Genome-Wide Analysis of the Oat (Avena sativa) HSP90 Gene Family Reveals Its Identification, Evolution, and Response to Abiotic Stress
Abstract
:1. Introduction
2. Results
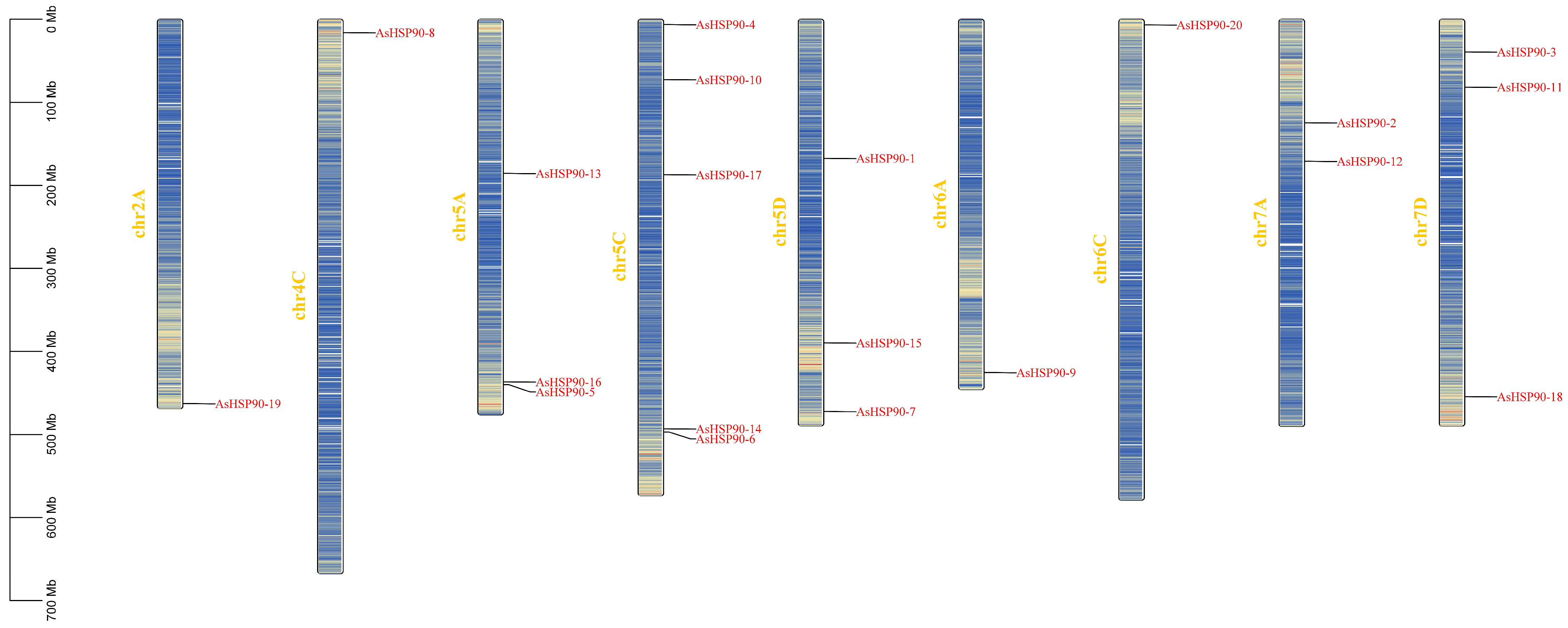
2.1. Identification of Oat HSP90 Genes and Chromosomal Distribution
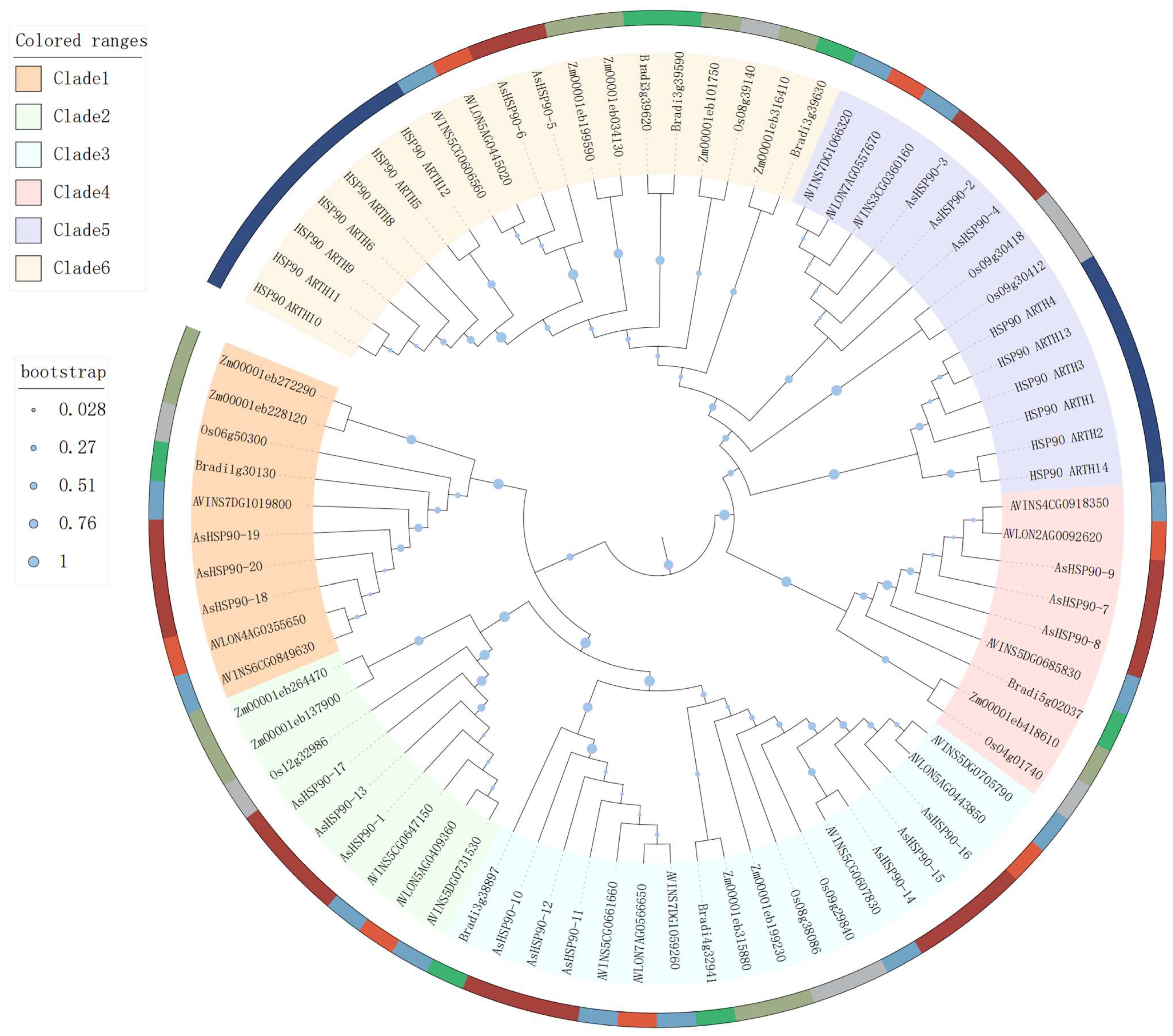
2.2. Phylogenetic Analysis of AsHSP90 Genes
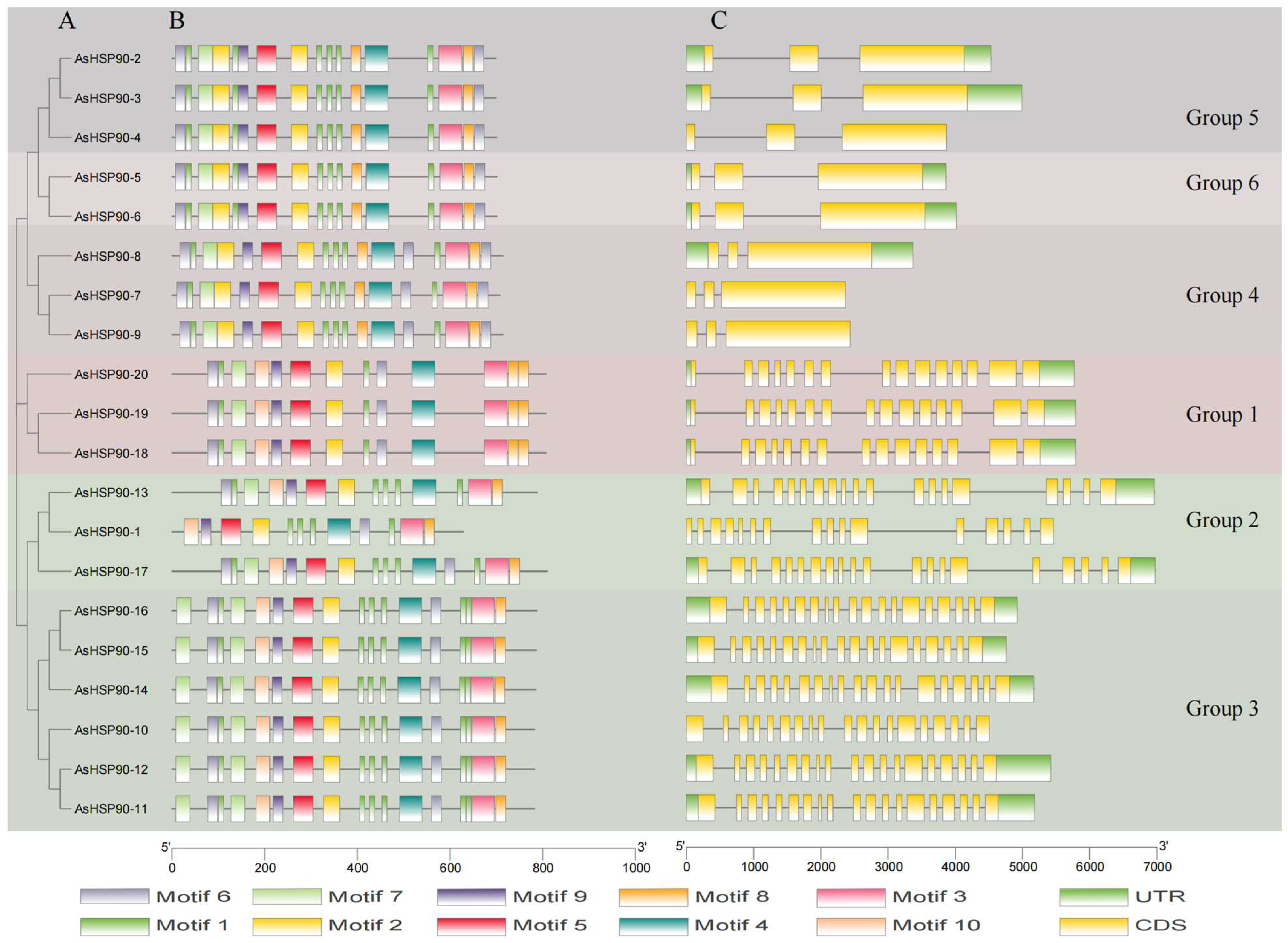
2.3. Motif Pattern and Gene Structure Analyses of AsHSP90 Members
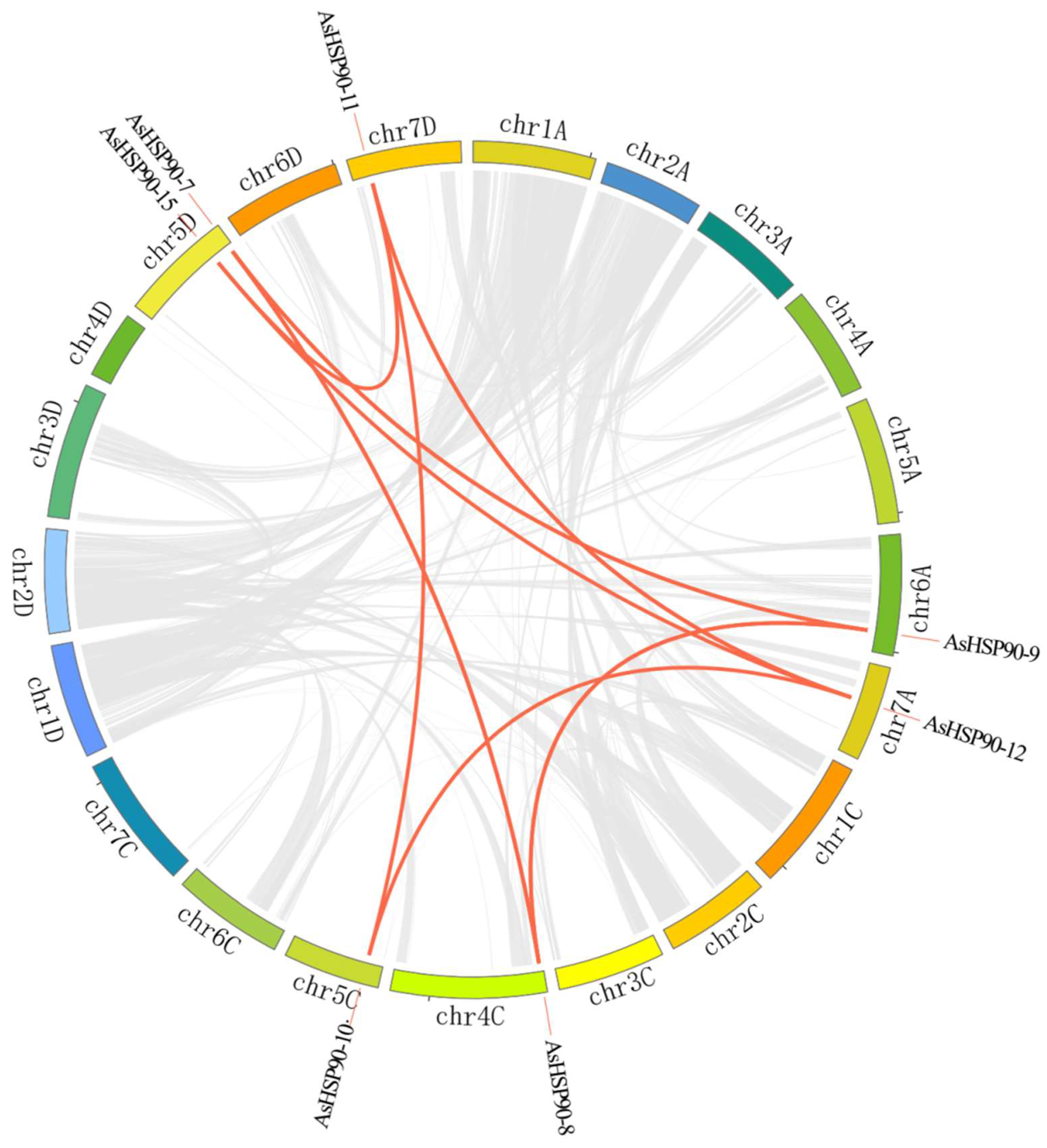
2.4. Duplication Analysis of AsHSP90 Members
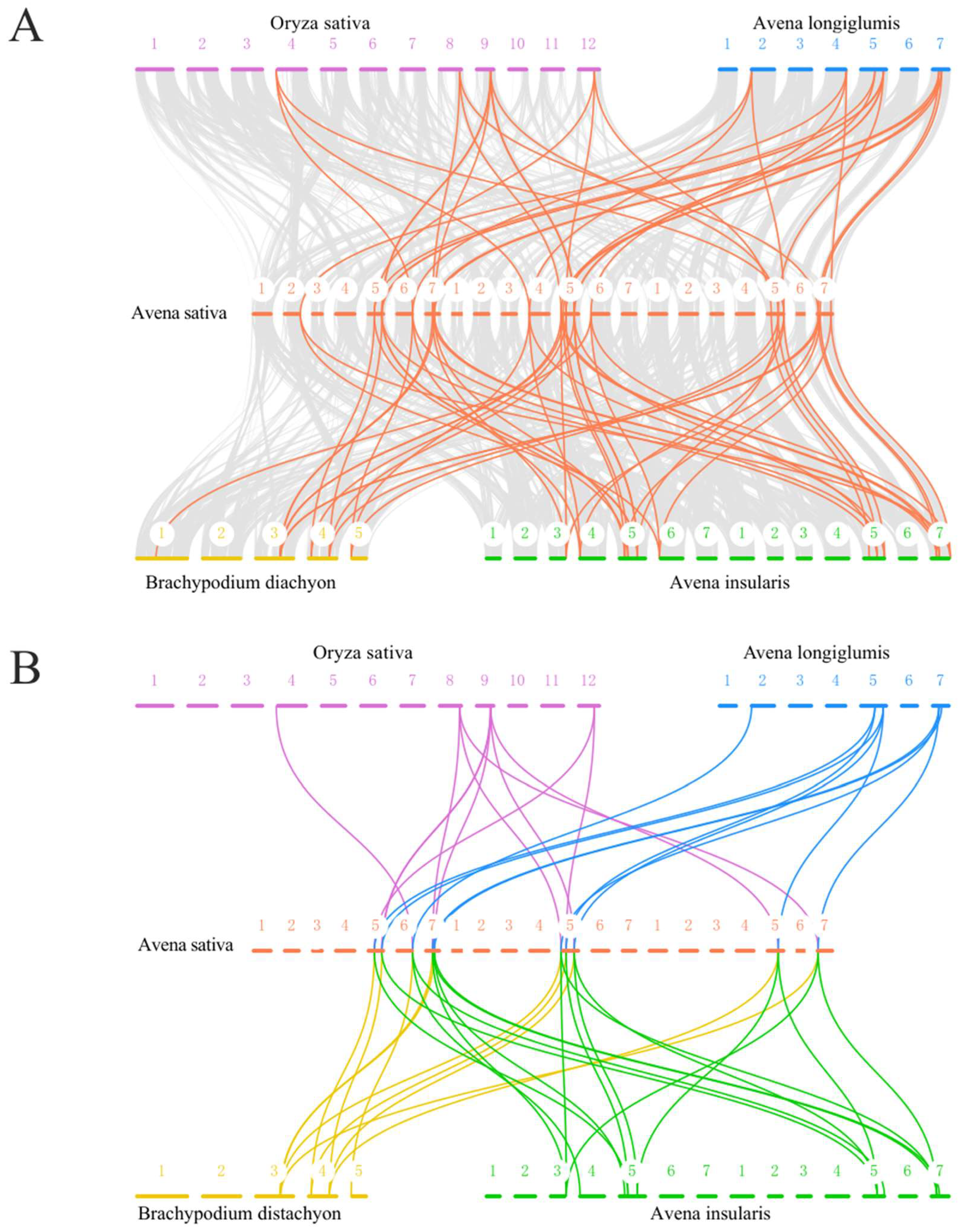
2.5. Collinearity Analysis of HSP90 Genes
2.6. Cis-Element Analysis of AsHSP90 Gene Promoters
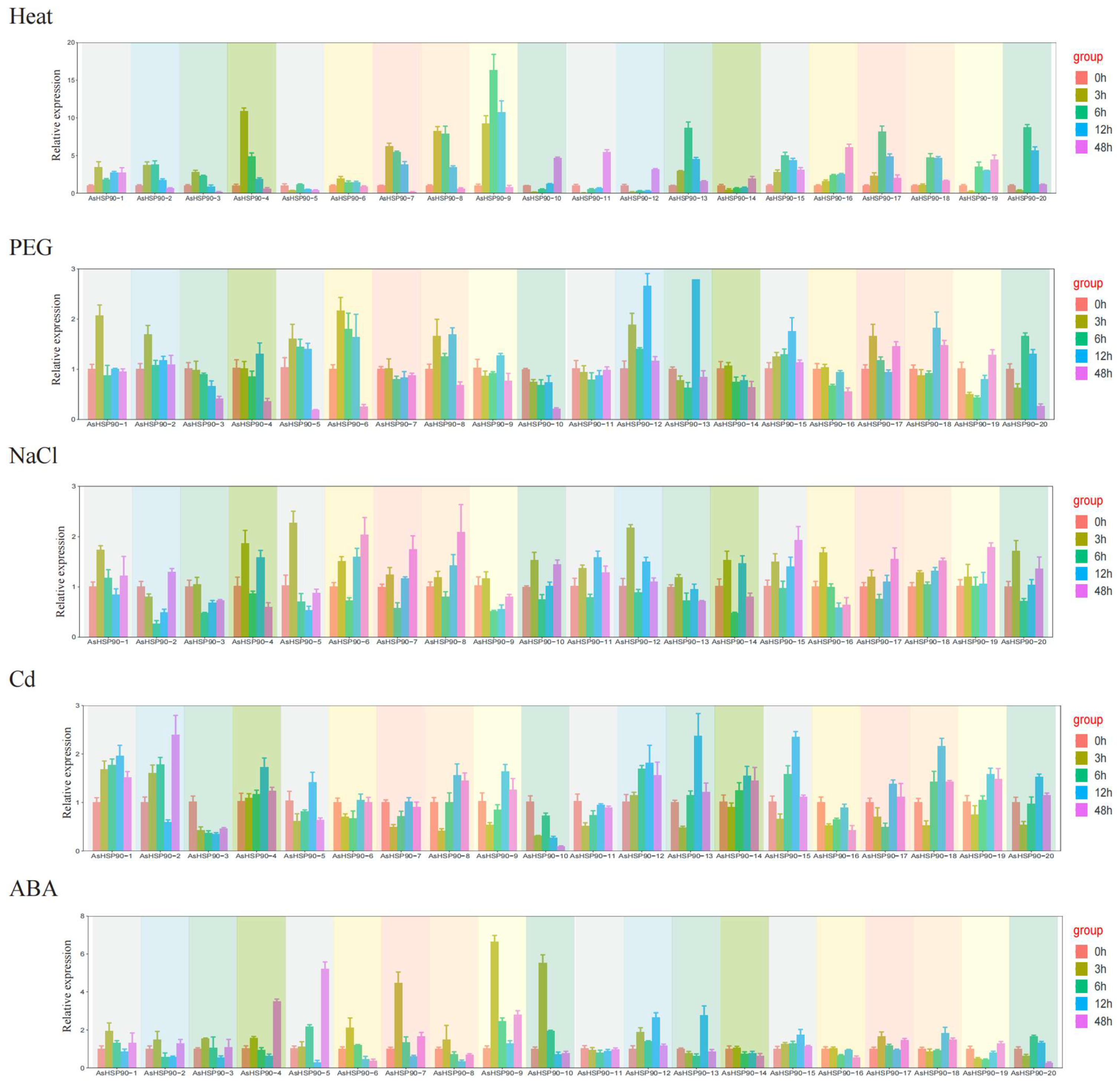
2.7. Expression Analysis of AsHSP90s in Oats under Five Abiotic Stresses
2.8. Three-Dimensional Structure Prediction and Protein–Protein Interaction Network
3. Discussion
4. Materials and Methods
4.1. HSP90 Gene Family Identification in Avena sativa, Avena insularis, and Avena longiglumis
4.2. Phylogenetic Analysis of HSP90 Genes
