
- Dioguardi, M.; Spirito, F.; Sovereto, D.; Alovisi, M.; Aiuto, R.; Garcovich, D.; Crincoli, V.; Laino, L.; Cazzolla, A.P.; Caloro, G.A.; et al. The Prognostic Role of miR-31 in Head and Neck Squamous Cell Carcinoma: Systematic Review and Meta-Analysis with Trial Sequential Analysis. Int. J. Environ. Res. Public Health 2022, 19, 5334. [Google Scholar] [CrossRef] [PubMed]
- Dioguardi, M.; Spirito, F.; Sovereto, D.; La Femina, L.; Campobasso, A.; Cazzolla, A.P.; Di Cosola, M.; Zhurakivska, K.; Cantore, S.; Ballini, A.; et al. Biological Prognostic Value of miR-155 for Survival Outcome in Head and Neck Squamous Cell Carcinomas: Systematic Review, Meta-Analysis and Trial Sequential Analysis. Biology 2022, 11, 651. [Google Scholar] [CrossRef] [PubMed]
- Dioguardi, M.; Cantore, S.; Sovereto, D.; La Femina, L.; Caloro, G.A.; Spirito, F.; Scacco, S.; Di Cosola, M.; Muzio, L.L.; Troiano, G.; et al. Potential Role of miR-196a and miR-196b as Prognostic Biomarkers of Survival in Head and Neck Squamous Cell Carcinoma: A Systematic Review, Meta-Analysis and Trial Sequential Analysis. Life 2022, 12, 1269. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Polpaya, K.; Kunale, M.; Muthukaliannan, G.K.; Shetty, S.; Baxi, S.; Mani, R.R.; Paranjothy, C.; Purushothaman, V.; Kayarohanam, S.; et al. Clinical Investigation of Chemotherapeutic Resistance and miRNA Expressions in Head and Neck Cancers: A Thorough PRISMA Compliant Systematic Review and Comprehensive Meta-Analysis. Genes 2022, 13, 2325. [Google Scholar] [CrossRef]
- Zhuang, Z.; Hu, F.; Hu, J.; Wang, C.; Hou, J.; Yu, Z.; Wang, T.T.; Liu, X.; Huang, H. MicroRNA-218 promotes cisplatin resistance in oral cancer via the PPP2R5A/Wnt signaling pathway. Oncol. Rep. 2017, 38, 2051–2061. [Google Scholar] [CrossRef]
- Chikuda, J.; Otsuka, K.; Shimomura, I.; Ito, K.; Miyazaki, H.; Takahashi, R.-U.; Nagasaki, M.; Mukudai, Y.; Ochiya, T.; Shimane, T.; et al. CD44s Induces miR-629-3p Expression in Association with Cisplatin Resistance in Head and Neck Cancer Cells. Cancers 2020, 12, 856. [Google Scholar] [CrossRef]
- Zheng, X.; Li, J.; Peng, C.; Zhao, J.; Chi, J.; Meng, X.; Yun, X.; Li, D.; Yu, Y.; Gao, M.; et al. MicroRNA-24 induces cisplatin resistance by targeting PTEN in human tongue squamous cell carcinoma. Oral Oncol. 2015, 51, 998–1003. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, H.; Yao, B.; Helms, J. miR-15b inhibits cancer-initiating cell phenotypes and chemoresistance of cisplatin by targeting TRIM14 in oral tongue squamous cell cancer. Oncol. Rep. 2017, 37, 2720–2726. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Cao, G.; Dong, Z.; Guo, T. Effect of microRNA-27b on cisplatin chemotherapy sensitivity of oral squamous cell carcinoma via FZD7 signaling pathway. Oncol. Lett. 2019, 18, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Sayyed, A.A.; Gondaliya, P.; Mali, M.; Pawar, A.; Bhat, P.; Khairnar, A.; Arya, N.; Kalia, K. MiR-155 Inhibitor-Laden Exosomes Reverse Resistance to Cisplatin in a 3D Tumor Spheroid and Xenograft Model of Oral Cancer. Mol. Pharm. 2021, 18, 3010–3025. [Google Scholar] [CrossRef] [PubMed]
- Coon, J.; Kingsley, K. Assessment of MicroRNA (miR)-365 Effects on Oral Squamous Carcinoma Cell Line Phenotypes. Biomolecules 2021, 11, 874. [Google Scholar] [CrossRef] [PubMed]
- Huni, K.C.; Cheung, J.; Sullivan, M.; Robison, W.T.; Howard, K.M.; Kingsley, K. Chemotherapeutic Drug Resistance Associated with Differential miRNA Expression of miR-375 and miR-27 among Oral Cancer Cell Lines. Int. J. Mol. Sci. 2023, 24, 1244. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Y.; Peng, C.-Y.; Lee, S.-S.; Chou, M.-Y.; Yu, C.-C.; Chang, Y.-C. Acquisition cancer stemness, mesenchymal transdifferentiation, and chemoresistance properties by chronic exposure of oral epithelial cells to arecoline. Oncotarget 2016, 7, 84072–84081. [Google Scholar] [CrossRef]
- Abdolrahmani, A.; Khoozestani, N.K.; Azmoudeh-Ardalan, F.; Shamshiri, A.R. Prognostic impact of MUC1 and potential regulatory miR-145 and miR-21 expression in salivary mucoepidermoid carcinoma. Head Neck Pathol. 2022, 16, 1134–1145. [Google Scholar] [CrossRef]
- Melling, G.E.; Flannery, S.E.; Abidin, S.A.; Clemmens, H.; Prajapati, P.; Hinsley, E.E.; Hunt, S.; Catto, J.W.F.; Coletta, R.D.; Mellone, M.; et al. A miRNA-145/TGF-β1 negative feedback loop regulates the cancer-associated fibroblast phenotype. Carcinogenesis 2018, 39, 798–807, Erratum in Carcinogenesis 2018, 39, 1094. [Google Scholar] [CrossRef]
- Zhou, J.; Jin, S. Circ_0058063 Contributed to Oral Squamous Cell Carcinoma Development by Sponging miR-145 and Regulating PI3K/AKT Pathway. Mol. Biotechnol. 2023, 65, 2049–2060. [Google Scholar] [CrossRef]
- Shao, Y.; Qu, Y.; Dang, S.; Yao, B.; Ji, M. MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int. 2013, 13, 51. [Google Scholar] [CrossRef]
- Gao, L.; Ren, W.; Chang, S.; Guo, B.; Huang, S.; Li, M.; Guo, Y.; Li, Z.; Song, T.; Zhi, K.; et al. Downregulation of miR-145 Expression in Oral Squamous Cell Carcinomas and Its Clinical Significance. Oncol. Res. Treat. 2013, 36, 194–199. [Google Scholar] [CrossRef]
