Cytokine Signaling in Pediatric Kidney Tumor Cell Lines WT-CLS1, WT-3ab and G-401
Abstract
1. Introduction
2. Results
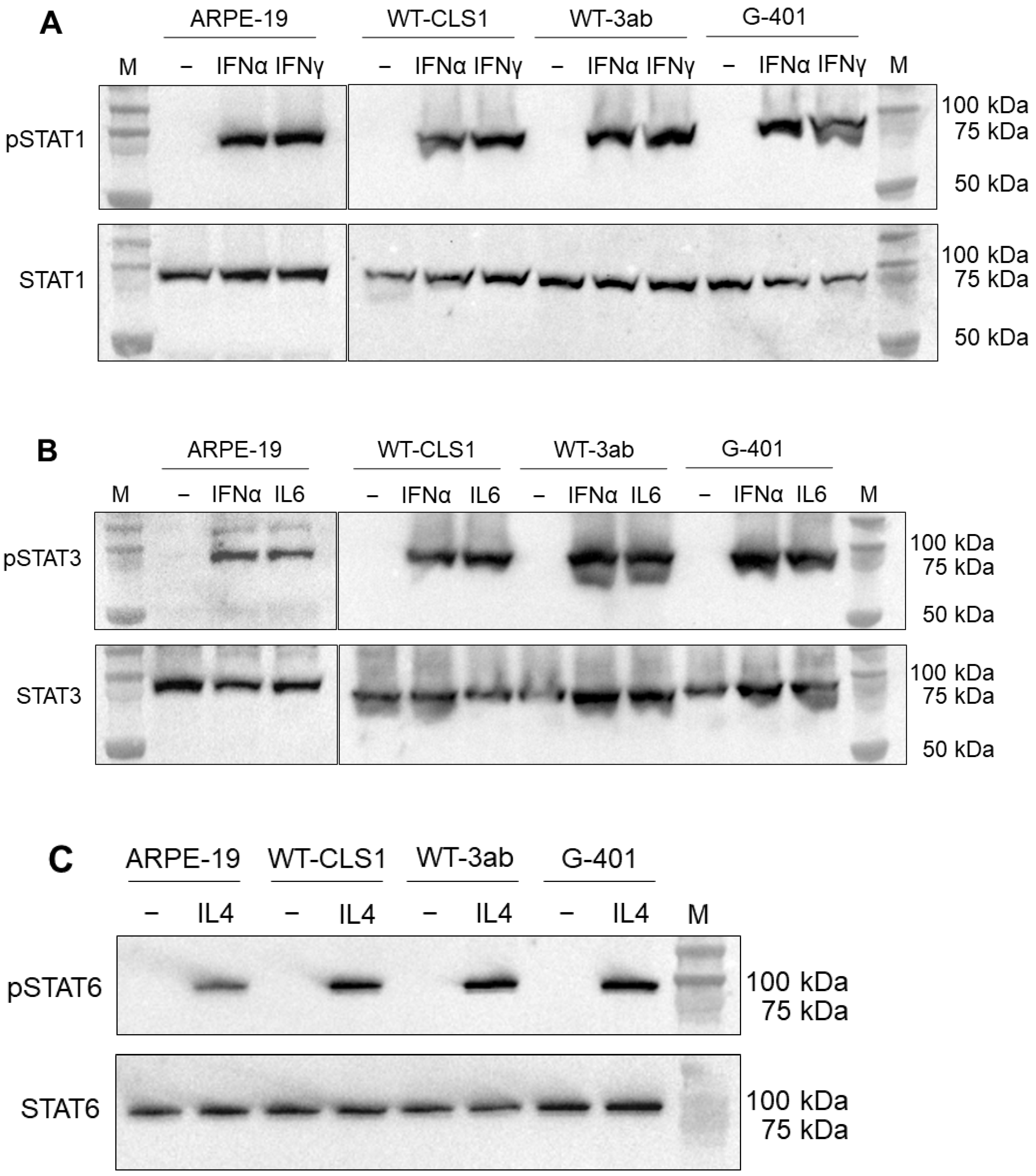
2.1. Western Blot Analysis of STAT Proteins
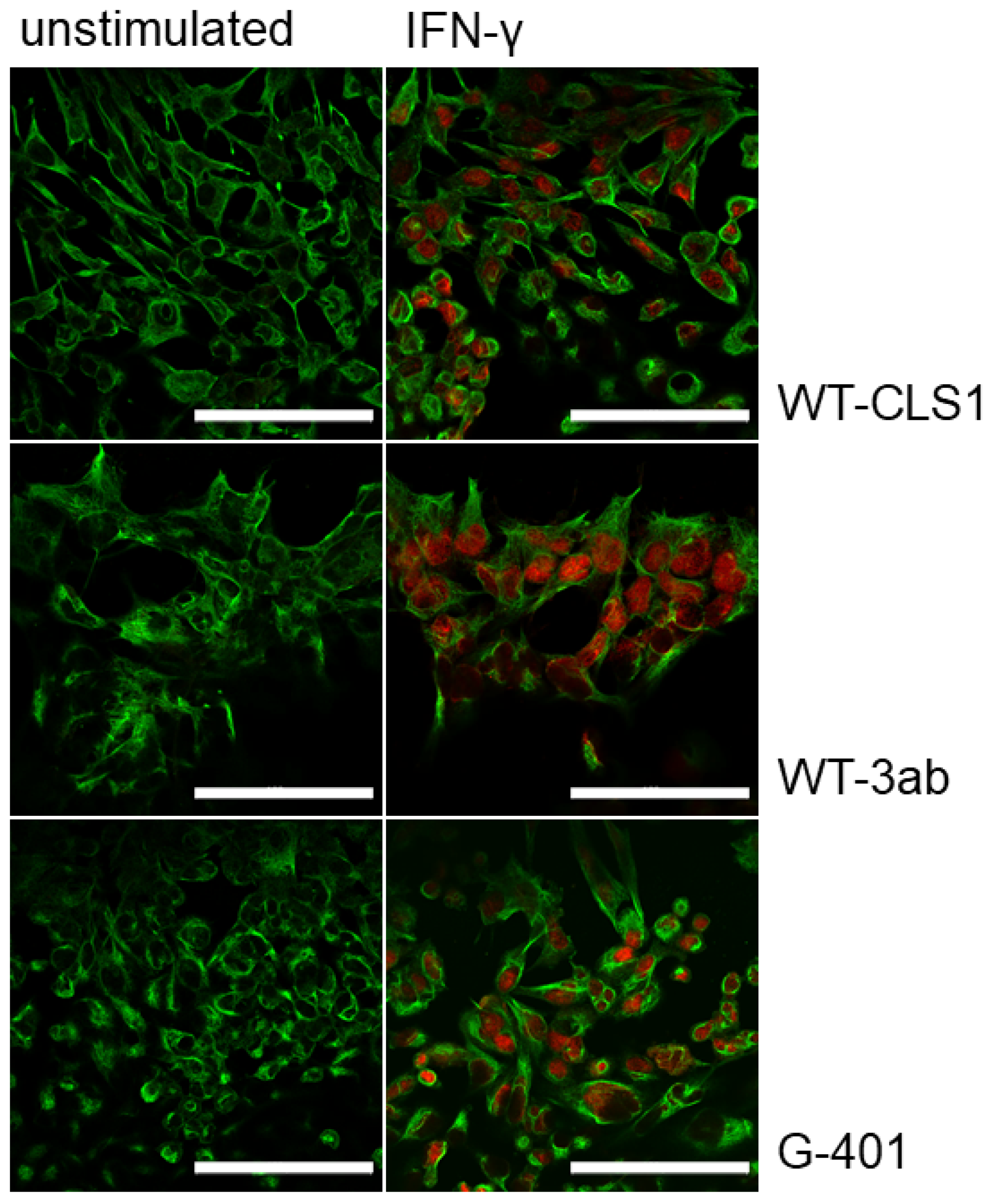
2.2. Immunofluorescence Analysis of Phosphorylated STAT Proteins
2.3. Flow Cytometry Analysis of STAT Proteins and MHC Modulation
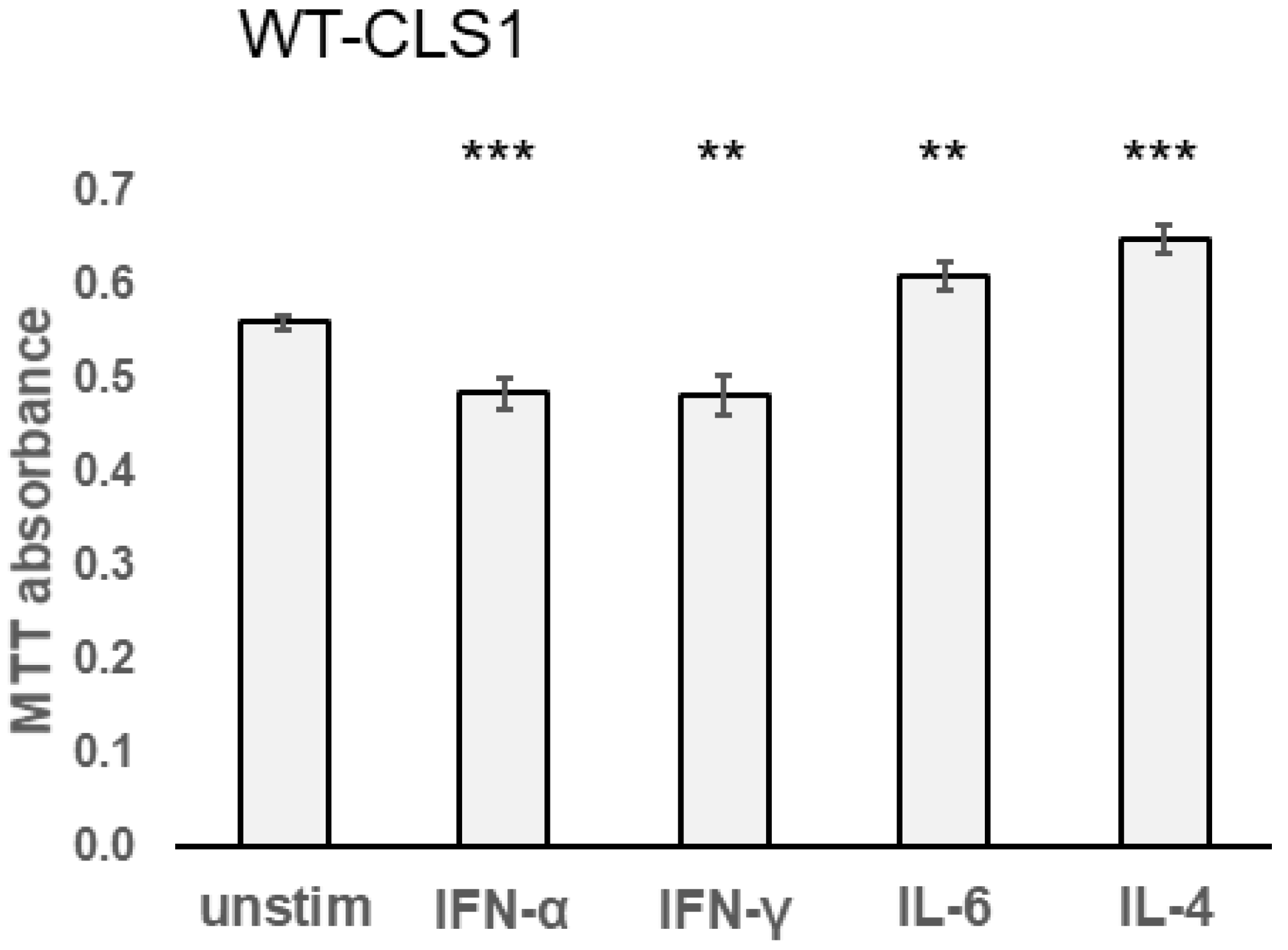
2.4. Effect of Cytokines in MTT Assay
2.5. Karyotyping Analysis of Pediatric Kidney Tumor Cells
2.6. Nuclear Factor Kappa B Signaling Pathway Activation in Pediatric Kidney Tumor Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Western Blot
4.4. Immunofluorescence Staining
4.5. Analysis of Activated STAT Proteins by Flow Cytometry
4.6. MHC Class I and Class II Modulation Assay
4.7. MTT Assay
4.8. Chromosome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ooms, A.H.A.G.; Vujanić, G.M.; D’Hooghe, E.; Collini, P.; L’Herminé-Coulomb, A.; Vokuhl, C.; Graf, N.; Heuvel-Eibrink, M.M.V.D.; de Krijger, R.R. Renal Tumors of Childhood-A Histopathologic Pattern-Based Diagnostic Approach. Cancers 2020, 12, 729. [Google Scholar] [CrossRef] [PubMed]
- Vujanić, G.M.; Gessler, M.; Ooms, A.H.A.G.; Collini, P.; Coulomb-l’Hermine, A.; D’Hooghe, E.; de Krijger, R.R.; Perotti, D.; Pritchard-Jones, K.; Vokuhl, C.; et al. International Society of Paediatric Oncology–Renal Tumour Study Group (SIOP–RTSG). The UMBRELLA SIOP-RTSG 2016 Wilms tumour pathology and molecular biology protocol. Nat. Rev. Urol. 2018, 15, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Hoot, A.C.; Russo, P.; Judkins, A.R.; Perlman, E.J.; Biegel, J.A. Immunohistochemical analysis of hSNF5/INI1 distinguishes renal and extra-renal malignant rhabdoid tumors from other pediatric soft tissue tumors. Am. J. Surg. Pathol. 