Comparative Analysis of Chloroplast Genome of Meconopsis (Papaveraceae) Provides Insights into Their Genomic Evolution and Adaptation to High Elevation
Abstract
1. Introduction
2. Results and Discussion
2.1. Characterization of the CP Genome Structure of Meconopsis Species
2.2. IR Boundary Analysis
2.3. Genomic Sequence Divergence
2.4. Codon Usage Analysis
2.5. Repeat Sequence Analysis
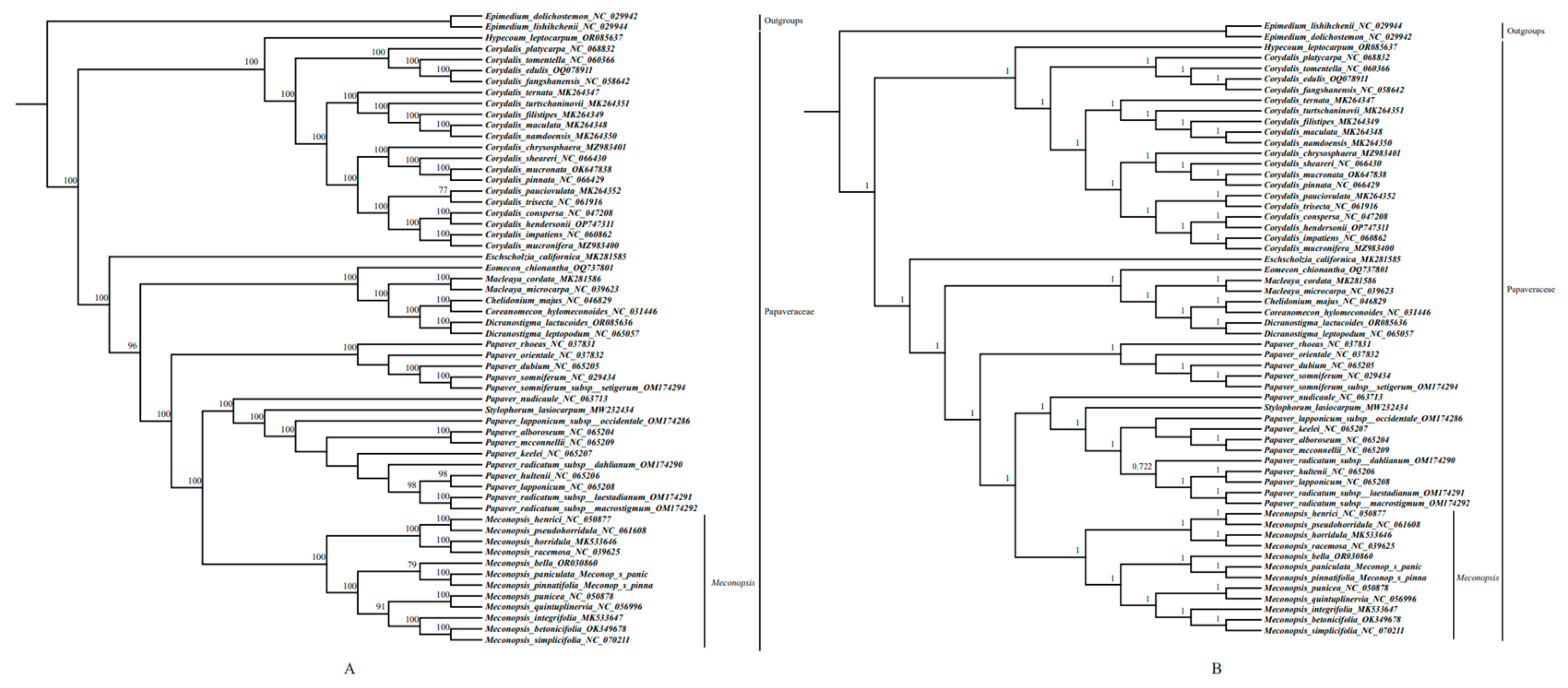
2.6. Phylogenetic Analysis
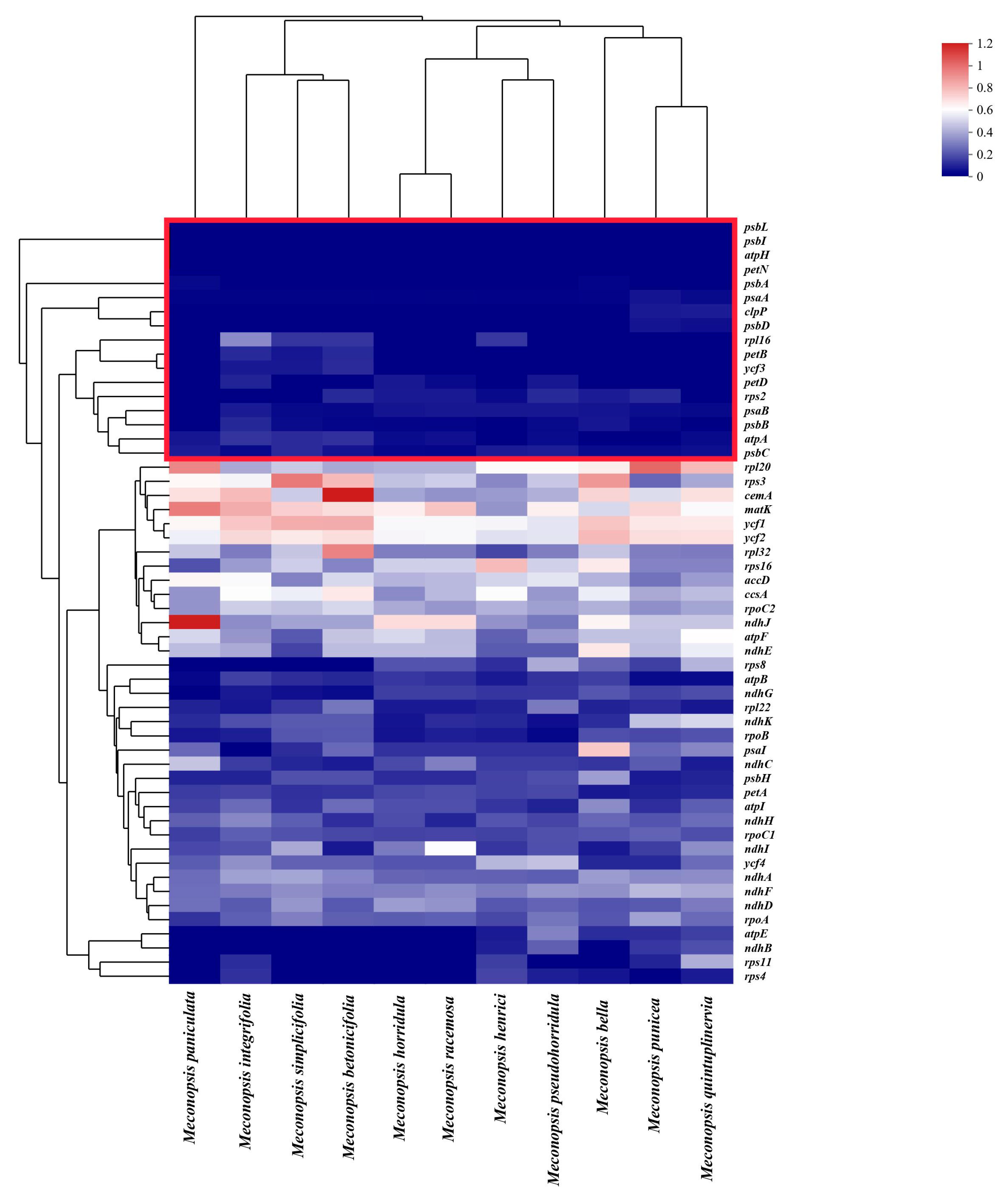
2.7. Selection and Adaptation Analyses
3. Materials and Methods
3.1. Plant Material Sampling, DNA Extracting, and DNA Sequencing
3.2. Chloroplast Genome De Novo Assembly and Annotation
3.3. Chloroplast Genome Visualization and Sequence Divergence Analysis
3.4. Analysis of Codon Usage
3.5. Analysis of Repeat Sequences in Organelle Genomes
3.6. Phylogenetic Analysis
3.7. Selective Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neuhaus, H.E.; Emes, M.J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 111–140. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Meger, J.; Ulaszewski, B.; Vendramin, G.G.; Burczyk, J. Using reduced representation libraries sequencing methods to identify cpDNA polymorphisms in European beech (Fagus sylvatica L). Tree Genet. Genomes 2019, 15, 7. [Google Scholar] [CrossRef]
- Schloss, P.D.; Jenior, M.L.; Koumpouras, C.C.; Westcott, S.L.; Highlander, S.K. Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. PeerJ 2016, 4, e1869. [Google Scholar] [CrossRef]
- Xiao, W.; Simpson, B.B. A New Infrageneric Classification of Meconopsis (Papaveraceae) Based on a Well-supported Molecular Phylogeny. Syst. Bot. 2017, 42, 226–233. [Google Scholar] [CrossRef]
- Guo, Q.; Bai, R.F.; Zhao, B.S.; Feng, X.; Zhao, Y.F.; Tu, P.F.; Chai, X.Y. An Ethnopharmacological, Phytochemical and Pharmacological Review of the Genus Meconopsis. Am. J. Chin. Med. 2016, 44, 439–462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Chen, Y.; Liu, S.; Yao, X.; Wang, Y. In vitro and in vivo hepatoprotective and antioxidant activity of ethanolic extract from Meconopsis integrifolia (Maxim.) Franch. J. Ethnopharmacol. 2013, 148, 664–670. [Google Scholar] [CrossRef]
