Isolation, Characterization, Genome Annotation, and Evaluation of Tyrosinase Inhibitory Activity in Secondary Metabolites of Paenibacillus sp. JNUCC32: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation
Abstract
1. Introduction
2. Results and Discussion
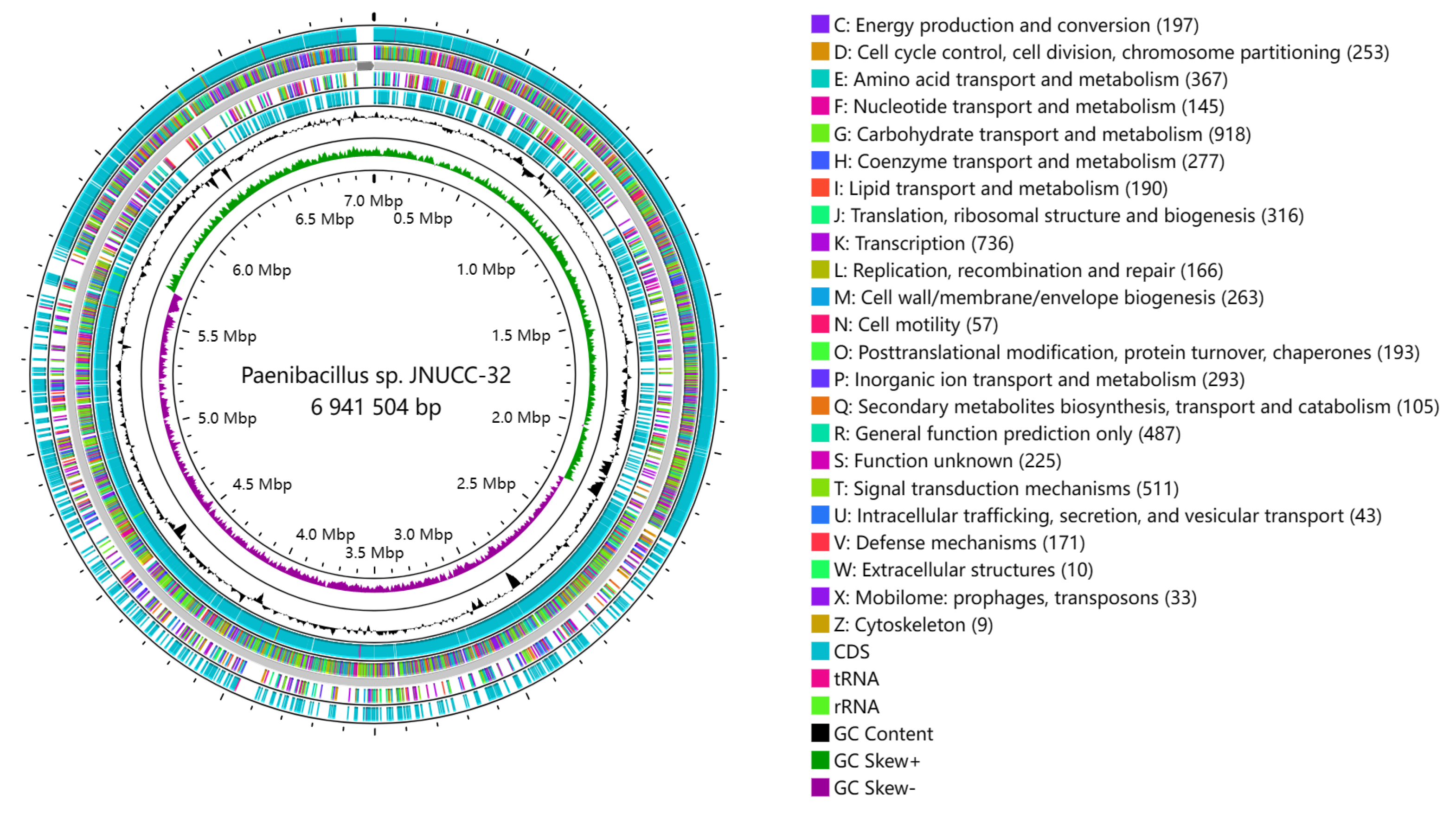
2.1. General Features of Paenibacillus sp. JNUCC32 Genome
2.2. Genome Annotation
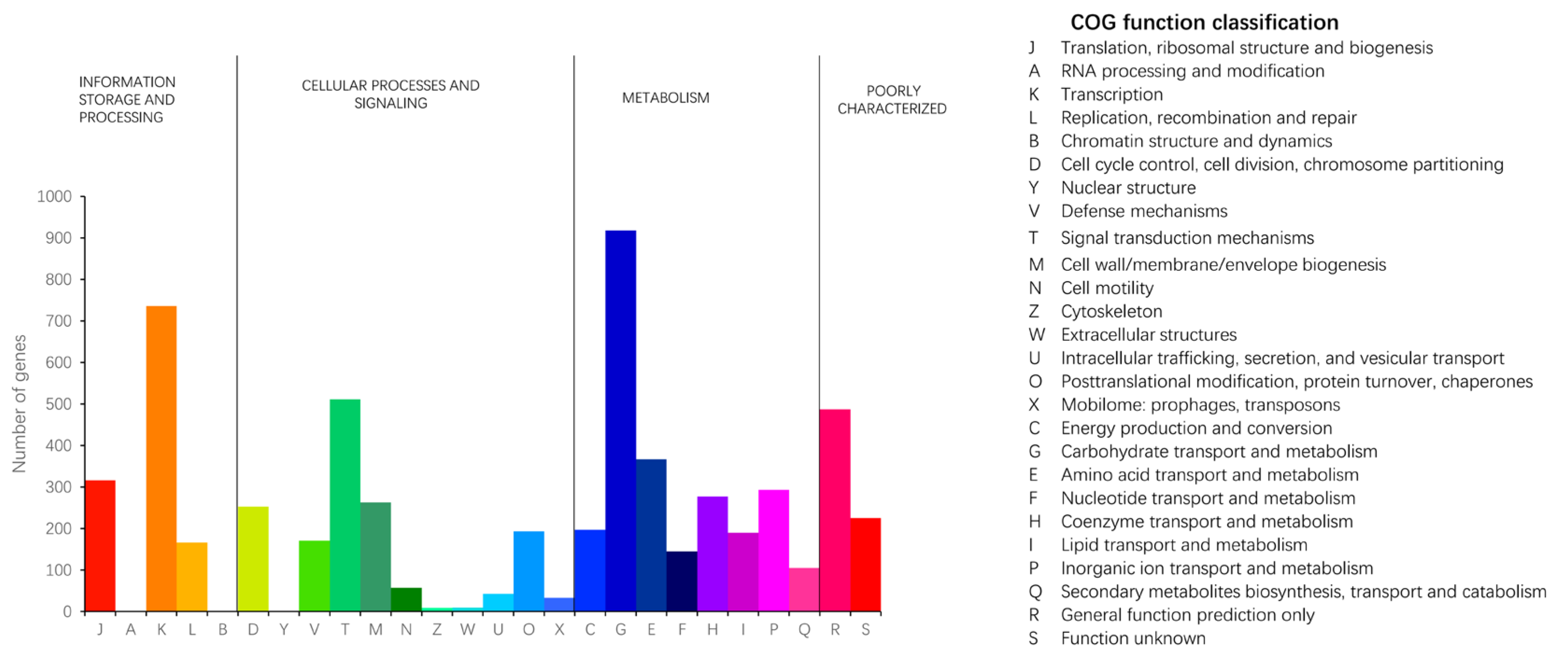
2.2.1. COG Database Annotations
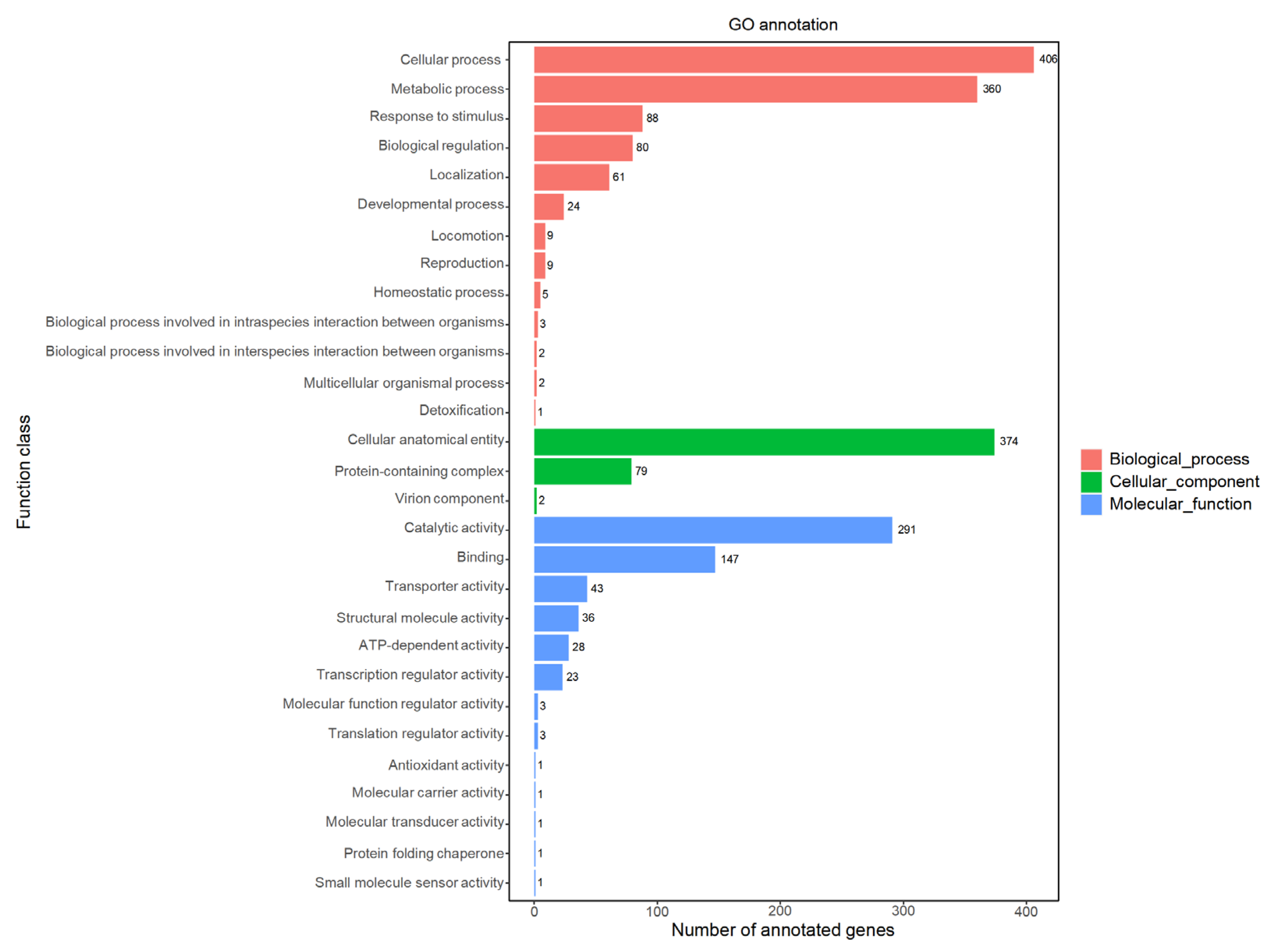
2.2.2. GO Database Annotations
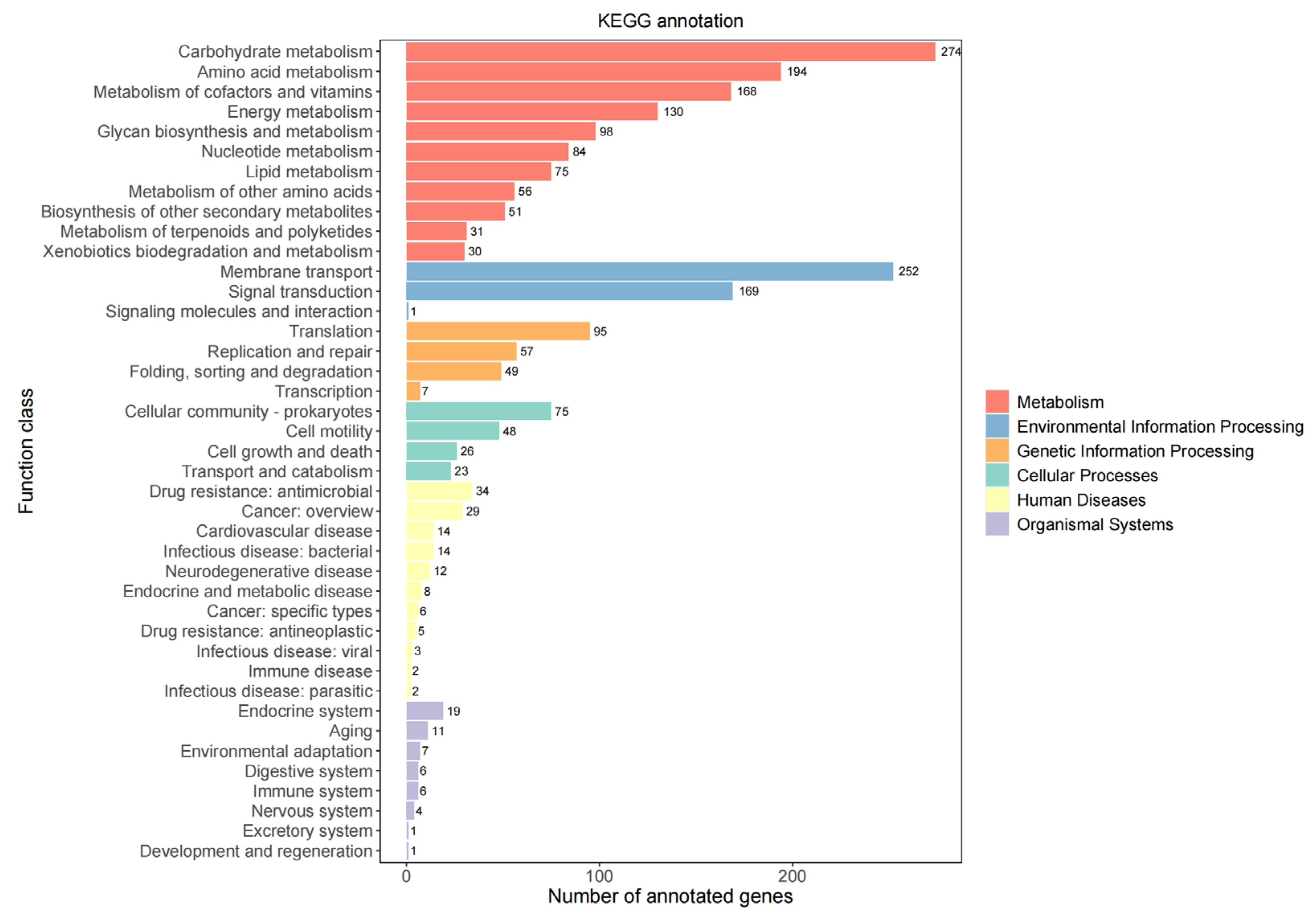
2.2.3. KEGG Database Annotations
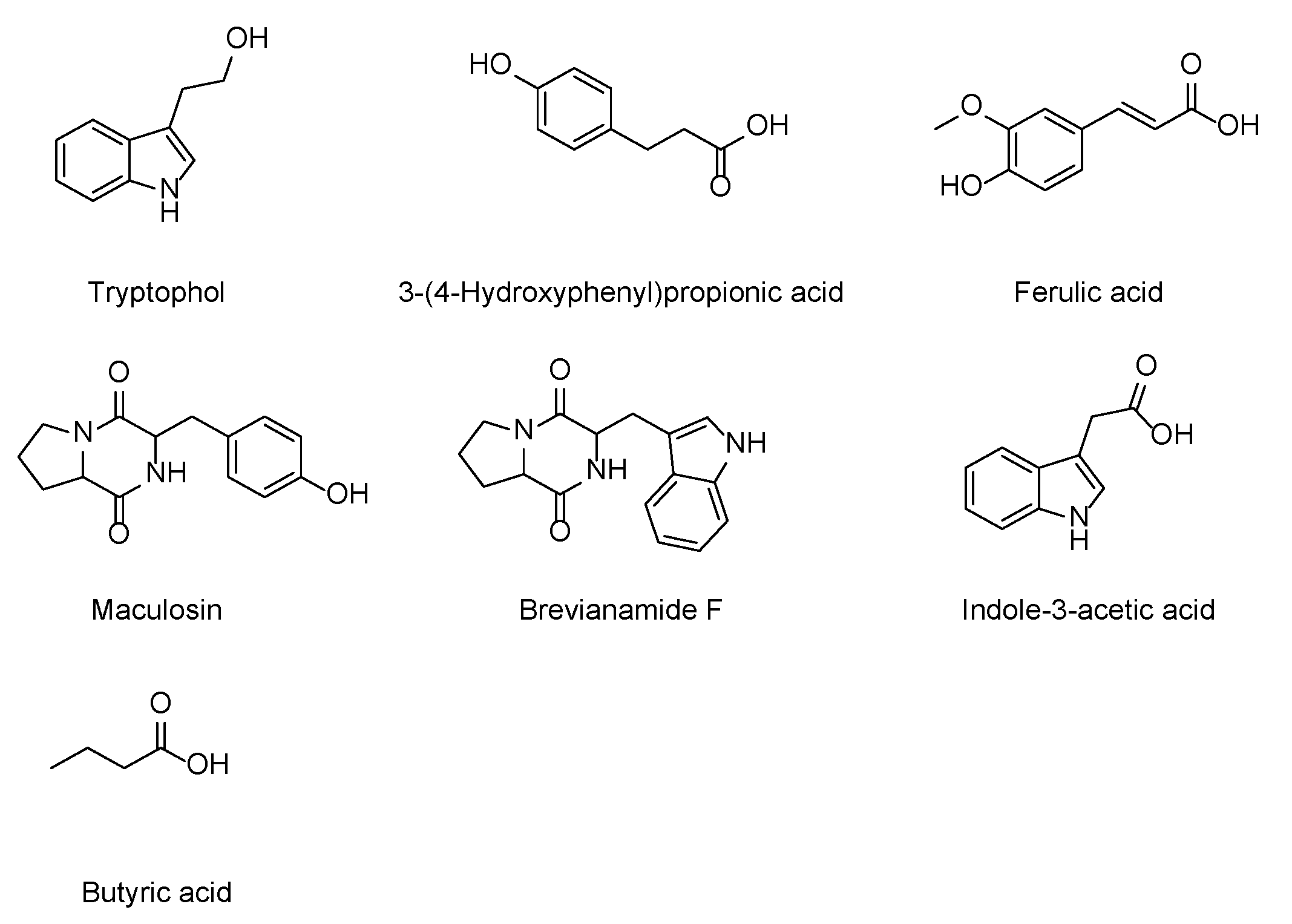
2.3. Secondary Metabolites Isolated from Paenibacillus sp. JNUCC32
2.4. Biological Activities of the Isolated Secondary Metabolites
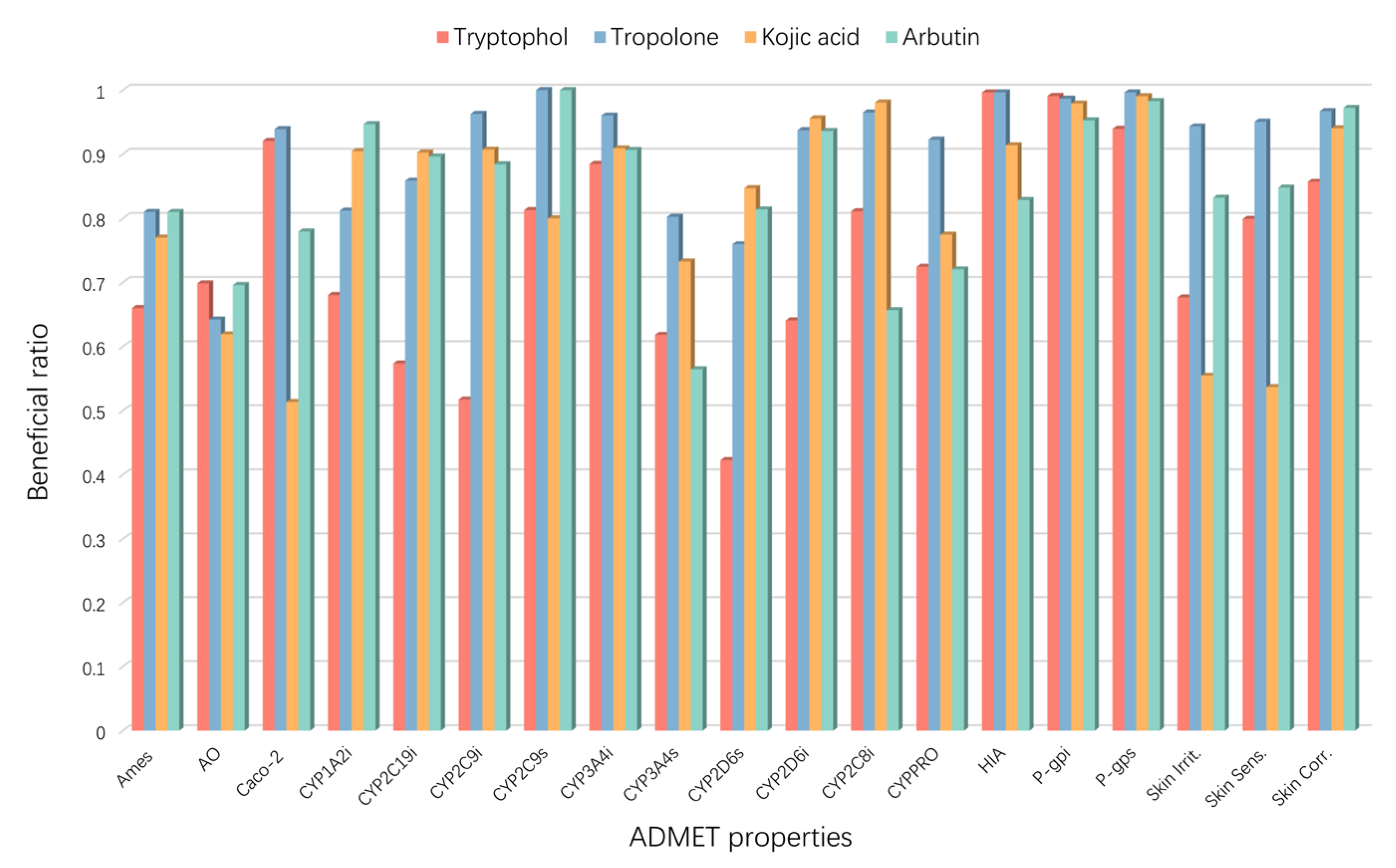
2.5. Molecular Properties and Drug-Likeness
2.6. Docking and Molecular Dynamics (MD) Simulations
2.6.1. Molecular Docking
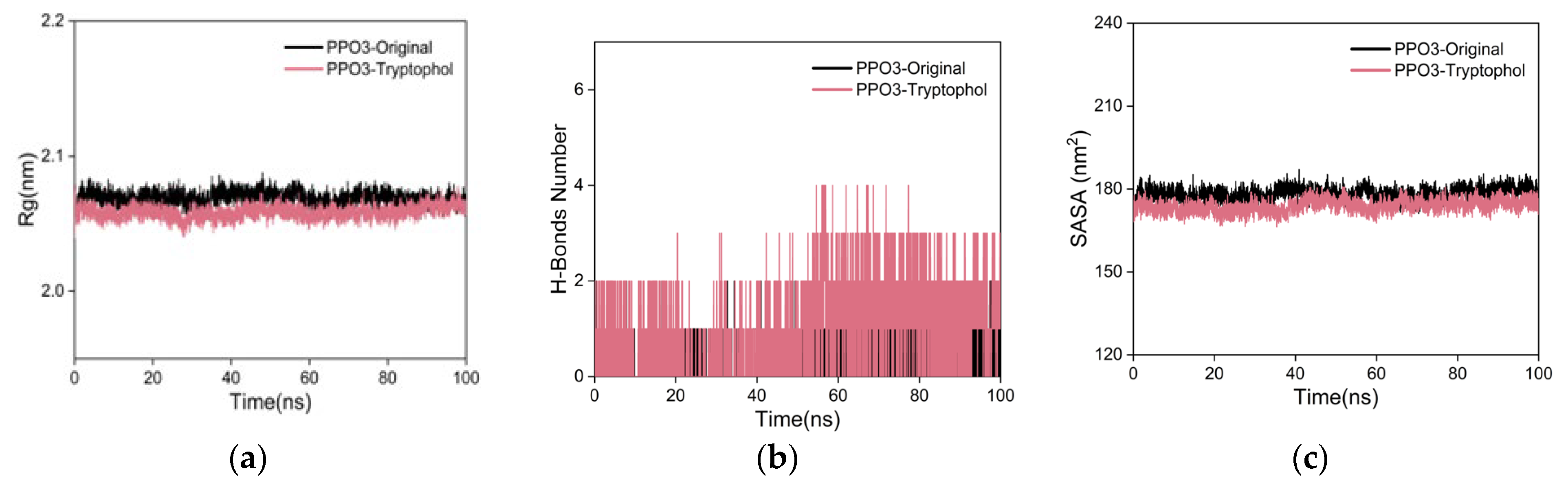
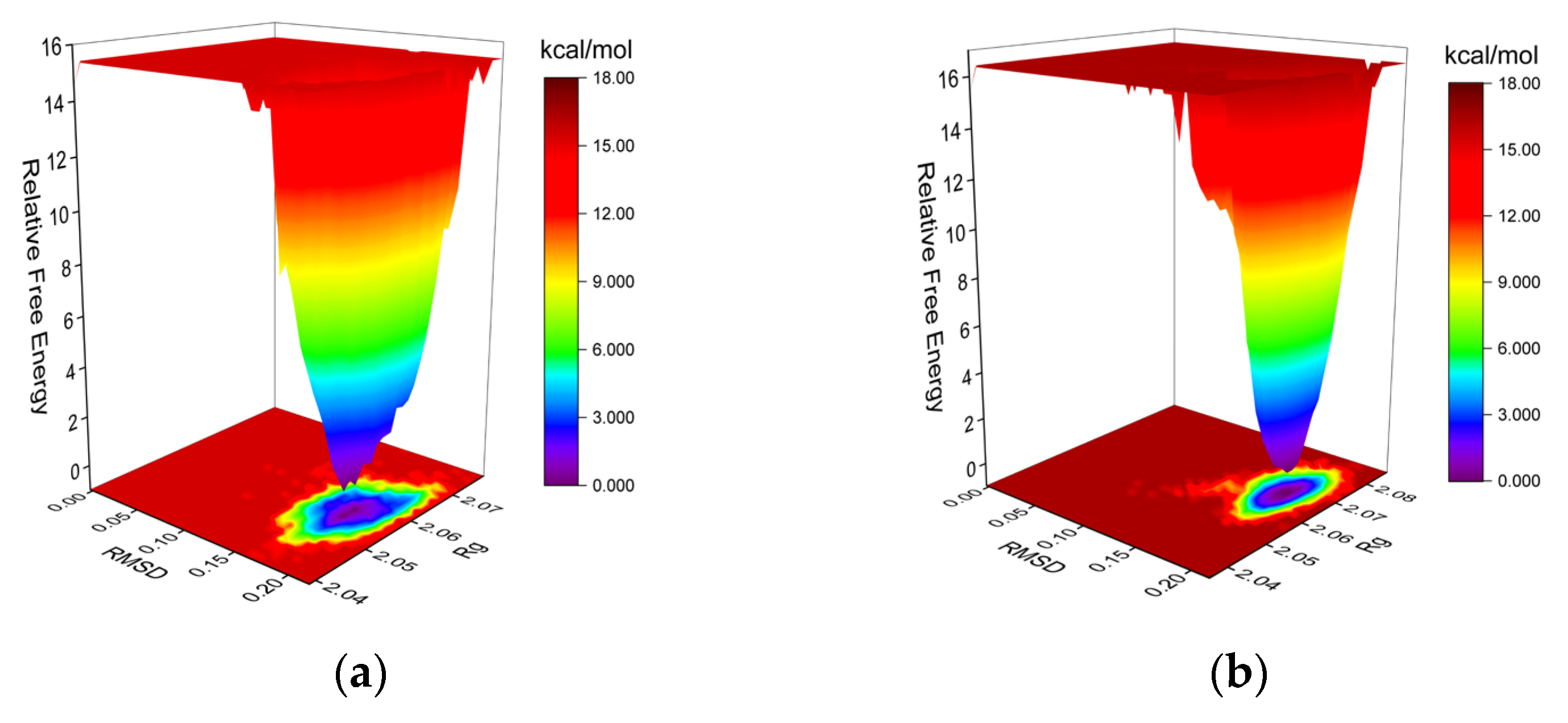
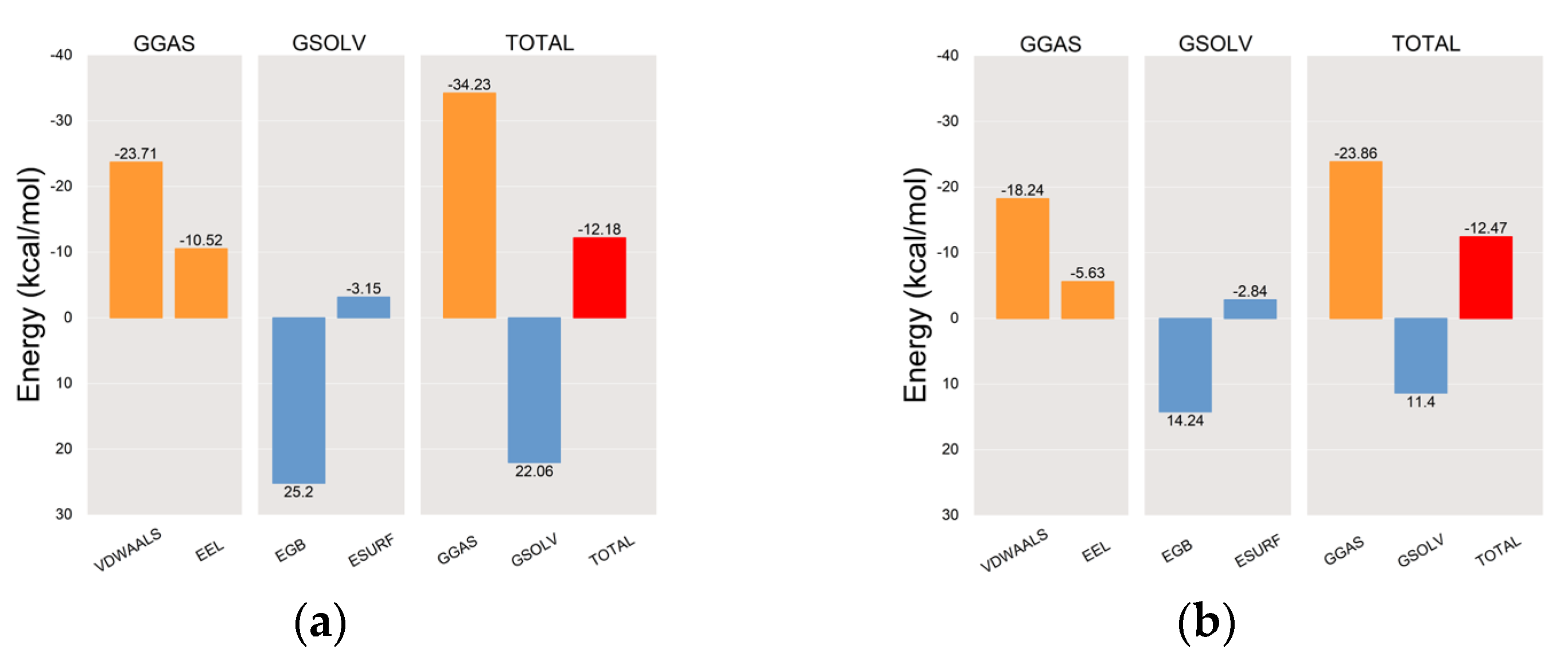
2.6.2. Molecular Dynamics (MD) Simulations
3. Materials and Methods
3.1. Bacterial Isolation
3.2. Genomic Analysis of Strain JNUCC32
3.3. General Experimental Procedures
3.4. Fermentation, Extraction, and Isolation
3.4.1. Tryptophol
3.4.2. 3-(4-Hydroxyphenyl)propionic Acid
3.4.3. Ferulic Acid
3.4.4. Maculosin
3.4.5. Brevianamide F
3.4.6. Indole-3-acetic Acid
3.4.7. Butyric Acid
3.5. Biological Activity of Secondary Metabolites
Enzymatic Assay
3.6. Molecular Properties and Drug-Likeness
3.7. Molecular Docking Simulation
3.8. Molecular Dynamics (MD) Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Priest, F.G.; Goodfellow, M.; Todd, C. A numerical classification of the genus Bacillus. J. Gen. Microbiol. 1988, 134, 1847–1882. [Google Scholar] [CrossRef]
- Ash, C.; Farrow, J.; Wallbanks, S.; Collins, M. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 1991, 13, 202–206. [Google Scholar] [CrossRef]
- Ash, C.; Fergus, G. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie van Leeuwenhoek 1993, 64, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Pichard, B.; Larue, J.P.; Thouvenot, D. Gavaserin and saltavalin, new peptide antibiotics produced by Bacillus polymyxa. FEMS Microbiol. Lett. 1955, 133, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Raza, W.; Yang, W.; Shen, Q.R. Paenibacillus polymyxa: Antibiotics, hydrolytic enzymes and hazard assessment. Plant Pathol. J. 2008, 90, 419–430. [Google Scholar]
- Yoshio, K.; Miyuki, K. Fusaricidin A, a New Depsipeptide Antibiotic Produced by Bacillus polymyxa KT-8. J. Antibiot. 1996, 49, 129–135. [Google Scholar]
- Singh, A.K.; Ghodke, I.; Chhatpar, H.S. Pesticide tolerance of Paenibacillus sp. D1 and its chitinase. J. Environ. Manag. 2009, 91, 358–362. [Google Scholar] [CrossRef]