2004, 28, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- López, J.I. Renal tumors with clear cells. A review. Pathol. Res. Pract. 2013, 209, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Nakata, K.; Colombet, M.; Stiller, C.A.; Pritchard-Jones, K.; Steliarova-Foucher, E.; IICC-3 Contributors. Incidence of childhood renal tumours: An international population-based study. Int. J. Cancer 2020, 147, 3313–3327. [Google Scholar] [CrossRef] [PubMed]
- Royer-Pokora, B.; Wruck, W.; Adjaye, J.; Beier, M. Gene expression studies of WT1 mutant Wilms tumor cell lines in the frame work of published kidney development data reveals their early kidney stem cell origin. PLoS ONE 2023, 18, e0270380. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E., Jr. STATs and gene regulation. Science 1997, 277, 1630–1635. [Google Scholar] [CrossRef]
- Schindler, C. Cytokines and JAK-STAT signaling. Exp. Cell Res. 1999, 253, 7–14. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Sig. Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. JAK-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef]
- Frank, D.A. STAT signaling in the pathogenesis and treatment of cancer. Mol. Med. 1999, 5, 432–456. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Darnell, J.E., Jr. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Bousoik, E.; Montazeri Aliabadi, H. “Do we know Jack” about JAK? A closer look at JAK-STAT Signaling pathway. Front. Oncol. 2018, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Ihle, J.N. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 2001, 13, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.; Jove, R. Activation of STAT transcription factors in oncogenic tyrosine kinase signaling. J. Biomed. Sci. 1998, 5, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Zhong, Z.; Darnell, J.E., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 1995, 82, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Jiang, M.; Pernis, A.B. IFN-alpha activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells. J. Immunol. 1999, 163, 3834–3841. [Google Scholar] [CrossRef]