4.3. Structure and Conserved-Motif Analysis of AsHSP90s
4.4. Gene Duplication and Ka and Ks Calculation
4.5. Synteny Analysis of AsHSP90 Genes and Selected Plants
4.6. Identification and Analysis of cis-Elements in the Promoter Region of AsHSP90
4.7. Three-Dimensional Protein Structure Prediction
4.8. Protein–Protein Interaction Network
4.9. Plant Material, Growth Conditions, and Treatment
4.10. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR Expression Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guihur, A.; Rebeaud, M.E.; Goloubinoff, P. How Do Plants Feel the Heat and Survive? Trends Biochem. Sci. 2022, 47, 824–838. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Li, B.; Gao, K.; Ren, H.; Tang, W. Molecular Mechanisms Governing Plant Responses to High Temperatures. J. Integr. Plant Biol. 2018, 60, 757–779. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Leyser, O. A Plant’s Diet, Surviving in a Variable Nutrient Environment. Science 2020, 368, eaba0196. [Google Scholar] [CrossRef]
- Tigchelaar, M.; Battisti, D.S.; Naylor, R.L.; Ray, D.K. Future Warming Increases Probability of Globally Synchronized Maize Production Shocks. Proc. Natl. Acad. Sci. USA 2018, 115, 6644–6649. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.B.; Brady, S.M. Plant Developmental Responses to Climate Change. Dev. Biol. 2016, 419, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Qin, Z.; Sun, L.; Zhang, Y.; Wang, D.; Peng, H.; Yao, Y.; Hu, Z.; Ni, Z.; Sun, Q.; et al. Alternative Splicing of TaHSFA6e Modulates Heat Shock Protein-Mediated Translational Regulation in Response to Heat Stress in Wheat. New Phytol. 2023, 239, 2235–2247. [Google Scholar] [CrossRef] [PubMed]
- di Donato, M.; Geisler, M. HSP90 and Co-Chaperones: A Multitaskers’ View on Plant Hormone Biology. FEBS Lett. 2019, 593, 1415–1430. [Google Scholar] [CrossRef]
- Tichá, T.; Samakovli, D.; Kuchařová, A.; Vavrdová, T.; Šamaj, J. Multifaceted Roles of HEAT SHOCK PROTEIN 90 Molecular Chaperones in Plant Development. J. Exp. Bot. 2020, 71, 3966–3985. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Singh, J.; Ranjan, K.; Kumar, P.; Khanna, S.; Gupta, M.; Kumar, V.; Wani, S.; Sirohi, A. Heat Shock Proteins: Master Players for Heat-Stress Tolerance in Plants during Climate Change. In Heat Stress Tolerance in Plants: Physiological, Molecular and Genetic Perspectives; Wiley: New York, NY, USA, 2020; pp. 189–211. ISBN 978-1-119-43236-4. [Google Scholar]
- Genest, O.; Wickner, S.; Doyle, S.M. Hsp90 and Hsp70 Chaperones: Collaborators in Protein Remodeling. J. Biol. Chem. 2019, 294, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Chiosis, G.; Digwal, C.S.; Trepel, J.B.; Neckers, L. Structural and Functional Complexity of HSP90 in Cellular Homeostasis and Disease. Nat. Rev. Mol. Cell Biol. 2023, 24, 797–815. [Google Scholar] [CrossRef] [PubMed]
- Dutta, R.; Inouye, M. GHKL, an Emergent ATPase/Kinase Superfamily. Trends Biochem. Sci. 2000, 25, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Picard, D. Heat-Shock Protein 90, a Chaperone for Folding and Regulation. Cell. Mol. Life Sci. CMLS 2002, 59, 1640–1648. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.