- Chandra, S.P.; Rafal, S.; Andrzej, T. Sequencing technologies and genome sequencing. J. Appl. Genet. 2011, 52, 413–435. [Google Scholar]
- Ralf, D. Friedrich Miescher and the discovery of DNA. Dev. Biol. 2005, 278, 274–288. [Google Scholar]
- Götz, S.; García-Gómez, J.M.; Terol, J. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Michael, Y.G.; Kira, S.M.; Yuri, I.W.; Eugene, V.K. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015, 43, 261–269. [Google Scholar]
- Evelyn, C.; Michele, M.; Daniel, B.; Vivian, L.; Emily, D.; John, M.; David, B.; Nicola, H.; Rodrigo, L.; Rolf, A. The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004, 32, 262–266. [Google Scholar]
- Minoru, K.; Yoko, S. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2019, 29, 28–35. [Google Scholar]
- Eva, G.G.; Sebastian, M.; Gillian, H.; Nina, H.; Roderich, D.S.; Elke, G. Biological effects of paenilamicin, a secondary metabolite antibiotic produced by the honey bee pathogenic bacterium Paenibacillus larvae. MicrobiologyOpen 2014, 3, 642–656. [Google Scholar]
- Delker, C.; Raschke, A.; Quint, M. Auxin dynamics: The dazzling complexity of a small molecule’s message. Planta 2008, 227, 929–941. [Google Scholar] [CrossRef]
- Lorentz, R.H.; Artico, S.; Da, S.A.B.; Einsfeld, A. Evaluation of antimicrobial activity in Paenibacillus sp. strains isolated from natural environment. Lett. Appl. Microbiol. 2006, 43, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Sang, G.K.; Zakaullah, K.; Yong, H.J.; Young, H.K. Inhibitory Effect of Paenibacillus polymyxa GBR-462 on Phytophthora capsici Causing Phytophthora Blight in Chili Pepper. J. Phytopathol. 2009, 157, 329–337. [Google Scholar]
- Qin, H.; Shang, Z.; Jantan, I. Molecular docking studies and biological evaluation of chalcone based pyrazolines as tyrosinase inhibitors and potential anticancer agents. RSC Adv. 2015, 5, 46330–46338. [Google Scholar] [CrossRef]
- Quispe, Y.N.; Hwang, S.H.; Wang, Z. Screening of peruvian medicinal plants for tyrosinase inhibitory properties: Identification of tyrosinase inhibitors in Hypericum laricifolium Juss. Molecules 2017, 22, 402. [Google Scholar] [CrossRef]
- Arianayagam, S.; Ryan, T.J. Human pigmentation: A side effect adapted from a primitive organism’s survival: Part 2: The melanocyte as mentor of the keratinocye. Indian Dermatol. Online 2014, 5, 328–333. [Google Scholar]
- Rivas, R.; Mateos, P.F.; Martínez-Molina, E. Paenibacillus xylanilyticus sp. nov., an airborne xylanolytic bacterium. Int. J. Syst. Evol. Microbiol. 2005, 55, 405–408. [Google Scholar] [CrossRef] [PubMed]
- Zitouni, M.; Fortin, M.; Scheerle, R.K. Biochemical and molecular characterization of a thermostable chitosanase produced by the strain Paenibacillus sp. 1794 newly isolated from compost. Appl. Microbiol. Biotechnol. 2013, 97, 5801–5813. [Google Scholar] [CrossRef] [PubMed]