- Fan, J.P.; Wang, Y.Q.; Wang, X.B.; Wang, P.; Tang, W.; Yuan, W.J.; Kong, L.L.; Liu, Q.H. The Antitumor Activity of Meconopsis horridula Hook, a Traditional Tibetan Medical Plant, in Murine Leukemia L1210 Cells. Cell. Physiol. Biochem. 2015, 37, 1055–1065. [Google Scholar] [CrossRef]
- Xie, H.Y.; Ash, J.E.; Linde, C.C.; Cunningham, S.; Nicotra, A. Himalayan-Tibetan Plateau Uplift Drives Divergence of Polyploid Poppies: Meconopsis viguier (Papaveraceae). PLoS ONE 2014, 9, e99177. [Google Scholar] [CrossRef]
- Egan, P.A. Meconopsis autumnalis and M. manasluensis (Papaveraceae), two new species of Himalayan poppy endemic to central Nepal with sympatric congeners. Phytotaxa 2011, 20, 47–56. [Google Scholar] [CrossRef]
- Yang, F.-S.; Qin, A.-L.; Li, Y.-F.; Wang, X.-Q. Great Genetic Differentiation among Populations of Meconopsis integrifolia and Its Implication for Plant Speciation in the Qinghai-Tibetan Plateau. PLoS ONE 2012, 7, e37196. [Google Scholar] [CrossRef] [PubMed]
- Favre, A.; Päckert, M.; Pauls, S.U.; Jähnig, S.C.; Uhl, D.; Michalak, I.; Muellner-Riehl, A.N. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 2014, 90, 236–253. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Wang, Z.; Shrestha, N.; Liu, J. Species divergence with gene flow and hybrid speciation on the Qinghai–Tibet Plateau. New Phytol. 2022, 234, 392–404. [Google Scholar] [CrossRef] [PubMed]
- An Account of the Genus Meconopsis. Nature 1934, 133, 777–778. [CrossRef][Green Version]
- Sonah, H.; Deshmukh, R.K.; Sharma, A.; Singh, V.P.; Gupta, D.K.; Gacche, R.N.; Rana, J.C.; Singh, N.K.; Sharma, T.R. Genome-Wide Distribution and Organization of Microsatellites in Plants: An Insight into Marker Development in Brachypodium. PLoS ONE 2011, 6, e21298. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Liu, Y.N.; Yang, F.S.; Wang, X.Q. Molecular Phylogeny of Asian Meconopsis Based on Nuclear Ribosomal and Chloroplast DNA Sequence Data. PLoS ONE 2014, 9, e104823. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, X.; Yang, F. Mechanisms underlying flower color variation in Asian species of Meconopsis: A preliminary phylogenetic analysis based on chloroplast DNA and anthocyanin biosynthesis genes. J. Syst. Evol. 2014, 52, 125–133. [Google Scholar] [CrossRef]