-D. Crosstalk between Hsp90 and Hsp70 Chaperones and Heat Stress Transcription Factors in Tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef]
- Li, W.; Chen, Y.; Ye, M.; Wang, D.; Chen, Q. Evolutionary History of the Heat Shock Protein 90 (Hsp90) Family of 43 Plants and Characterization of Hsp90s in Solanum Tuberosum. Mol. Biol. Rep. 2020, 47, 6679–6691. [Google Scholar] [CrossRef]
- Krishna, P.; Gloor, G. The Hsp90 Family of Proteins in Arabidopsis Thaliana. Cell Stress Chaperones 2001, 6, 238. [Google Scholar] [CrossRef]
- Zhang, M.; Shen, Z.; Meng, G.; Lu, Y.; Wang, Y. Genome-Wide Analysis of the Brachypodium Distachyon (L.) P. Beauv. Hsp90 Gene Family Reveals Molecular Evolution and Expression Profiling under Drought and Salt Stresses. PLoS ONE 2017, 12, e0189187. [Google Scholar] [CrossRef]
- Song, Z.; Pan, F.; Yang, C.; Jia, H.; Jiang, H.; He, F.; Li, N.; Lu, X.; Zhang, H. Genome-Wide Identification and Expression Analysis of HSP90 Gene Family in Nicotiana Tabacum. BMC Genet. 2019, 20, 35. [Google Scholar] [CrossRef]