- Saruno, R.; Kato, F.; Ikeno, T. Kojic acid, a tyrosinase inhibitor from Aspergillus albus. Agric. Biol. Chem. 1979, 43, 1337–1338. [Google Scholar] [CrossRef]
- Wei, C.I.; Huang, T.S.; Chen, J.S. Production of kojic acid by Aspergillus candidus in three culture media. J. Food Prot. 1991, 54, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Vasantha, K.Y.; Murugesh, C.S.; Sattur, A.P. A tyrosinase inhibitor from Aspergillus niger. J. Food Sci. Technol. 2014, 51, 2877–2880. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jeong, J.H.; Lee, K.T. γ-Pyrone derivatives, kojic acid methyl ethers from a marine-derived fungus Altenaria sp. Arch. Pharm. Res. 2003, 26, 532–534. [Google Scholar] [CrossRef]
- Chang, T.S.; Ding, H.Y.; Tai, S.S.K. Mushroom tyrosinase inhibitory effects of isoflavones isolated from soygerm koji fermented with Aspergillus oryzae BCRC 32288. Food Chem. 2007, 105, 1430–1438. [Google Scholar] [CrossRef]
- Takahashi, S.; Iwai, H.; Kosaka, K. Byelyankacin: A novel melanogenesis inhibitor produced by Enterobacter sp. B20. J. Antibiot. 2007, 60, 717–720. [Google Scholar] [CrossRef]
- Arai, N.; Shiomi, K.; Takamatsu, S. Amphistin, a new melanogenesis inhibitor, produced by an actinomycete. J. Antibiot. 1997, 50, 808–814. [Google Scholar] [CrossRef][Green Version]
- Kawagishi, H.; Somoto, A.; Kuranari, J. A novel cyclotetrapeptide produced by Lactobacillus helveticus as a tyrosinase inhibitor. Tetrahedron Lett. 1993, 34, 3439–3440. [Google Scholar] [CrossRef]
- Ann, K.; John, P. Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science 1982, 217, 1163–1165. [Google Scholar]
- Olivares, C.; Solano, F. New insights into the active site structure and catalytic mechanism of tyrosinase and its related proteins. Pigment Cell Melanoma Res. 2009, 22, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, Z.R.M.; Seo, S.Y. Kinetic and in silico studies of novel hydroxy-based thymol analogues as inhibitors of mushroom tyrosinase. Eur. J. Med. Chem. 2015, 98, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.Y.; Wang, G.C.; Zeng, Q.H.; Li, Y.F. A systematic review of synthetic tyrosinase inhibitors and their structure-activity relationship. Crit. Rev. Food Sci. Nutr. 2022, 62, 4053–4094. [Google Scholar] [CrossRef] [PubMed]
- Kahn, V.; Andrawis, A. Inhibition of mushroom tyrosinase by tropolone. Phytochemistry 1985, 24, 905–908. [Google Scholar] [CrossRef]
- Espin, J.C.; Wichers, H.J. Slow-binding inhibition of mushroom (Agaricus bisporus) tyrosinase isoforms by tropolone. J. Agric. Food Chem. 1999, 47, 2638–2644. [Google Scholar] [CrossRef]
- Mayer, A.M. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry 2006, 67, 2318–2331. [Google Scholar] [CrossRef]
- Lin, X.H.; Sun, D.W. Research advances in browning of button mushroom (Agaricus bisporus): Affecting factors and controlling methods. Trends Food Sci. Technol. 2019, 90, 63–75. [Google Scholar] [CrossRef]