- Morinobu, A.; Gadina, M.; Strober, W.; Visconti, R.; Fornace, A.; Montagna, C.; Feldman, G.M.; Nishikomori, R.; O’Shea, J.J. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc. Natl. Acad. Sci. USA 2002, 99, 12281–12286. [Google Scholar] [CrossRef]
- Wang, K.S.; Ritz, J.; Frank, D.A. IL-2 induces STAT4 activation in primary NK cells and NK cell lines, but not in T cells. J. Immunol. 1999, 162, 299–304. [Google Scholar] [CrossRef]
- Maurer, B.; Kollmann, S.; Pickem, J.; Hoelbl-Kovacic, A.; Sexl, V. STAT5A and STAT5B-Twins with Different Personalities in Hematopoiesis and Leukemia. Cancers 2019, 11, 1726. [Google Scholar] [CrossRef]
- Meinke, A.; Barahmand-Pour, F.; Wöhrl, S.; Stoiber, D.; Decker, T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol. Cell Biol. 1996, 16, 6937–6944. [Google Scholar] [CrossRef]
- Demoulin, J.B.; Van Roost, E.; Stevens, M.; Groner, B.; Renauld, J.C. Distinct roles for STAT1, STAT3, and STAT5 in differentiation gene induction and apoptosis inhibition by interleukin-9. J. Biol. Chem. 1999, 274, 25855–25861. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Schindler, U.; Henzel, W.J.; Ho, T.C.; Brasseur, M.; McKnight, S.L. An interleukin-4-induced transcription factor: IL-4 Stat. Science 1994, 265, 1701–1706. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Tanaka, T.; Shi, W.; Matsumoto, M.; Minami, M.; Kashiwamura, S.; Nakanishi, K.; Yoshida, N.; Kishimoto, T.; Akira, S. Essential role of Stat6 in IL-4 signalling. Nature 1996, 380, 627–630. [Google Scholar] [CrossRef] [PubMed]
- Quelle, F.W.; Shimoda, K.; Thierfelder, W.; Fischer, C.; Kim, A.; Ruben, S.M.; Cleveland, J.L.; Pierce, J.H.; Keegan, A.D.; Nelms, K. Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol. Cell Biol. 1995, 15, 3336–3343. [Google Scholar] [CrossRef] [PubMed]