- Patel, A.; Patel, P.; Mandlik, D.; Patel, K.; Malaviya, P.; Johar, K.; Swamy, K.B.; Patel, S.; Tanavde, V. A novel 3-miRNA network regulates tumour progression in oral squamous cell carcinoma. Biomark. Res. 2023, 11, 64. [Google Scholar] [CrossRef]
- Singh, A.; Khan, D.-U.; Singh, P.; Singh, A.K.; Agarwal, P. Prognostic utility of microRNA-145 and CD 133 in oral squamous cell carcinoma: A pilot study from Northern India. J. Oral Biol. Craniofacial Res. 2023, 13, 92–95. [Google Scholar] [CrossRef]
- Lu, Q.; Che, H.; Che, Y.; Hu, M. CircZNF236 facilitates malignant progression in oral squamous cell carcinoma by sequestering miR-145-5p. Clin. Transl. Oncol. 2023, 25, 1690–1701. [Google Scholar] [CrossRef]
- Luo, Y.; Liu, F.; Guo, J.; Gui, R. Upregulation of circ_0000199 in circulating exosomes is associated with survival outcome in OSCC. Sci. Rep. 2020, 10, 13739. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Huang, F.; Zhang, Y.; Zhou, Z. Knockdown of circGOLPH3 inhibits cell progression and glycolysis by targeting miR-145-5p/lysine demethylase 2A (KDM2A) axis in oral squamous cell carcinoma. Head Neck 2023, 45, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Song, J.; Wei, H.; Tang, Z.; Li, X.; Lv, X.; Luo, H.; Wu, S.; Zou, C. circ_0001461 promotes oral squamous cell carcinoma progression through miR-145/TLR4/NF-κB axis. Biochem. Biophys. Res. Commun. 2021, 566, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lou, Y.; Hou, M.; Ma, X.; Wang, L. Circ_0058063 contributes to oral squamous cell carcinoma development by sponging miR-145-5p and upregulating SERPINE1. J. Oral Pathol. Med. 2022, 51, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Guo, M.; Yao, L.; Deng, Z. Circular RNA hsa_circ_0033144 (CircBCL11B) regulates oral squamous cell carcinoma progression via the miR-579/LASP1 axis. Bioengineered 2021, 12, 4111–4122. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Hu, M.; Gao, Y.; Wang, Y.; Yuan, X.; Yang, Y.; Song, W.; Yin, W.; Gong, P.; Wei, L.; et al. LncRNA MIR17HG Suppresses Breast Cancer Proliferation and Migration as ceRNA to Target FAM135A by Sponging miR-454-3p. Mol. Biotechnol. 2023, 65, 2071–2085. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Ye, Z.; Xu, X.; Jiang, M.; Lu, R.; Jing, D.; Zhang, W.; Gao, H.; Wang, F.; Zhang, Y.; et al. Establishment and characterization of the third non-functional human pancreatic neuroendocrine tumor cell line. Hum. Cell 2022, 35, 1248–1261. [Google Scholar] [CrossRef] [PubMed]