- Wu, J.J.; Chen, H.B.; Gao, J.Y.; Liu, X.; Cheng, W. Cloning, characterization and expression of two new polyphenol oxidase cDNAs from Agaricus bisporus. Biotechnol. Lett. 2010, 32, 1439–1447. [Google Scholar] [CrossRef]
- Reem, I.A.; Ehsan, U.M.; Nafeesa, N.; Meshari, A.A.; Amina, S.; Anser, A.; Rabab, S.J. Flavone-based hydrazones as new tyrosinase inhibitors: Synthetic imines with emerging biological potential, SAR, molecular docking and drug-likeness studies. J. Mol. Struct. 2022, 1251, 131933. [Google Scholar]
- Yasir, N.; Aamer, S.; Muhammad, R.; Samina, A.; Anser, A. Hydroxyl substituted benzoic acid/cinnamic acid derivatives: Tyrosinase inhibitory kinetics, anti-melanogenic activity and molecular docking studies. Bioorg Med. Chem. Lett. 2020, 30, 126722. [Google Scholar]
- Francisco, G.M.; Alexander, N.P.H.; Lorena, G.F.; José, N.R.L.; Pedro, A.G.R. Mushroom Tyrosinase: Catalase Activity, Inhibition, and Suicide Inactivation. J. Agric. Food Chem. 2005, 53, 3702–3709. [Google Scholar]
- Zhao, J. Plant Troponoids: Chemistry, Biological Activity, and Biosynthesis. Curr. Med. Chem. 2007, 14, 2597–2621. [Google Scholar] [CrossRef] [PubMed]
- Paweł, B.; Beata, C.G.; Tomasz, G.; Sebastian, L. In vitro and in vivo biological activities of azulene derivatives with potential applications in medicine. Med. Chem. Res. 2021, 30, 834–846. [Google Scholar]
- Wangsa, T.I.; Henriëtte, J.R.; Amrah, W.; Jurriaan, J.M.; Fabrizia, F.; Harry, J.W.; Bauke, W.D. Crystal Structure of Agaricus bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar]
- Mor, S.; Margarita, K.; Vered, S.B.Y.; Noam, A.; Ayelet, F. First Structures of an Active Bacterial Tyrosinase Reveal Copper Plasticity. J. Mol. Biol. 2011, 405, 227–237. [Google Scholar]
- Daungkamon, N.; Lalida, S.; Vannajan, S.L.; Piyarat, N. Estimation of Inhibitory Effect against Tyrosinase Activity through Homology Modeling and Molecular Docking. Enzym. Res. 2015, 2015, 12. [Google Scholar]
- Safithri, M.; Andrianto, D.; Arda, A.G. The effect of red betel (Piper crocatum) water fraction as tyrosinase inhibitors: In vitro, molecular docking, and molecular dynamics studies. J. King Saud. Univ. Sci. 2023, 35, 102933. [Google Scholar] [CrossRef]
- Kang, S.M.; Heo, S.J.; Kim, K.N.; Lee, S.H.; Yang, H.M.; Kim, A.D. Molecular docking studies of a phlorotannin, dieckol isolated from Ecklonia cava with tyrosinase inhibitory activity. Bioorg. Med. Chem. 2012, 20, 311–316. [Google Scholar] [CrossRef]
- Chen, J.S.; Wei, C.I.; Marshall, M.R. Inhibition mechanism of kojic acid on polyphenol oxidase. J. Agric. Food Chem. 1991, 39, 1897–1901. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.Q.; He, X.F.; Teng, B. Valonea Tannin: Tyrosinase Inhibition Activity, Structural Elucidation and Insights into the Inhibition Mechanism. Molecules 2021, 26, 2747. [Google Scholar] [CrossRef]