- Appiah, C.; Yang, Z.-F.; He, J.; Wang, Y.; Zhou, J.; Xu, W.-Z.; Nie, G.; Zhu, Y.-Q. Genome-Wide Identification of Hsp90 Gene Family in Perennial Ryegrass and Expression Analysis under Various Abiotic Stresses. Plants 2021, 10, 2509. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhao, R.; Fan, P.; Wang, X.; Chen, X.; Li, Y. Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis Thaliana Enhances Plant Sensitivity to Salt and Drought Stresses. Planta 2009, 229, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Burch-Smith, T.; Schiff, M.; Feng, S.; Dinesh-Kumar, S.P. Molecular Chaperone Hsp90 Associates with Resistance Protein N and Its Signaling Proteins SGT1 and Rar1 to Modulate an Innate Immune Response in Plants. J. Biol. Chem. 2004, 279, 2101–2108. [Google Scholar] [CrossRef]
- Hubert, D.A.; Tornero, P.; Belkhadir, Y.; Krishna, P.; Takahashi, A.; Shirasu, K.; Dangl, J.L. Cytosolic HSP90 Associates with and Modulates the Arabidopsis RPM1 Disease Resistance Protein. EMBO J. 2003, 22, 5679–5689. [Google Scholar] [CrossRef] [PubMed]