- Mathur, Y.; Shafie, A.; Alharbi, B.; Ashour, A.A.; Abu Al-Soud, W.; Alhassan, H.H.; Alharethi, S.H.; Anjum, F. Genome-Wide Analysis of Kidney Renal Cell Carcinoma: Exploring Differentially Expressed Genes for Diagnostic and Therapeutic Targets. OMICS 2023, 27, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, G.; Jia, Y.; Sun, T. SRGAP2 controls colorectal cancer chemosensitivity via regulation of mitochondrial complex I activity. Hum. Cell 2022, 35, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qiao, L.; Bai, Y.; Xiao, C.; Wu, J.; Gao, X.; Qiao, C.; Shi, Y.; Hou, W.; Wang, J.; et al. Identification of SRGAP2 as a potential oncogene and a prognostic biomarker in hepatocellular carcinoma. Life Sci. 2021, 277, 119592. [Google Scholar] [CrossRef] [PubMed]
- Lucas, B.; Hardin, J. Mind the (sr)GAP—Roles of Slit–Robo GAPs in neurons, brains and beyond. J. Cell Sci. 2017, 130, 3965–3974, Erratum in J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Tao, X.; Huang, F.; Wu, T.; Wang, J.; Jiang, X.; Kuang, Z.; Cheng, B. Overexpression of miR-155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/cyclin D2. Int. J. Mol. Med. 2016, 37, 1274–1280. [Google Scholar] [CrossRef]
- Eslami, M.; Khazeni, S.; Khanaghah, X.M.; Asadi, M.H.; Ansari, M.A.; Garjan, J.H.; Lotfalizadeh, M.H.; Bayat, M.; Taghizadieh, M.; Taghavi, S.P.; et al. MiRNA-related metastasis in oral cancer: Moving and shaking. Cancer Cell Int. 2023, 23, 182. [Google Scholar] [CrossRef]
- Liu, B.; Hu, J.; Zhao, H.; Zhao, L.; Pan, S. MicroRNA-155-5p Contributes to 5-Fluorouracil Resistance Through Down-Regulating TP53INP1 in Oral Squamous Cell Carcinoma. Front. Oncol. 2022, 11, 706095. [Google Scholar] [CrossRef]
- Kirave, P.; Gondaliya, P.; Kulkarni, B.; Rawal, R.; Garg, R.; Jain, A.; Kalia, K. Exosome mediated miR-155 delivery confers cisplatin chemoresistance in oral cancer cells via epithelial-mesenchymal transition. Oncotarget 2020, 11, 1157–1171. [Google Scholar] [CrossRef]
- Geretto, M.; Pulliero, A.; Rosano, C.; Zhabayeva, D.; Bersimbaev, R.; Izzotti, A. Resistance to cancer chemotherapeutic drugs is determined by pivotal microRNA regulators. Am. J. Cancer Res. 2017, 7, 1350–1371. [Google Scholar]
- Law, Z.-J.; Khoo, X.H.; Lim, P.T.; Goh, B.H.; Ming, L.C.; Lee, W.-L.; Goh, H.P. Extracellular Vesicle-Mediated Chemoresistance in Oral Squamous Cell Carcinoma. Front. Mol. Biosci. 2021, 8, 629888. [Google Scholar] [CrossRef] [PubMed]