- Byars, S.G.; Papst, W.; Hoffmann, A.A. Local Adaptation And Cogradient Selection in The Alpine Plant, Poa Hiemata, Along a Narrow Altitudinal Gradient. Evolution 2007, 61, 2925–2941. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, Z.; Zhang, H.; Cui, C.; Qi, X.; Luo, X.; Tao, X.; Wu, T.; Ouzhuluobu; Basang; et al. Genetic Variations in Tibetan Populations and High-Altitude Adaptation at the Himalayas. Mol. Biol. Evol. 2010, 28, 1075–1081. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Casati, P.; Stapleton, A.E.; Blum, J.E.; Walbot, V. Genome-wide analysis of high-altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in response to UV-B. Plant J. 2006, 46, 613–627. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, Y.; Yan, L.; Zhang, G.; Wang, X.; Zeng, Y.; Zhang, J.; Ma, X.; Tan, Y.; Long, N.; et al. Genome of Plant Maca (Lepidium meyenii ) Illuminates Genomic Basis for High-Altitude Adaptation in the Central Andes. Mol. Plant 2016, 9, 1066–1077. [Google Scholar] [CrossRef]
- Guo, X.; Hu, Q.; Hao, G.; Wang, X.; Zhang, D.; Ma, T.; Liu, J. The genomes of two Eutrema species provide insight into plant adaptation to high altitudes. DNA Res. 2018, 25, 307–315. [Google Scholar] [CrossRef]
- Hu, S.; Sablok, G.; Wang, B.; Qu, D.; Barbaro, E.; Viola, R.; Li, M.; Varotto, C. Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genom. 2015, 16, 306. [Google Scholar] [CrossRef]
- Zhao, D.-N.; Ren, Y.; Zhang, J.-Q. Conservation and innovation: Plastome evolution during rapid radiation of Rhodiola on the Qinghai-Tibetan Plateau. Mol. Phylogenet. Evol. 2019, 144, 106713. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, X.; Landis, J.B.; Zhang, H.; Deng, T.; Sun, H.; Wang, H. Plastome Evolution in Dolomiaea (Asteraceae, Cardueae) Using Phylogenomic and Comparative Analyses. Front. Plant Sci. 2020, 11, 376. [Google Scholar] [CrossRef]
- Zhou, J.; Cui, Y.; Chen, X.; Li, Y.; Xu, Z.; Duan, B.; Li, Y.; Song, J.; Yao, H. Complete Chloroplast Genomes of Papaver rhoeas and Papaver orientale: Molecular Structures, Comparative Analysis, and Phylogenetic Analysis. Molecules 2018, 23, 437. [Google Scholar] [CrossRef] [PubMed]