- Hu, W.J.; Yan, L.; Park, D. Kinetic, structural and molecular docking studies on the inhibition of tyrosinase induced by arabinose. Int. J. Biol. Macromol. 2012, 50, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; He, M.; Zhai, Y. Inhibitory activity and mechanism of trilobatin on tyrosinase: Kinetics, interaction mechanism and molecular docking. Food Funct. 2021, 12, 2569–2579. [Google Scholar] [CrossRef] [PubMed]
- Kaya, E.D.; Türkhan, A.; Gür, F. A novel method for explaining the product inhibition mechanisms via molecular docking: Inhibition studies for tyrosinase from Agaricus bisporus. J. Biomol. Struct. Dyn. 2022, 40, 7926–7939. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, G.; Zeng, Q.H. Synthesis, antioxidant and anti-tyrosinase activity of 1, 2, 4-triazole hydrazones as antibrowning agents. Food Chem. 2021, 341, 128265. [Google Scholar] [CrossRef] [PubMed]
- Karakaya, G.; Türe, A.; Ercan, A. Synthesis, computational molecular docking analysis and effectiveness on tyrosinase inhibition of kojic acid derivatives. Bioorg. Chem. 2019, 88, 102950. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.R.; Silva, J.R.A. Combined kinetic studies and computational analysis on kojic acid analogs as tyrosinase inhibitors. Molecules 2014, 19, 9591–9605. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.C.; Chang, C.S.; Chien, Y.C. Tyrosol and its analogues inhibit alpha-melanocyte-stimulating hormone induced melanogenesis. Int. J. Mol. Sci. 2013, 14, 23420–23440. [Google Scholar] [CrossRef]
- Scott, A.H.; Ron, O.D. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar]
- Aminpour, M.; Montemagno, C.; Tuszynski, J.A. An overview of molecular modeling for drug discovery with specific illustrative examples of applications. Molecules 2019, 24, 1693. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Chen, F.; Ye, J.; Jiang, F. Applications of molecular dynamics simulation in structure prediction of peptides and proteins. Comput. Struct. Biotechnol. J. 2019, 17, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Hospital, A.; Goñi, J.R.; Orozco, M.; Gelpí, J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. [Google Scholar]
- Jason, R.G.; Paul, S. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar]
- Kaleem, S.; Ge, H.J.; Yi, W.W.; Zhang, Z.Z.; Wu, B. Isolation, structural elucidation, and antibacterial evaluation of the metabolites from a marine-derived fungus Penicillium sp. ZZ1283. Nat. Prod. Res. 2019, 35, 2498–2506. [Google Scholar] [CrossRef] [PubMed]
- Blanca, C.L.; Samuel, E.S.; Fabiola, C.S.; Abraham, G.H.; Litzia, C.R. Design, Synthesis and in Combo Antidiabetic Bioevaluation of Multitarget Phenylpropanoic Acids. Molecules 2018, 23, 340. [Google Scholar]
- Wu, Y.; Zhang, Y.F.; Zhang, H.T.; Ma, X.H. A new cyclopeptide alkaloid from Clematis florida. Nat. Prod. Res. 2020, 36, 1693–1699. [Google Scholar] [CrossRef]