- Cristaldi, M.; Mauceri, R.; Di Fede, O.; Giuliana, G.; Campisi, G.; Panzarella, V. Salivary Biomarkers for Oral Squamous Cell Carcinoma Diagnosis and Follow-Up: Current Status and Perspectives. Front. Physiol. 2019, 10, 1476. [Google Scholar] [CrossRef] [PubMed]
- Manzano-Moreno, F.J.; Costela-Ruiz, V.J.; García-Recio, E.; Olmedo-Gaya, M.V.; Ruiz, C.; Reyes-Botella, C. Role of Salivary MicroRNA and Cytokines in the Diagnosis and Prognosis of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2021, 22, 12215. [Google Scholar] [CrossRef]
- Holjencin, C.; Jakymiw, A. MicroRNAs and Their Big Therapeutic Impacts: Delivery Strategies for Cancer Intervention. Cells 2022, 11, 2332. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Loong, H.; Winquist, E.; Waldron, J.; Chen, E.; Kim, J.; Palma, D.; Read, N.; Razak, A.; Diaz-Padilla, I.; Chan, K.; et al. Phase 1 study of nab-paclitaxel, cisplatin and 5-fluorouracil as induction chemotherapy followed by concurrent chemoradiotherapy in locoregionally advanced squamous cell carcinoma of the oropharynx. Eur. J. Cancer 2014, 50, 2263–2270. [Google Scholar] [CrossRef] [PubMed]
- Bauman, J.; Langer, C.; Quon, H.; Algazy, K.; Lin, A.; Desai, A.; Mutale, F.; Weiss, J. Induction chemotherapy with cetuximab, carboplatin and paclitaxel for the treatment of locally advanced squamous cell carcinoma of the head and neck. Exp. Ther. Med. 2013, 5, 1247–1253. [Google Scholar] [CrossRef]
- Bassett, C.; Triplett, H.; Lott, K.; Howard, K.M.; Kingsley, K. Differential Expression of MicroRNA (MiR-27, MiR-145) among Dental Pulp Stem Cells (DPSCs) Following Neurogenic Differentiation Stimuli. Biomedicines 2023, 11, 3003. [Google Scholar] [CrossRef]

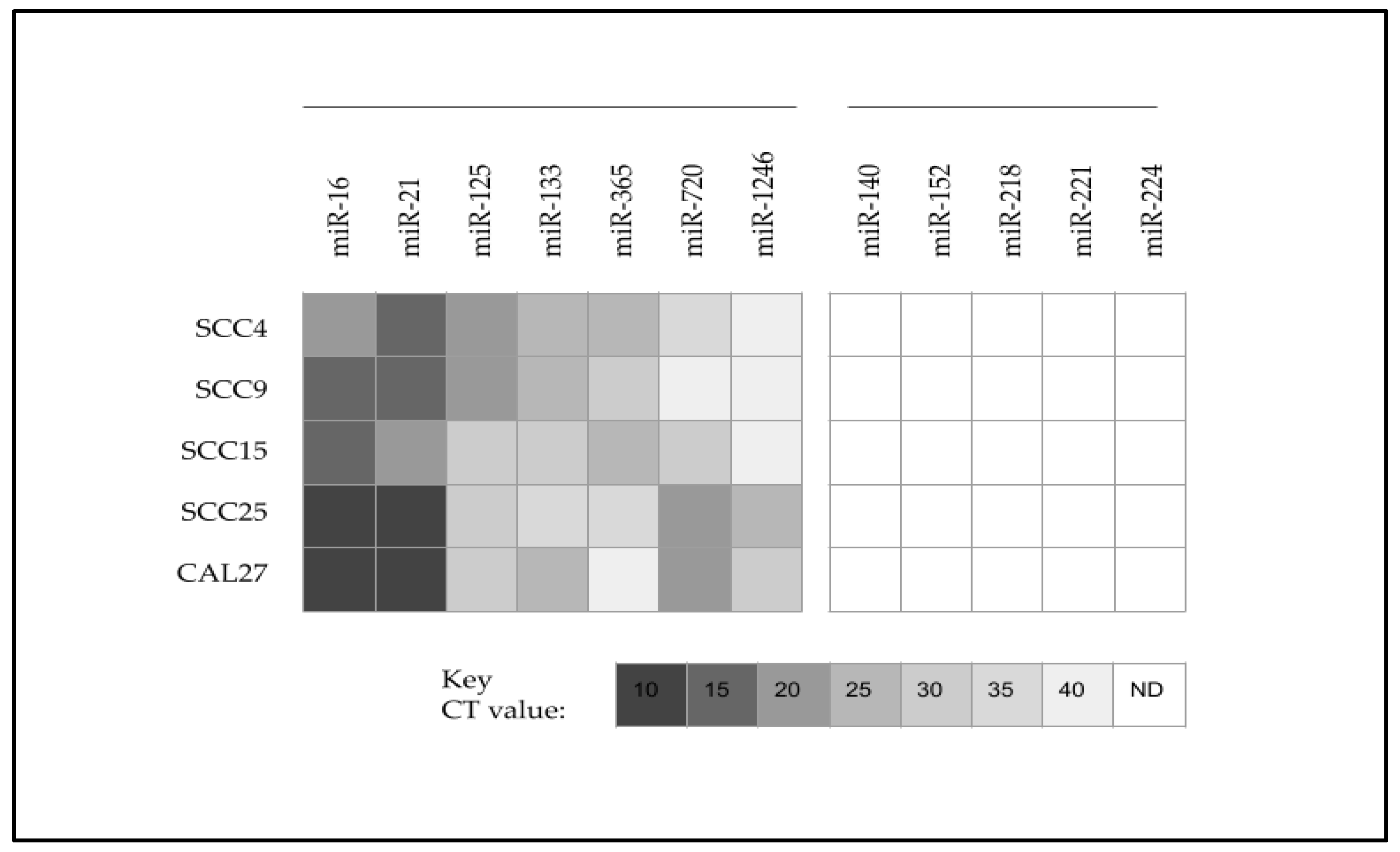
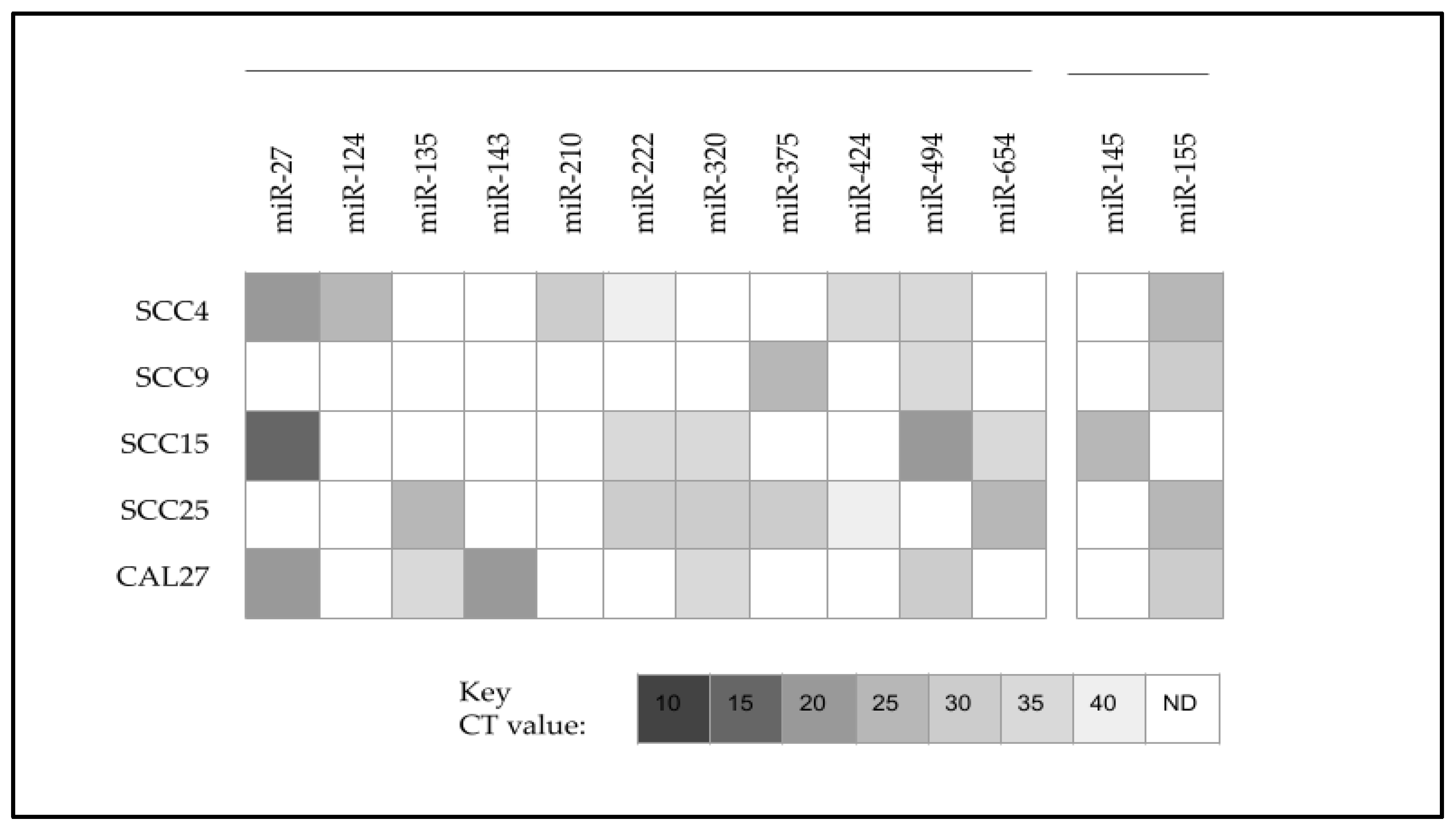
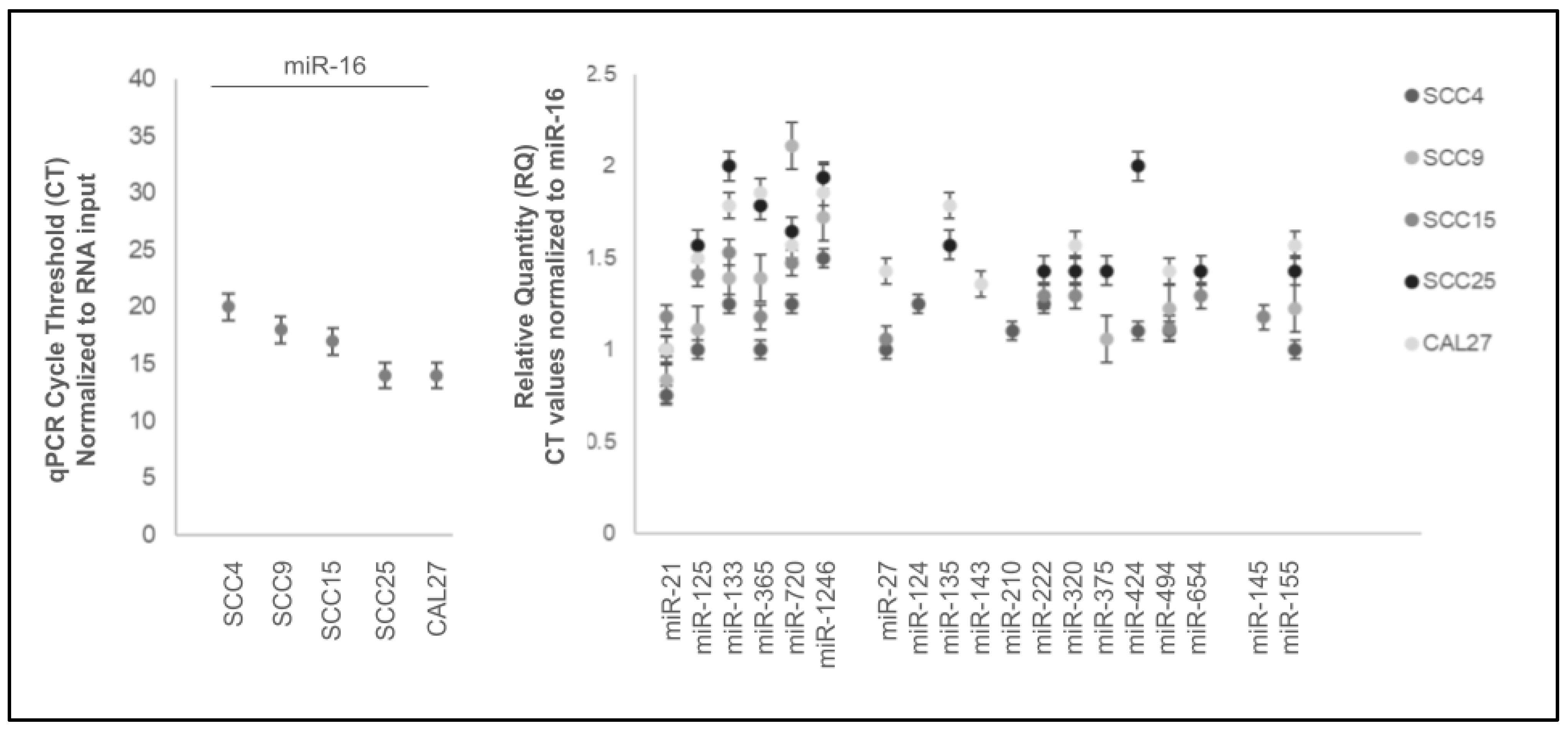
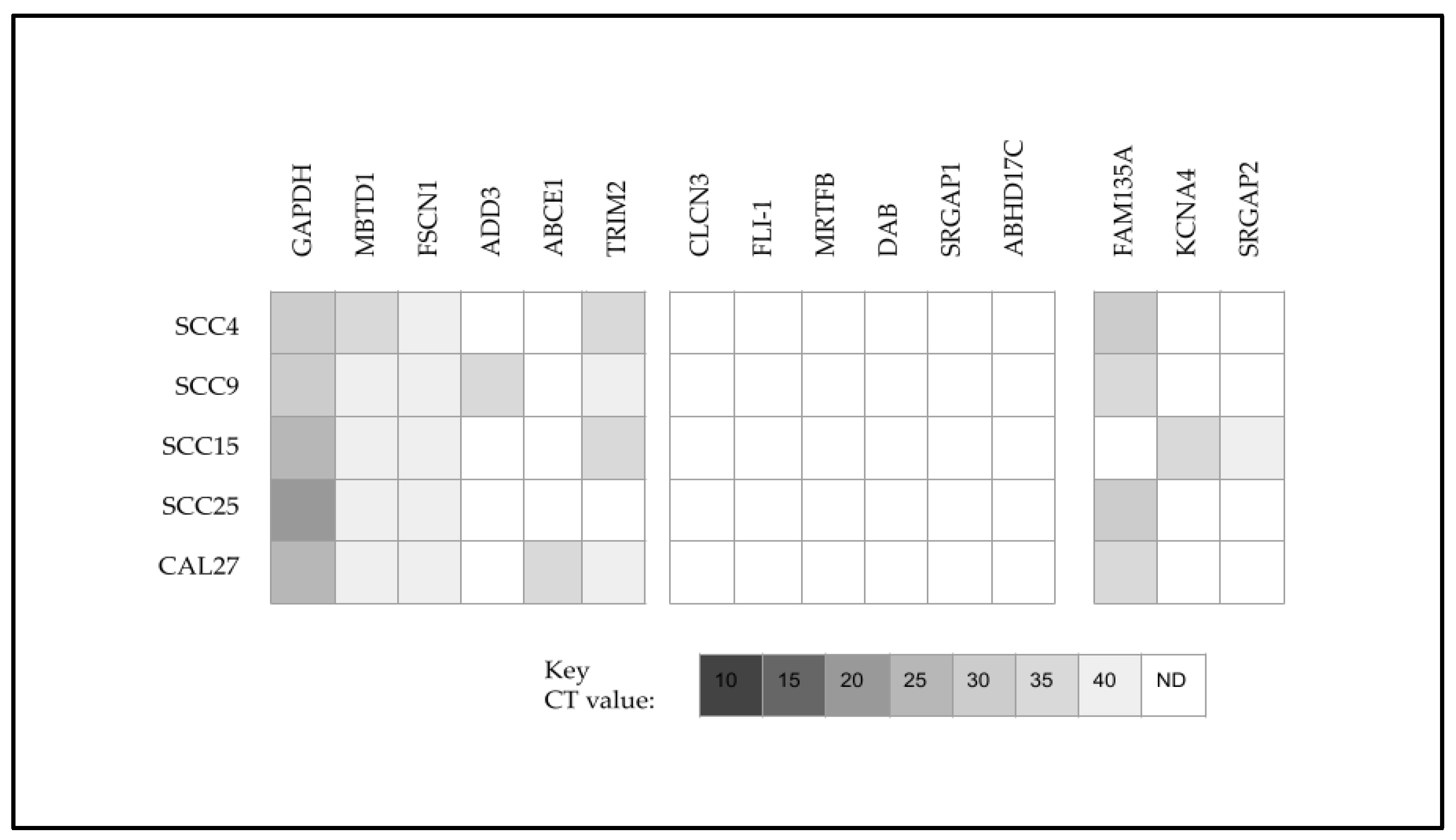

| Cell Line | Media | Designation Cell Type | STR Analysis | Sex | Age |
|---|---|---|---|---|---|
| CAL27 (CRL-2095) | DMEM | OSCC | 93% | Male | 56 years |
| SCC25 (CRL-1628) | DMEM:F12 | OSCC | 100% | Male | 70 years |
| SCC15 (CRL-1623) | DMEM:F12 | OSCC | 95% | Male | 55 years |
| SCC9 (CRL-1629) | DMEM:F12 | OSCC | 100% | Male | 25 years |
| SCC4 (CRL-1624) | DMEM:F12 | OSCC | 92% | Male | 55 years |
| Control primers | Primer sequence | Temp. |
| GAPDH forward | 5′ATCTTCCAGGAGCGAGATCC-3′ | Tm: 66 °C |
| GAPDH reverse | 5′ACCACTGACACGTTGGCAGT-3′ | Tm: 70 °C |
| Beta-actin forward | 5′-GTGGGGTCCTGTGGTGTG-3′ | Tm: 69 °C |
| Beta-actin reverse | 5′-GAAGGGGACAGGCAGTGA-3′ | Tm: 67 °C |