- Yan-Yan, L.; Sheng-Long, K.; Jun-Li, W.; Cao, Y.-N.; Jia-Mei, L. Complete chloroplast genome sequences of Corydalis edulis and Corydalis shensiana (Papaveraceae). Mitochondrial DNA Part B Resour. 2021, 6, 257–258. [Google Scholar]
- Huang, H.; Shi, C.; Liu, Y.; Mao, S.-Y.; Gao, L.-Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: Genome structure and phylogenetic relationships. BMC Evol. Biol. 2014, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Wang, L.; Li, Y.; Zhuo, W.; Xu, Z.; Guo, H.; Liu, Y.; Gao, R.; Song, J. Highly variable chloroplast genome from two endangered Papaveraceae lithophytes Corydalis tomentella and Corydalis saxicola. Ecol. Evol. 2021, 11, 4158–4171. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.; Chen, S. Plant DNA barcoding: From gene to genome. Biol. Rev. 2014, 90, 157–166. [Google Scholar] [CrossRef]
- Nock, C.J.; Waters, D.L.; Edwards, M.A.; Bowen, S.G.; Rice, N.; Cordeiro, G.M.; Henry, R.J. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J. 2010, 9, 328–333. [Google Scholar] [CrossRef]
- Group, C.P.W. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar]
- Yang, J.; Feng, L.; Yue, M.; He, Y.-L.; Zhao, G.-F.; Li, Z.-H. Species delimitation and interspecific relationships of the endangered herb genus Notopterygium inferred from multilocus variations. Mol. Phylogenet. Evol. 2019, 133, 142–151. [Google Scholar] [CrossRef]
- Jia, J.; Xue, Q. Codon Usage Biases of Transposable Elements and Host Nuclear Genes in Arabidopsis thaliana and Oryza sativa. Genom. Proteom. Bioinform. 2009, 7, 175–184. [Google Scholar] [CrossRef] [PubMed]
- McClellan, D.A. The Codon-Degeneracy Model of Molecular Evolution. J. Mol. Evol. 2000, 50, 131–140. [Google Scholar] [CrossRef]
- Zhang, P.; Xu, W.; Lu, X.; Wang, L. Analysis of codon usage bias of chloroplast genomes in Gynostemma species. Physiol. Mol. Biol. Plants 2021, 27, 2727–2737. [Google Scholar] [CrossRef]
- Hu, J.; Zhao, M.; Hou, Z.; Shang, J. The complete chloroplast genome sequence of Salvia miltiorrhiza, a medicinal plant for preventing and treating vascular dementia. Mitochondrial DNA B Resour 2020, 5, 2460–2462. [Google Scholar] [CrossRef]