- Banilas, G.; Korkas, E.; Englezos, V.; Nisiotou, A.A.; Hatzopoulos, P. Genome-Wide Analysis of the Heat Shock Protein 90 Gene Family in Grapevine (Vitis Vinifera L.). Aust. J. Grape Wine Res. 2012, 18, 29–38. [Google Scholar] [CrossRef]
- Samakovli, D.; Roka, L.; Dimopoulou, A.; Plitsi, P.K.; Žukauskait, A.; Georgopoulou, P.; Novák, O.; Milioni, D.; Hatzopoulos, P. HSP90 Affects Root Growth in Arabidopsis by Regulating the Polar Distribution of PIN1. New Phytol. 2021, 231, 1814–1831. [Google Scholar] [CrossRef]
- Baum, B.R. Oats: Wild and Cultivated. A Monograph of the Genus Avena L. (Poaceae); Minister of Supply and Services: Ottawa, ON, Canada, 1977. [Google Scholar]
- Fu, J.; Zhang, Y.; Hu, Y.; Zhao, G.; Tang, Y.; Zou, L. Concise Review: Coarse Cereals Exert Multiple Beneficial Effects on Human Health. Food Chem. 2020, 325, 126761. [Google Scholar] [CrossRef] [PubMed]
- Oats|Diseases and Pests, Description, Uses, Propagation. Available online: https://plantvillage.psu.edu/topics/oats/infos (accessed on 10 December 2023).