| miRNA primers | Primer sequence | Temp. |
| miR-16 forward | 5′-TAGCAGCACGTAAATATTGGCG-3 | Tm: 65 °C |
| miR-16 reverse | 5′-TGCGTGTCGTGGAGTC-3′ | Tm: 65 °C |
| miR-21 forward | 5′-GCCACCACACCAGCTAATTT-3′ | Tm: 66 °C |
| miR-21 reverse | 5′-CTGAAGTCGCCATGCAGATA-3′ | Tm: 65 °C |
| miR-27 forward | 5′-ATATGAGAAAAGAGCTTCCCTGTG-3′ | Tm: 61 °C |
| miR-27 reverse | 5′-CAAGGCCAGAGGAGGTGAG-3′ | Tm: 67 °C |
| miR-124 forward | 5′-TTCACAGCGGACCTTGA-3′ | Tm: 64 °C |
| miR-124 reverse | 5′-GAACATGTCTGCGTATCTC-3′ | Tm: 60 °C |
| miR-125 forward | 5′-GCCCTCCCTGAGACCTCAA-3′ | Tm: 69 °C |
| miR-125 reverse | 5′-GTGCAGGGTCCGAGGT-3′ | Tm: 68 °C |
| miR-133 forward | 5′-CCGGTTAACTCGAGCTCTGTGAGAG-3′ | Tm: 71 °C |
| miR-133 reverse | 5′-CTAGCTAGGAATTCTGTGACCTGTG-’3′ | Tm: 66 °C |
| miR-135 forward | 5′-CGATATGGCTTTTTATTCCTA -3′ | Tm: 56 °C |
| miR-135 reverse | 5′-GAGCAGGGTCCGAGGT -3′ | Tm: 67 °C |
| miR-140 forward | 5′-GGGCAGTGGTTTTACCCTA -3′ | Tm: 64 °C |
| miR-140 reverse | 5′-CAGTGCGTGTCGTGGAGT -3′ | Tm: 68 °C |
| miR-143 forward | 5′-AGTGCGTGTCGTGGAGTC-3′ | Tm: 68 °C |
| miR-143 reverse | 5′-GCCTGAGATGAAGCACTGT-3′ | Tm: 65 °C |
| miR-145 forward | 5′-AGAGAACTCCAGCTG-3′ | Tm: 56 °C |
| miR-145 reverse | 5′-GGCAACTGTGGGGTG-3′ | Tm: 64 °C |
| miR-152 forward | 5′-GGTTCAAGACAGTACGTGACT-3′ | Tm: 64 °C |
| miR-152 reverse | 5′-CCAAGTTCTGTATGCACTGA-3′ | Tm: 62 °C |
| miR-155 forward | 5′-TTAATGCTAATTGTGATAGGGGT-3′ | Tm: 61 °C |
| miR-155 reverse | 5′-CCTATCACAATTAGCATTAATT-3′ | Tm: 55 °C |
| miR-210 forward | 5′-CATAGATAGCCACTGCCCACA-3′ | Tm: 67 °C |
| miR-210 reverse | 5′-GTGCAGGGTCCGAGGTATTC-3′ | Tm: 68 °C |
| miR-218 forward | 5′-TCGGGCTTGTGCTTGATC T-3′ | Tm: 65 °C |
| miR-218 reverse | 5′-GTGCAGGGTCCGAGTG-3′ | Tm: 66 °C |
| miR-221 forward | 5′-CCCAGCATTTCTGACTGTTG-3′ | Tm: 64 °C |
| miR-221 reverse | 5′-TGTGAGACCATTTGGGTGAA-3′ | Tm: 64 °C |
| miR-222 forward | 5′-CGCAGCTACATCTGGCTACTG-3′ | Tm: 68 °C |
| miR-222 reverse | 5′-GTGCAGGGTCCGAGGT-3′ | Tm: 68 °C |
| miR-224 forward | 5′-GCGAGGTCAAGTCACTAGTGGT-3′ | Tm: 69 °C |
| miR-224 reverse | 5′-CGAGAAGCTTGCATCACCAGAGAACG-3′ | Tm: 72 °C |
| miR-320 forward | 5′-AACGGAGAGTTGGGTCGAAA-3′ | Tm: 66 °C |
| miR-320 reverse | 5′-TTGCCTCTCAACCCAGCTTT-3′ | Tm: 67 °C |
| miR-365 forward | 5′-ATAGGATCCTGAGGTCCCTTTCGTG-3′ | Tm: 70 °C |
| miR-365 reverse | 5′-GCGAAGCTTAAAAACAGCGGAAGAGTTT-3′ | Tm: 72 °C |
| miR-375 forward | 5′-GGCTCTAGAGGGGACGAAGC-3′ | Tm: 70 °C |
| miR-375 reverse | 5′-GGCAAGCTTTTTCCACACCTCAGCCTTG-3′ | Tm: 74 °C |
| miR-424 forward | 5′-AGGACGAAACACCCCCTATTCCTTGC-3′ | Tm: 73 °C |