- Tuler, A.C.; Carrijo, T.T.; Nóia, L.R.; Ferreira, A.; Peixoto, A.L.; Ferreira, M.F.d.S. SSR markers: A tool for species identification in Psidium (Myrtaceae). Mol. Biol. Rep. 2015, 42, 1501–1513. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lin, F.; Huang, P.; Guo, W.; Zheng, Y. Development of nuclear SSR and chloroplast genome markers in diverse Liriodendron chinense germplasm based on low-coverage whole genome sequencing. Biol. Res. 2020, 53, 1–12. [Google Scholar] [CrossRef]
- Rajendrakumar, P.; Biswal, A.K.; Balachandran, S.M.; Srinivasarao, K.; Sundaram, R.M. Simple sequence repeats in organellar genomes of rice: Frequency and distribution in genic and intergenic regions. Bioinformatics 2006, 23, 1–4. [Google Scholar] [CrossRef]
- Mehmood, F.; Abdullah, S.I.; Ahmed, I.; Waheed, M.T.; Mirza, B. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics 2020, 112, 1522–1530. [Google Scholar] [CrossRef]
- Xuan, Z. Phylogenetic and geographic distribution of Meconopsis. Plant Divers. 1981, 3, 1–3. [Google Scholar]
- Xie, D.-F.; Tan, J.-B.; Yu, Y.; Gui, L.-J.; Su, D.-M.; Zhou, S.-D.; He, X.-J. Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann. Bot. 2020, 125, 1039–1055. [Google Scholar] [CrossRef]
- Fu, X.; Xie, D.-F.; Zhou, Y.-Y.; Cheng, R.-Y.; Zhang, X.-Y.; Zhou, S.-D.; He, X.-J. Phylogeny and adaptive evolution of subgenus Rhizirideum (Amaryllidaceae, Allium) based on plastid genomes. BMC Plant Biol. 2023, 23, 70. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Li, J.-L.; Ma, X.-R. Exogenous hydrogen peroxide enhanced the thermotolerance of Festuca arundinacea and Lolium perenne by increasing the antioxidative capacity. Acta Physiol. Plant. 2014, 36, 2915–2924. [Google Scholar] [CrossRef]
- Drescher, A.; Ruf, S.; Calsa, T.; Carrer, H.; Bock, R.; Jr, T.C. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 2000, 22, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Asakura, Y.; Imai, M.; Nakahira, Y.; Kotani, Y.; Hashiguchi, Y.; Nakai, Y.; Takafuji, K.; Bédard, J.; Hirabayashi-Ishioka, Y.; et al. A Ycf2-FtsHi Heteromeric AAA-ATPase Complex Is Required for Chloroplast Protein Import. Plant Cell 2018, 30, 2677–2703. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-L.; Fan, W.-B.; Wang, N.; Dong, P.-B.; Zhang, T.-T.; Yue, M.; Li, Z.-H. Evolutionary Analysis of Plastid Genomes of Seven Lonicera L. Species: Implications for Sequence Divergence and Phylogenetic Relationships. Int. J. Mol. Sci. 2018, 19, 4039. [Google Scholar] [CrossRef] [PubMed]