- Fasler-Kan, E.; Pansky, A.; Wiederkehr, M.; Battegay, M.; Heim, M.H. Interferon-α activates signal transducers and activators of transcription 5 and 6 in Daudi cells. Eur. J. Biochem. 1998, 254, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, R.; Fimiani, G.; Monfregola, J.; Sanzari, E.; De Felice, N.; Salerno, M.C.; Pignata, C.; D’Urso, M.; Ursini, M.V. The structure of human STAT5A and B genes reveals two regions of nearly identical sequence and an alternative tissue specific STAT5B promoter. Gene 2002, 285, 311–318. [Google Scholar] [CrossRef]
- van Loo, G.; Bertrand, M.J.M. Death by TNF: A road to inflammation. Nat. Rev. Immunol. 2023, 23, 289–303. [Google Scholar] [CrossRef]
- Jang, D.I.; Lee, A.H.; Shin, H.Y.; Song, H.R.; Park, J.H.; Kang, T.B.; Lee, S.R.; Yang, S.H. The Role of Tumor Necrosis Factor Alpha (TNF-α) in Autoimmune Disease and Current TNF-α Inhibitors in Therapeutics. Int. J. Mol. Sci. 2021, 22, 2719. [Google Scholar] [CrossRef]
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Baltimore, D. NF-kappa B: Ten years after. Cell 1996, 87, 13–20. [Google Scholar] [CrossRef]
- Whiteside, S.T.; Epinat, J.C.; Rice, N.R.; Israël, A. I kappa B epsilon, a novel member of the I kappa B family, controls RelA and cRel NF-kappa B activity. EMBO J. 1997, 16, 1413–1426. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ma, C.; Zhang, Z.; Zhang, H.; Hu, H. NF-κB signaling in inflammation and cancer. Med. Comm. 2021, 2, 618–653. [Google Scholar] [CrossRef] [PubMed]
- Philips, R.L.; Wang, Y.; Cheon, H.; Kanno, Y.; Gadina, M.; Sartorelli, V.; Horvath, C.M.; Darnell, J.E., Jr.; Stark, G.R.; O’Shea, J.J. The JAK-STAT pathway at 30: Much learned, much more to do. Cell 2022, 185, 3857–3876. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Bian, Q.; Rong, D.; Wang, L.; Song, J.; Huang, H.S.; Zeng, J.; Mei, J.; Wang, P.Y. JAK/STAT pathway: Extracellular signals, diseases, immunity, and therapeutic regimens. Front. Bioeng. Biotechnol. 2023, 11, 1110765. [Google Scholar] [CrossRef] [PubMed]