- Nishanth, K.; Mohandas, C.; Bala, N.; Kumar, D.R.S.; Lankalapalli, R.S. Isolation of proline-based cyclic dipeptides from Bacillus sp. N strain associated with rhabitid entomopathogenic nematode and its antimicrobial properties. World J. Microb. Biot. 2013, 29, 355–364. [Google Scholar]
- Maria, H.; Eleni, K.; Panagiota, G.; Vassilios, R.; Efstathia, I. New Chlorinated 2,5-Diketopiperazines from Marine-Derived Bacteria Isolated from Sediments of the Eastern Mediterranean Sea. Molecules 2020, 25, 1509. [Google Scholar]
- Tsui, K.Y.; Tombari, R.J.; Olson, D.E.; Tantillo, D.J. Reconsidering the Structure of Serlyticin-A. J. Nat. Prod. 2019, 82, 3464–3468. [Google Scholar] [CrossRef]
- Zhang, S.J.; Chang, H.B.; Lv, W.Q.; Yu, D.; Li, Y.F. Chemical Components of Fallen Leaves of Salix matsudana Koidz. Chem. Ind. For. Prod. 2013, 33, 97–101. [Google Scholar]
- Paul, S.M.; Mytelka, D.S.; Dunwiddie, C.T. How to improve R&D productivity: The pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010, 9, 203–214. [Google Scholar]
- Xu, C.; Cheng, F.; Chen, L. In silico prediction of chemical Ames mutagenicity. J. Chem. Inf. Model. 2012, 52, 2840–2847. [Google Scholar] [CrossRef]
- Manikandan, P.; Nagini, S. Cytochrome P450 structure, function and clinical significance: A review. Curr. Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef]
- Thorn, C.F.; Aklillu, E.; Klein, T.E. PharmGKB summary: Very important pharmacogene information for CYP1A2. Pharmacogenet. Genom. 2012, 22, 73. [Google Scholar] [CrossRef]
- Rosemary, J.; Adithan, C. The pharmacogenetics of CYP2C9 and CYP2C19: Ethnic variation and clinical significance. Curr. Clin. Pharmacol. 2007, 2, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Ingelman-Sundberg, M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharmacogenom. J. 2005, 5, 6–13. [Google Scholar] [CrossRef]
- Lagorce, D.; Douguet, D.; Miteva, M.A. Computational analysis of calculated physicochemical and ADMET properties of protein-protein interaction inhibitors. Sci. Rep. 2017, 7, 46277. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Doak, B.C.; Over, B.; Giordanetto, F. Oral druggable space beyond the rule of 5: Insights from drugs and clinical candidates. Chem. Biol. 2014, 21, 1115–1142. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Daniel, S.; Bert, L.D.G. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar]
- Mariana, P.B.; Cleydson, B.R.S.; Leonardo, B.F. Pharmacophore and structure-based drug design, molecular dynamics and admet/tox studies to design novel potential pad4 inhibitors. J. Biomol. Struct. Dyn. 2018, 37, 966–981. [Google Scholar]