- D’agostino, N.; Tamburino, R.; Cantarella, C.; De Carluccio, V.; Sannino, L.; Cozzolino, S.; Cardi, T.; Scotti, N. The Complete Plastome Sequences of Eleven Capsicum Genotypes: Insights into DNA Variation and Molecular Evolution. Genes 2018, 9, 503. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, U357–U359. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Michael, T.; Pascal, L.; Tommaso, P.; Ulbricht-Jones, E.S.; Axel, F.; Ralph, B.; Stephan, G. GeSeq—versatile and accurate an-notation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32 (Suppl. S2), W273–W279. [Google Scholar] [CrossRef]
- Brudno, M.; Do, C.B.; Cooper, G.M.; Kim, M.F.; Davydov, E.; Green, E.D.; Sidow, A.; Batzoglou, S. LAGAN and Mul-ti-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 2003, 13, 721–731. [Google Scholar] [CrossRef]
- Li, H.; Guo, Q.; Xu, L.; Gao, H.; Liu, L.; Zhou, X. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 2023, 11, e15326. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Zhao, Y.; Zheng, H.; Xu, A.; Yan, D.; Jiang, Z.; Qi, Q.; Sun, J. Analysis of codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) and its relation to evolution. BMC Genom. 2016, 17, 677. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, D. Complete chloroplast genome sequences of two species used for Tibetan medicines, Meconopsis punicea vig. and M. henrici vig. (Papaveraceae). Mitochondrial DNA Part B 2019, 5, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, J.; Wu, J.; Zhang, H.; Liu, G.; Wang, X.; Dai, L. ParaAT: A parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 2012, 419, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-Coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
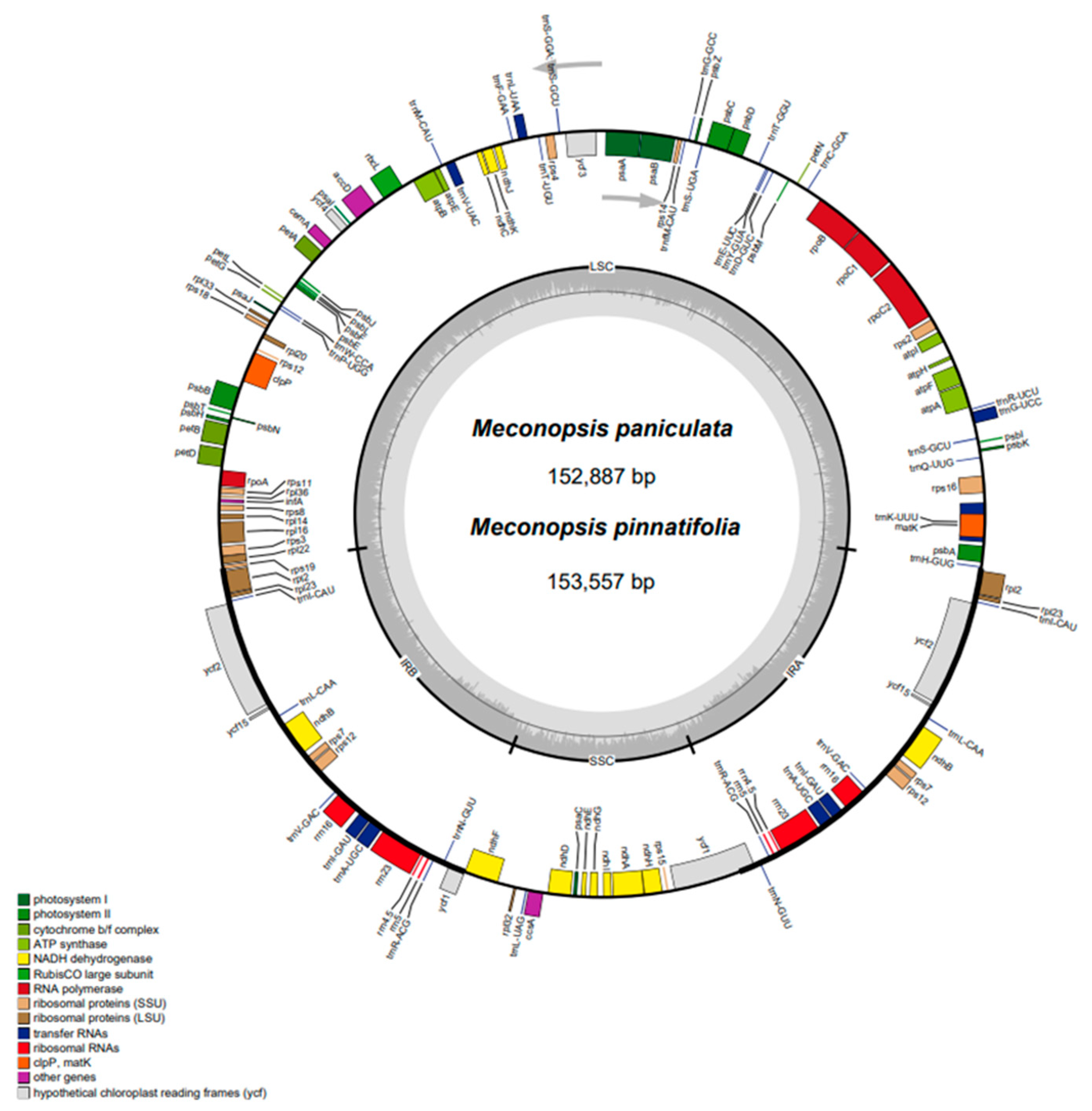

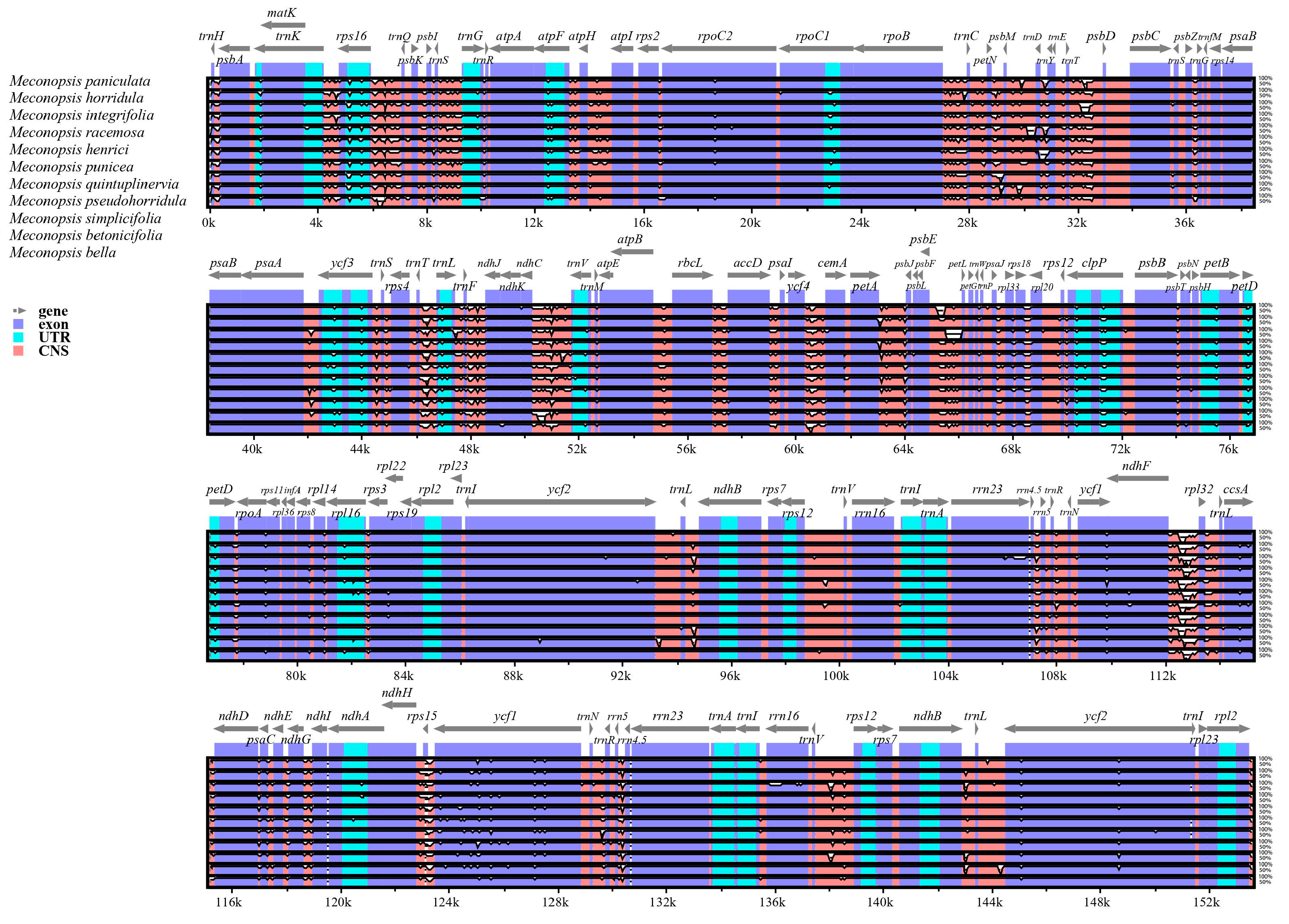
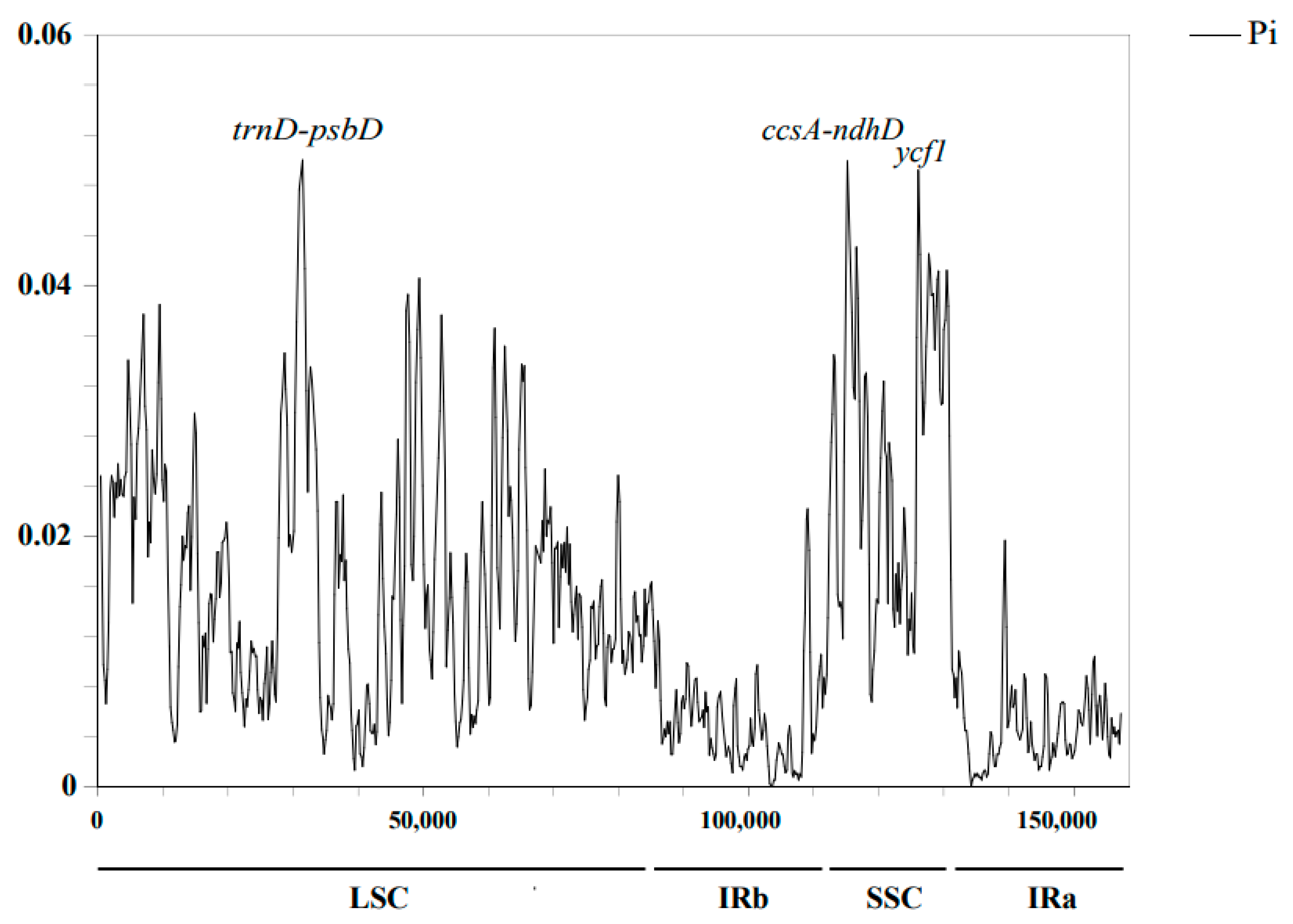
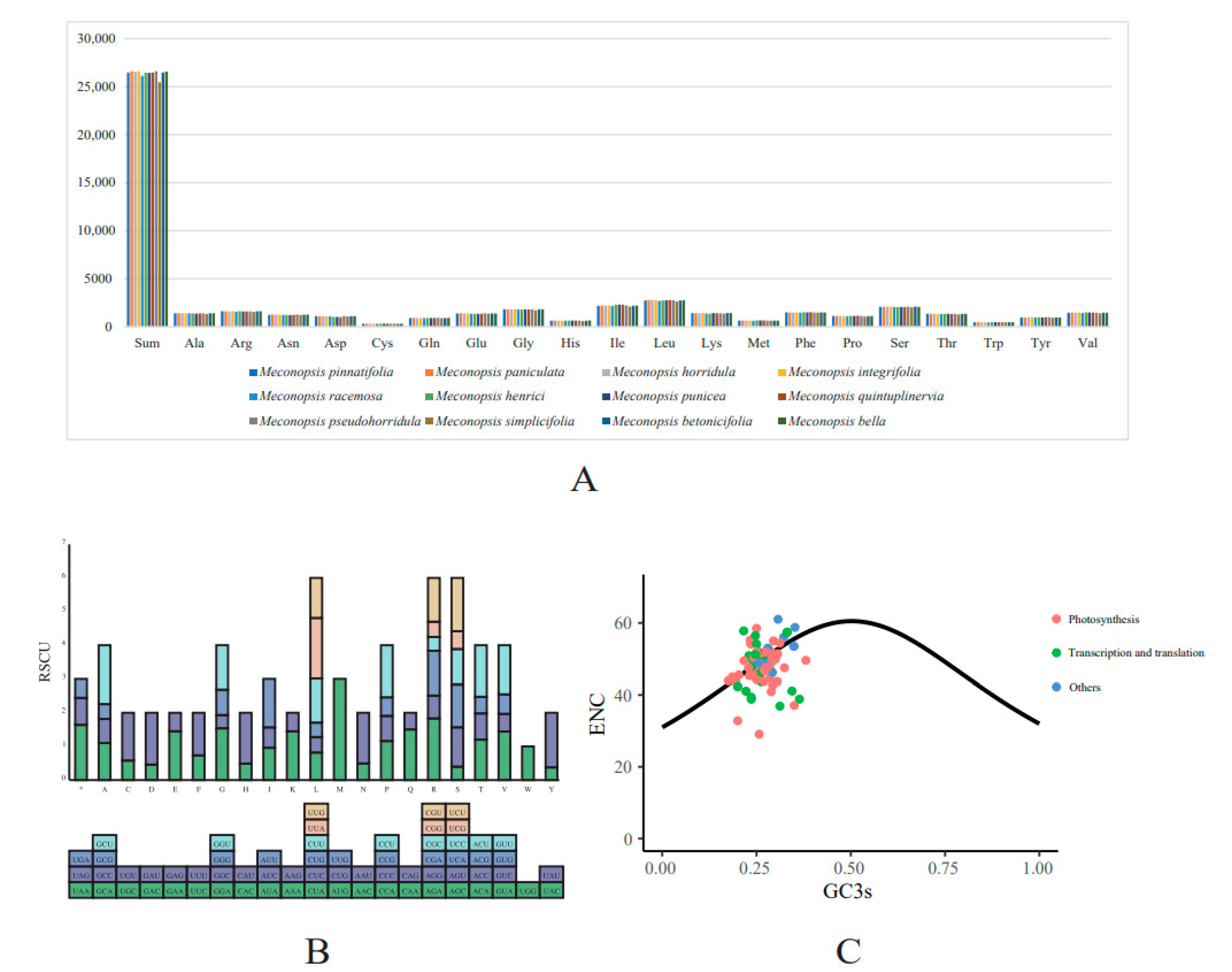

| Category | Group | Genes |
|---|---|---|
| Photosynthesis related genes | Rubisco | rbcL |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Photosystem II | psbA, psbB, psbT, psbK, psbI, psbH, psbM, psbN, psbD, psbC, psbZ, psbJ, psbL, psbE, psbF | |