- Fasler-Kan, E.; Barteneva, N.; Ketterer, S.; Wunderlich, K.; Huwyler, J.; Gygax, D.; Flammer, J.; Meyer, P. Activation of the JAK-STAT intracellular pathway in human retinal pigment epithelial cell line ARPE-19. Int. J. Interferon Cytokine and Mediat. Res. 2010, 2, 127–136. [Google Scholar] [CrossRef]
- Stock, C.; Ambros, I.M.; Lion, T.; Zoubek, A.; Amann, G.; Gadner, H.; Ambros, P.F. Genetic changes of two Wilms tumors with anaplasia and a review of the literature suggesting a marker profile for therapy resistance. Cancer Genet. Cytogenet. 2002, 135, 128–138. [Google Scholar] [CrossRef]
- Fasler-Kan, E.; Aliu, N.; Häcker, F.-M.; Ruggiero, S.; Maltsev, N.; Cholewa, D.; Milosevic, M.; Bartenstein, A.; Berger, S.M. Chromosomal heterogeneity of the G-401 rhabdoid tumor cell line: Unusual partial trisomy 7p. Front. Med. 2019, 6, 187. [Google Scholar] [CrossRef]
- Taylor, B.C.; Balko, J.M. Mechanisms of MHC-I Downregulation and Role in Immunotherapy Response. Front. Immunol. 2022, 13, 844866. [Google Scholar] [CrossRef] [PubMed]
- Casey, D.L.; Cheung, N.V. Immunotherapy of Pediatric Solid Tumors: Treatments at a Crossroads, with an Emphasis on Antibodies. Cancer Immunol. Res. 2020, 8, 161–166. [Google Scholar] [CrossRef]
- von Locquenghien, M.; Rozalén, C.; Celià-Terrassa, T. Interferons in cancer immunoediting: Sculpting metastasis and immunotherapy response. J. Clin. Investig. 