- Sargsyan, K.; Grauffel, C.; Lim, C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Benson, N.C.; Daggett, V. A comparison of multiscale methods for the analysis of molecular dynamics simulations. J. Phys. Chem. B 2012, 116, 8722–8731. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, E.; Akimoto, T.; Mitsutake, A. Universal relation between instantaneous diffusivity and radius of gyration of proteins in aqueous solution. Phys. Rev. Lett. 2021, 126, 128101. [Google Scholar] [CrossRef]
- Martı, J. Analysis of the hydrogen bonding and vibrational spectra of supercritical model water by molecular dynamics simulations. J. Chem. Phys. 1999, 110, 6876–6886. [Google Scholar] [CrossRef]
- Kleinjung, J.; Fraternali, F. Design and application of implicit solvent models in biomolecular simulations. Curr. Opin. Struct. Biol. 2014, 25, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Ausaf, A.S.; Hassan, I.; Islam, A. A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr. Protein Pept. Sci. 2014, 15, 456–476. [Google Scholar]
- Deng, Y.; Roux, B. Computations of standard binding free energies with molecular dynamics simulations. J. Phys. Chem. B 2009, 113, 2234–2246. [Google Scholar] [CrossRef]
- Fu, H.; Chen, H.; Blazhynska, M. Accurate determination of protein: Ligand standard binding free energies from molecular dynamics simulations. Nat. Protoc. 2022, 17, 1114–1141. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.K.; Kumar, T.R.; Prasad, O.R. Recent advances in protein-ligand interactions: Molecular dynamics simulations and binding free energy. Curr. Comput. Aided Drug Des. 2013, 9, 518–531. [Google Scholar]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Fu, T.; Jin, Z.; Xiu, Z. Binding free energy estimation for protein-ligand complex based on MM-PBSA with various partial charge models. Curr. Pharm. Des. 2013, 19, 2293–2307. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, Z.; Zhang, J. Molecular dynamics simulation of the interaction of food proteins with small molecules. Food Chem. 2023, 405, 134824. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, 5–14. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Yang, H.; Cai, Y. ADMET-score–a comprehensive scoring function for evaluation of chemical drug-likeness. MedChemComm 2019, 10, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Noel, M.O.B.; Michael, B.; Craig, A.J.; Chris, M.; Tim, V.; Geoffrey, R.H. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar]
- Garrett, M.M.; Ruth, H.; Arthur, J.O. Using AutoDock for Ligand-Receptor Docking. Curr. Protoc. Bioinform. 2008, 24, 8–14. [Google Scholar]
- Feldstein, A.; Chang, F.H.; Kucharski, J.M. Tryptophol, 5-hydroxytryptophol and 5-methoxytryptophol induced sleep in mice. Life Sci. 1970, 9, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Gong, J.; Kim, J. Adiponectin-Secretion-Promoting Cyclic Peptide–Polyketide Hybrids from a Halophyte-Associated Fungus, Colletotrichum gloeosporioides JS0417. J. Nat. Prod. 2022, 85, 501–510. [Google Scholar] [CrossRef]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 2016, 167, 1125–1136. [Google Scholar] [CrossRef]