| ATP synthase | atpA, atpB, atpE, atpF a, atpH, atpI | |
| Cytochrome b/f complex | petA, petB a, petD a, petN, petL, petG | |
| Cytochrome C synthesis | ccsA | |
| NADPH dehydrogenase | ndhA a, ndhB a,c (×2), ndhC, ndhD, ndhE, ndhF, ndhH, ndhG, ndhJ, ndhK, ndhI | |
| Transcription- and translation-related genes | Transcription | rpoA, rpoB, rpoC2, rpoC1 a |
| Ribosomal proteins | rps2, rps3, rps4, rps7 c (×2), rps8, rps11, rps12 a,c (×2), rps14, rps15, rps16 a, rps18, rps19, rpl2 a,c (×2), rpl14, rpl16 a, rpl20, rpl22, rpl23 c (×2), rpl32, rpl33, rpl36 | |
| Translation initiation factor | infA | |
| RNA genes | Ribosomal RNA | rrn16 c (×2), rrn23 c (×2), rrn4.5 c (×2), rrn5 c (×2) |
| Transfer RNA | trnH-GUG, trnK-UUU a, trnQ-UUG, trnS-GCU, trnS-UGA, trnS-GGA, trnG-GCC a, trnR-UCU, trnR-ACG c (×2), trnC-GCA, trnD-GUC, trnY-GUA, trnE-UUC, trnT-UGU, trnG-UCC, trnfM-CAU, trnL-CAA c (×2), trnL-UAA a, trnL-UAG, trnF-GAA, trnV-GAC c (×2), trnV-UAC a, trnM-CAU, trnT-GGU, trnW-CCA, trnP-UGG, trnI-CAU c (×2), trnI-GAU a,c (×2), trnA-UGC a,c (×2), trnN-GUU c (×2) | |
| Other genes | RNA processing | matK |
| Carbon metabolism | cemA | |
| Fatty acid synthesis | accD | |
| Proteolysis | ClpP b | |
| Conserved ORFs | ycf1c (×2), ycf2 c (×2), ycf3 b, ycf4, ycf15 c,d (×2) |
| Genome Feature | M. paniculata | M. pinnatifolia | M. racemosa | M. henrici | M. punicea | M. quintuplinervia | M. pseudohorridula | M. simplicifolia | M. betonicifolia | M. horridula | M. integrifolia | M. bella |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome size (bp) | 152,887 | 153,557 | 153,763 | 153,788 | 153,281 | 154,997 | 154,190 | 152,772 | 151,935 | 153,785 | 151,864 | 153,073 |
| LSC size (bp) | 83.366 | 84,067 | 83,868 | 83,644 | 83,999 | 85,153 | 84,064 | 83,778 | 83,147 | 83,901 | 82,809 | 83,562 |
| SSC size (bp) | 17,857 | 17,894 | 17,905 | 17,822 | 17,728 | 17,876 | 17,770 | 17,646 | 17,746 | 17,898 | 17,753 | 17,833 |
| IR size (bp) | 25,832 | 25,798 | 25,995 | 26,161 | 25,777 | 25,984 | 26,178 | 25,674 | 25,521 | 25,993 | 25,649 | 25,839 |
| Number of genes | 133 | 131 | 129 | 133 | 133 | 133 | 134 | 131 | 131 | 127 | 127 | 133 |