| miR-424 reverse | 5′-TAATGGATCCGAATACCTGCTCCTGA-3′ | Tm: 69 °C |
| miR-494 forward | 5′-GAAGATCTACGTCTGGTCTACCCAGTGC-3′ | Tm: 72 °C |
| miR-494 reverse | 5′-GGGGTACCACCGAGAGTGGAGCCGGCAA-3′ | Tm: 82 °C |
| miR-654 forward | 5′-GGGATGTCTGCTGACCA-3′ | Tm: 64 °C |
| miR-654 reverse | 5′-CAGTGCGTGTCGTGGA-3′ | Tm: 65 °C |
| miR-720 forward | 5′-GCGTGCTCTCGCTGGGG-3′ | Tm: 73 °C |
| miR-720 reverse | 5′-GTGCAGGGTCCGAGGT-3′ | Tm: 68 °C |
| miR-1246 forward | 5′-TGAAGTAGGACTGGGCAGAGA-3′ | Tm: 67 °C |
| miR-1246 reverse | 5′-TTTGGGTCAGGTGTCCACTC-3′ | Tm: 67 °C |
| miR-145 targets | Primer sequence | Temp. |
| FSCN1 forward | 5′-CCAGGGTATGGACCTGTCTG-3′ | Tm: 65 °C |
| FSCN1 reverse | 5′-GTGTGGGTACGGAAGGCAC-3′ | Tm: 65 °C |
| ABHD17C forward | 5′-CTACTCGGGATACGGCGTCA-3′ | Tm: 65 °C |
| ABHD17C reverse | 5′-AGAGGATAATGTTCTCGGGACTC-3′ | Tm: 63 °C |
| FLI forward | 5′-CAGCCCCACAAGATCAACCC-3′ | Tm: 65 °C |
| FLI reverse | 5′-CACCGGAGACTCCCTGGAT-3′ | Tm: 65 °C |
| MRTFB forward | 5′-ATGGATCACACAGGGGCGATA-3 | Tm: 63 °C |
| MRTFB reverse | 5′-CCGCTGGGCTCTTCAAAGG-3′ | Tm: 65 °C |
| DAB2 forward | 5′-GTAGAAACAAGTGCAACCAATGG-3′ | Tm: 61 °C |
| DAB2 reverse | 5′-GCCTTTGAACCTTGCTAAGAGA-3′ | Tm: 61 °C |
| SRGAP1 forward | 5′-ACCCCGAGCCGATTCAAGA-3′ | Tm: 62 °C |
| SRGAP1 reverse | 5′-GAACTCGCATCTCCGTTTGCT-3′ | Tm: 63 °C |
| SRGAP2 forward | 5′-TGAAGGAGAAAGCGTCAAGCC-3′ | Tm: 62 °C |
| SRGAP2 reverse | 5′-AAGGTCAGATAGGTCATGGATGT-3′ | Tm: 61 °C |
| CLCN3 forward | 5′-GGAGGCAGCATTAACAGTTCT-3′ | Tm: 61 °C |
| CLCN3 reverse | 5′-TCGCACCCAATCAATAGTATGGA-3′ | Tm: 61 °C |
| MBTD1 forward | 5′-GGCATGGCTACCTGTGAGATG-3′ | Tm: 65 °C |
| MBTD1 reverse | 5′-GGCCAAAATGCTTGCCTTCT-3′ | Tm: 61 °C |
| FAM135A forward | 5′-AGTAGCCGAACATTGAAGCTG-3′ | Tm: 61 °C |
| FAM135A reverse | 5′-TGGCTGGTGTAGTGCAACC-3′ | Tm: 62 °C |
| ABCE1 forward | 5′-GGAATGCAAAAAGAGTTGTCCTG-3′ | Tm: 61 °C |
| ABCE1 reverse | 5′-CGAGGGATAGGCAACCTGTG-3′ | Tm: 65 °C |
| KCNA4 forward | 5′-GTACCTCCCATGACCCTCAGA-3′ | Tm: 65 °C |
| KCNA4 reverse | 5′-CTGCCGGTAGTGGGCTTTC-3′ | Tm: 65 °C |
| ADD3 forward | 5′-CCAGCCAAGGCGTGATTAC′ | Tm: 62 °C |
| ADD3 reverse | 5′-TGAAGTCTTGTCGTAGATCAGGA-3′ | Tm: 61 °C |
| TRIM2 forward | 5′-TGCGCCAGATTGACAAGCA′; | Tm: 60 °C |
| TRIM2 reverse | 5′-GCACCTCTCGCAGAAAGTG-3′ | Tm: 62 °C |
| miR-155 targets | Primer sequence | Temp. |
| ZNF652 forward | 5′-GCTGGTTGAAAACTGTGCTGT-3′ | Tm: 61 °C |
| ZNF652 reverse | 5′-GAAGATGGCACTTGACCACGA-3′ | Tm: 63 °C |
| ZIC3 forward | 5′-CGGCGCACGATCTATCTTCAG-3′ | Tm: 65 °C |