- Dos, S.R.; Morais-Urano, R.P.; Marçal, R.M. Acetylcholinesterase and butyrylcholinesterase inhibition by nectriapyrone and tryptophol isolated from endophytic fungus Phomopsis sp. Nat. Prod. Res. 2022, 36, 153–4158. [Google Scholar]
- Inagaki, S.; Morimura, S.; Gondo, K. Isolation of tryptophol as an apoptosis-inducing component of vinegar produced from boiled extract of black soybean in human monoblastic leukemia U937 cells. Biosci. Biotechnol. Biochem. 2007, 71, 371–379. [Google Scholar] [CrossRef]
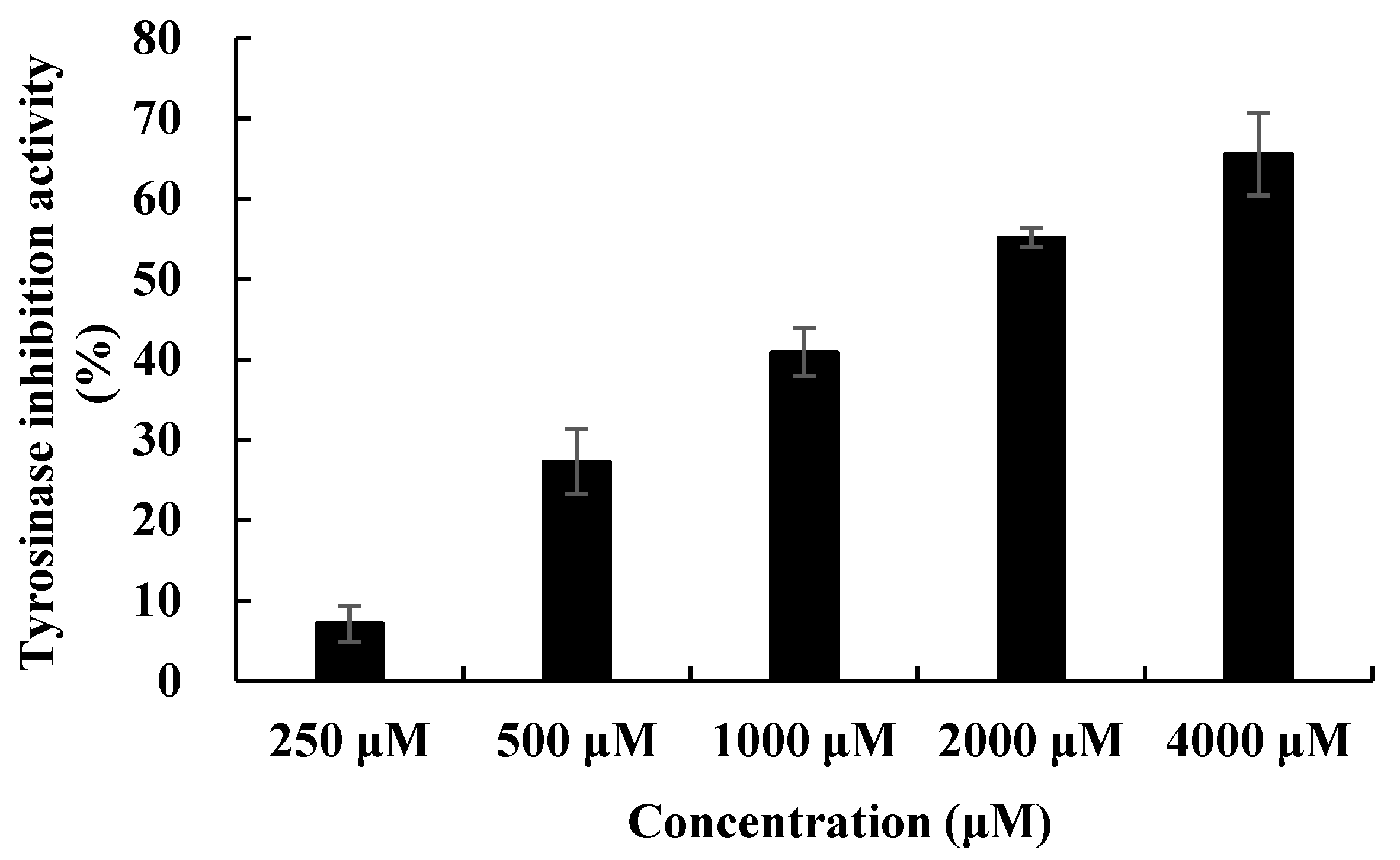




| Compounds | Tyrosinase Activity IC50 (μM) |
|---|---|
| Tryptophol | 999 |
| Maculosin | - |
| Brevianamide F | - |
| Indole-3-acetic acid | - |
| Kojic acid | 336.9 |
| Arbutin | 106.0 |
| Compound | MW | HBA | HBD | RB | TPSA (Å2) | Log p | MR | RO5 | Ghose Filter | Veber Rule | Egan Rule | Drug Likeness |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tryptophol | 161.2 | 2 | 2 | 2 | 36.02 | 1.661 | 49.23 | Yes | Yes | Yes | Yes | Yes |
| Tropolone | 122.1 | 2 | 1 | 0 | 37.30 | 0.565 | 34.74 | Yes | No | Yes | Yes | No |
| Kojic acid | 142.1 | 4 | 2 | 1 | 70.67 | −0.878 | 33.13 | Yes | No | Yes | Yes | No |
| Arbutin | 272.3 | 7 | 5 | 3 | 119.61 | −0.884 | 62.61 | Yes | No | Yes | Yes | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Liang, X.; Hyun, C.-G. Isolation, Characterization, Genome Annotation, and Evaluation of Tyrosinase Inhibitory Activity in Secondary Metabolites of Paenibacillus sp. JNUCC32: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation. Int. J. Mol. Sci. 2024, 25, 2213. https://doi.org/10.3390/ijms25042213
Xu Y, Liang X, Hyun C-G. Isolation, Characterization, Genome Annotation, and Evaluation of Tyrosinase Inhibitory Activity in Secondary Metabolites of Paenibacillus sp. JNUCC32: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation. International Journal of Molecular Sciences. 2024; 25(4):2213. https://doi.org/10.3390/ijms25042213
Chicago/Turabian StyleXu, Yang, Xuhui Liang, and Chang-Gu Hyun. 2024. "Isolation, Characterization, Genome Annotation, and Evaluation of Tyrosinase Inhibitory Activity in Secondary Metabolites of Paenibacillus sp. JNUCC32: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation" International Journal of Molecular Sciences 25, no. 4: 2213. https://doi.org/10.3390/ijms25042213
APA StyleXu, Y., Liang, X., & Hyun, C.-G. (2024). Isolation, Characterization, Genome Annotation, and Evaluation of Tyrosinase Inhibitory Activity in Secondary Metabolites of Paenibacillus sp. JNUCC32: A Comprehensive Analysis through Molecular Docking and Molecular Dynamics Simulation. International Journal of Molecular Sciences, 25(4), 2213. https://doi.org/10.3390/ijms25042213