| Protein genes | 88 | 86 | 84 | 88 | 88 | 88 | 88 | 84 | 86 | 87 | 88 | 88 |
| tRNA genes | 37 | 37 | 37 | 37 | 37 | 37 | 37 | 37 | 37 | 29 | 29 | 37 |
| rRNA genes | 8 | 8 | 6 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Duplicated genes in IRs | 19 | 18 | 17 | 19 | 19 | 19 | 20 | 19 | 18 | 19 | 19 | 19 |
| GC content (%) | 38.8% | 38.8% | 38.8% | 38.5% | 38.5% | 38.5% | 38.6% | 38.7% | 38.8% | 38.8% | 38.8% | 38.9% |
| GC content in LSC (%) | 37.3% | 37.3% | 37.3% | 37.0% | 37.0% | 37.1% | 37.0% | 37.3% | 37.3% | 37.2% | 37.4% | 37.5% |
| GC content in SSC (%) | 33.2% | 33.3% | 33.1% | 32.8% | 32.7% | 32.8% | 33.0% | 33.0% | 33.0% | 33.2% | 33.3% | 33.5% |
| GC content in IRs (%) | 43.1% | 43.1% | 43.1% | 43.0% | 42.9% | 43.0% | 43.0% | 43.1% | 43.1% | 43.1% | 43.1% | 43.2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Gao, X.; Yu, X.; Yuan, T.; Zhang, G.; Liu, C.; Li, X.; Wei, P.; Li, X.; Liu, X. Comparative Analysis of Chloroplast Genome of Meconopsis (Papaveraceae) Provides Insights into Their Genomic Evolution and Adaptation to High Elevation. Int. J. Mol. Sci. 2024, 25, 2193. https://doi.org/10.3390/ijms25042193
Zhao S, Gao X, Yu X, Yuan T, Zhang G, Liu C, Li X, Wei P, Li X, Liu X. Comparative Analysis of Chloroplast Genome of Meconopsis (Papaveraceae) Provides Insights into Their Genomic Evolution and Adaptation to High Elevation. International Journal of Molecular Sciences. 2024; 25(4):2193. https://doi.org/10.3390/ijms25042193
Chicago/Turabian StyleZhao, Shuqi, Xiaoman Gao, Xiaolei Yu, Tao Yuan, Guiyu Zhang, Chenlai Liu, Xinzhong Li, Pei Wei, Xiaoyan Li, and Xing Liu. 2024. "Comparative Analysis of Chloroplast Genome of Meconopsis (Papaveraceae) Provides Insights into Their Genomic Evolution and Adaptation to High Elevation" International Journal of Molecular Sciences 25, no. 4: 2193. https://doi.org/10.3390/ijms25042193
APA StyleZhao, S., Gao, X., Yu, X., Yuan, T., Zhang, G., Liu, C., Li, X., Wei, P., Li, X., & Liu, X. (2024). Comparative Analysis of Chloroplast Genome of Meconopsis (Papaveraceae) Provides Insights into Their Genomic Evolution and Adaptation to High Elevation. International Journal of Molecular Sciences, 25(4), 2193. https://doi.org/10.3390/ijms25042193