| ZIC3 reverse | 5′-TGCGGAACAGAAACTCGC-3′ | Tm: 62 °C |
| BACH1 forward | 5′-TCTGAGTGAGAACTCGGTTTTTG-3′ | Tm: 61 °C |
| BACH1 reverse | 5′-CGCTGGTCATTAAGGCTGAGTCC-3′ | Tm: 63 °C |
| JARID2 forward | 5′-ACCAGTCTAAGGGATTAGGACC-3′ | Tm: 63 °C |
| JARID2 reverse | 5′-TGCTGGGACTATTCGGCTGA-3′ | Tm: 62 °C |
| KDM5B forward | 5′-CCATAGCCGAGCAGACTGG-3′ | Tm: 65 °C |
| KDM5B reverse | 5′-GGATACGTGGCGTAAAATGAAGT-3′ | Tm: 61 °C |
| TBR1 forward | 5′-GCAGCAGCTACCCACATTCA-3′ | Tm: 62 °C |
| TBR1 reverse | 5′-AGGTTGTCAGTGGTCGAGATA-3′ | Tm: 61 °C |
| IRF2-BP2 forward | 5′-CCCATGACTCCTACATCCTCTT-3′ | Tm: 63 °C |
| IRF2-BP2 reverse | 5′-GAGGGCGGACTGTTGCTATTC-3′ | Tm: 65 °C |
| OLFML3 forward | 5′-TCCTTTTGTCATGGTCGGGAC-3′ | Tm: 63 °C |
| OLFML3 reverse | 5′-TAAAGCAGCTAGTCGGCGTTC-3′ | Tm: 63 °C |
| MPEG1 forward | 5′-CGGCAGCATGGGCTAAATCA-3′ | Tm: 62 °C |
| MPEG1 reverse | 5′-TGTCCACATTCCGCAGATTGT-3′ | Tm: 61 °C |
| CDX1 forward | 5′-GGTGGCAGCGGTAAGACTC-3′ | Tm: 65 °C |
| CDX1 reverse | 5′-TGTAACGGCTGTAATGAAACTCC-3′ | Tm: 61 °C |
| ACTL7A forward | 5′-TGGGTCCGCCATACGAGTT-3′ | Tm: 62 °C |
| ACTL7A reverse | 5′-GTCCACGACCACTGCTTTG-3′ | Tm: 62 °C |
| MARCH1 forward | 5′-CACTGGGACACTGCGCTTT-3′ | Tm: 62 °C |
| MARCH1 reverse | 5′-TCACAGCAGCGTGTATCTGAG-3′ | Tm: 63 °C |
| FOS forward | 5′-CCGGGGATAGCCTCTCTTACT-3 | Tm: 65 °C |
| FOS reverse | 5′-CCAGGTCCGTGCAGAAGTC-3′ | Tm: 65 °C |
| IKBIP forward | 5′-GCTCATCTAAAGCGTCTACAGG-3′ | Tm: 63 °C |
| IKBIP reverse | 5′-AAGCGTCGTCAGACTGTTGTT-3′ | Tm: 61 °C |
| CHAF1A forward | 5′-AGCCCGTCTGCCGTTTAAG-3′ | Tm: 62 °C |
| CHAF1A reverse | 5′-AGAAGTACCCTGATCGTCTGAC-3′ | Tm: 63 °C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belnap, C.; Divis, T.; Kingsley, K.; Howard, K.M. Differential Expression of MicroRNA MiR-145 and MiR-155 Downstream Targets in Oral Cancers Exhibiting Limited Chemotherapy Resistance. Int. J. Mol. Sci. 2024, 25, 2167. https://doi.org/10.3390/ijms25042167
Belnap C, Divis T, Kingsley K, Howard KM. Differential Expression of MicroRNA MiR-145 and MiR-155 Downstream Targets in Oral Cancers Exhibiting Limited Chemotherapy Resistance. International Journal of Molecular Sciences. 2024; 25(4):2167. https://doi.org/10.3390/ijms25042167
Chicago/Turabian StyleBelnap, Conner, Tyler Divis, Karl Kingsley, and Katherine M. Howard. 2024. "Differential Expression of MicroRNA MiR-145 and MiR-155 Downstream Targets in Oral Cancers Exhibiting Limited Chemotherapy Resistance" International Journal of Molecular Sciences 25, no. 4: 2167. https://doi.org/10.3390/ijms25042167