2021, 131, e143296. [Google Scholar] [CrossRef] [PubMed]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in tumor progression and regression: A review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Schlichtner, S.; Yasinska, I.M.; Lall, G.S.; Berger, S.M.; Ruggiero, S.; Cholewa, D.; Aliu, N.; Gibbs, B.F.; Fasler-Kan, E.; Sumbayev, V.V. T lymphocytes induce human cancer cells derived from solid malignant tumors to secrete galectin-9 which facilitates immunosuppression in cooperation with other immune checkpoint proteins. J. Immunother. Cancer 2023, 11, e005714. [Google Scholar] [CrossRef] [PubMed]
- Kallingal, A.; Olszewski, M.; Maciejewska, N.; Brankiewicz, W.; Baginski, M. Cancer immune escape: The role of antigen presentation machinery. J. Cancer Res. Clin. Oncol. 2023, 149, 8131–8141. [Google Scholar] [CrossRef]
- Luo, Y.; Alexander, M.; Gadina, M.; O’Shea, J.J.; Meylan, F.; Schwartz, D.M. JAK-STAT signaling in human disease: From genetic syndromes to clinical inhibition. J. Allergy Clin. Immunol. 2021, 148, 911–925. [Google Scholar] [CrossRef] [PubMed]
- Rah, B.; Rather, R.A.; Bhat, G.R.; Baba, A.B.; Mushtaq, I.; Farooq, M.; Yousuf, T.; Dar, S.B.; Parveen, S.; Hassan, R.; et al. JAK/STAT Signaling: Molecular Targets, Therapeutic Opportunities, and Limitations of Targeted Inhibitions in Solid Malignancies. Front. Pharmacol. 2022, 13, 821344. [Google Scholar] [CrossRef]
- Mukaddam, K.; Ruggiero, S.; Berger, S.M.; Cholewa, D.; Kühl, S.; Vegh, D.; Payer, M.; Bornstein, M.M.; Alhawasli, F.; Fasler-Kan, E. Cytokines Activate JAK–STAT Signaling Pathway in MG-63 Cells on Titanium and Zirconia. Materials 2022, 15, 5621. [Google Scholar] [CrossRef]
- Fasler-Kan, E.; Suenderhauf, C.; Barteneva, N.; Poller, B.; Gygax, D.; Huwyler, J. Cytokine signaling in the human brain capillary endothelial cell line hCMEC/D3. Brain Res. 2010, 1354, 15–22. [Google Scholar] [CrossRef]
- Mukaddam, K.; Astasov-Frauenhoffer, M.; Fasler-Kan, E.; Ruggiero, S.; Köser, J.; Bornstein, M.; Kühl, S. Piranha-etched titanium nanostructure reduces biofilm formation. J. Clin. Oral. Investig. 2023, 27, 6187–6197. [Google Scholar] [CrossRef]
- Fasler-Kan, E.; Aliu, N.; Wunderlich, K.; Ketterer, S.; Ruggiero, S.; Berger, S.M.; Meyer, P. The Retinal Pigment Epithelial Cell Line (ARPE-19) Displays Mosaic Structural Chromosomal Aberrations. Methods Mol. Biol. 2018, 1745, 305–314. [Google Scholar]


| Cytokine | Protein | WB | IF | FC |
|---|---|---|---|---|
| IFN-α | STAT1 | +++ | ++ | ++ |
| IFN-γ | STAT1 | +++ | +++ | +++ |
| IFN-α | STAT3 | +++ | ++ | + |
| IL-6 | STAT3 | +++ | ++ | ++ |
| IL-4 | STAT6 | +++ | ++ | ++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fasler-Kan, E.; Milošević, M.; Ruggiero, S.; Aliu, N.; Cholewa, D.; Häcker, F.-M.; Dekany, G.; Bartenstein, A.; Berger, S.M. Cytokine Signaling in Pediatric Kidney Tumor Cell Lines WT-CLS1, WT-3ab and G-401. Int. J. Mol. Sci. 2024, 25, 2281. https://doi.org/10.3390/ijms25042281
Fasler-Kan E, Milošević M, Ruggiero S, Aliu N, Cholewa D, Häcker F-M, Dekany G, Bartenstein A, Berger SM. Cytokine Signaling in Pediatric Kidney Tumor Cell Lines WT-CLS1, WT-3ab and G-401. International Journal of Molecular Sciences. 2024; 25(4):2281. https://doi.org/10.3390/ijms25042281
Chicago/Turabian StyleFasler-Kan, Elizaveta, Milan Milošević, Sabrina Ruggiero, Nijas Aliu, Dietmar Cholewa, Frank-Martin Häcker, Gabriela Dekany, Andreas Bartenstein, and Steffen M. Berger. 2024. "Cytokine Signaling in Pediatric Kidney Tumor Cell Lines WT-CLS1, WT-3ab and G-401" International Journal of Molecular Sciences 25, no. 4: 2281. https://doi.org/10.3390/ijms25042281
APA StyleFasler-Kan, E., Milošević, M., Ruggiero, S., Aliu, N., Cholewa, D., Häcker, F.-M., Dekany, G., Bartenstein, A., & Berger, S. M. (2024). Cytokine Signaling in Pediatric Kidney Tumor Cell Lines WT-CLS1, WT-3ab and G-401. International Journal of Molecular Sciences, 25(4), 2281. https://doi.org/10.3390/ijms25042281