- Kamal, N.; Tsardakas Renhuldt, N.; Bentzer, J.; Gundlach, H.; Haberer, G.; Juhász, A.; Lux, T.; Bose, U.; Tye-Din, J.A.; Lang, D.; et al. The Mosaic Oat Genome Gives Insights into a Uniquely Healthy Cereal Crop. Nature 2022, 606, 113–119. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a Plant Cis-Acting Regulatory Element Database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Liu, B.; Zhang, L.; Chen, J.; Lu, M. Genome-Wide Analysis of the Populus Hsp90 Gene Family Reveals Differential Expression Patterns, Localization, and Heat Stress Responses. BMC Genom. 2013, 14, 532. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yan, H.; Guo, L.; Deng, C.; Wang, C.; Wang, Y.; Kang, L.; Zhou, P.; Yu, K.; Dong, X.; et al. Reference Genome Assemblies Reveal the Origin and Evolution of Allohexaploid Oat. Nat. Genet. 2022, 54, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, L.; Yun, L.; Ji, L.; Li, G.; Ji, M.; Shi, Y.; Zheng, X. Catalase (CAT) Gene Family in Wheat (Triticum Aestivum L.): Evolution, Expression Pattern and Function Analysis. Int. J. Mol. Sci. 2022, 23, 542. [Google Scholar] [CrossRef] [PubMed]
- Delsuc, F.; Brinkmann, H.; Philippe, H. Phylogenomics and the Reconstruction of the Tree of Life. Nat. Rev. Genet. 2005, 6, 361–375. [Google Scholar] [CrossRef]
- Radoeva, T.; Vaddepalli, P.; Zhang, Z.; Weijers, D. Evolution, Initiation, and Diversity in Early Plant Embryogenesis. Dev. Cell 2019, 50, 533–543. [Google Scholar] [CrossRef]
- Pearl, L.H.; Prodromou, C. Structure and Mechanism of the Hsp90 Molecular Chaperone Machinery. Annu. Rev. Biochem. 2006, 75, 271–294. [Google Scholar] [CrossRef]
- Reddy, R.K.; Chaudhary, S.; Patil, P.; Krishna, P. The 90 kDa Heat Shock Protein (Hsp90) Is Expressed throughout Brassica Napus Seed Development and Germination. Plant Sci. 1998, 131, 131–137. [Google Scholar] [CrossRef]
- Ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat Shock Proteins: Dynamic Biomolecules to Counter Plant Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef]
- Blanc, G.; Wolfe, K.H. Widespread Paleopolyploidy in Model Plant Species Inferred from Age Distributions of Duplicate Genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA Signaling in Stress-Response and Seed Development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- O’Sullivan, H. GrainGenes. Methods Mol. Biol. 2007, 406, 301–314. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.-C.; Bouman, P.; Li, W.-H. Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-Coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Bhagwat, M.; Aravind, L. PSI-BLAST Tutorial. Methods Mol. Biol. 2007, 395, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, K.; Aziz, U.; Zhao, C.; Zhang, M. Evaluation of Duplicated Reference Genes for Quantitative Real-Time PCR Analysis in Genome Unknown Hexaploid Oat (Avena Sativa L.). Plant Methods 2020, 16, 138. [Google Scholar] [CrossRef]
- Pandey, A.; Khan, M.K.; Hamurcu, M.; Brestic, M.; Topal, A.; Gezgin, S. Insight into the Root Transcriptome of a Boron-Tolerant Triticum Zhukovskyi Genotype Grown under Boron Toxicity. Agronomy 2022, 12, 2421. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]

| Gene | ID | Length | MW | pI | Instability Index | Aliphatic Index | GRAVY | Predicted Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| AsHSP90-1 | AVESA.00010b.r2.5DG0985750.1 | 627 | 71,881.4 | 4.79 | 41.94 | 74.32 | −0.639 | Endoplasmic reticulum |
| AsHSP90-2 | AVESA.00010b.r2.7AG1217140.1 | 698 | 80,094.2 | 4.67 | 39.68 | 83.67 | −0.592 | Cytoplasm |
| AsHSP90-3 | AVESA.00010b.r2.7DG1391180.1 | 698 | 80,103.2 | 4.69 | 39.7 | 83.81 | −0.597 | Chloroplast |
| AsHSP90-4 | AVESA.00010b.r2.5CG0932690.1 | 699 | 80,214.3 | 4.69 | 39.72 | 83.98 | −0.602 | Chloroplast |
| AsHSP90-5 | AVESA.00010b.r2.5AG0850550.1 | 700 | 80,432.6 | 4.67 | 40.22 | 82.17 | −0.617 | Nucleus |
| AsHSP90-6 | AVESA.00010b.r2.5CG0883430.1 | 700 | 80,475.6 | 4.67 | 40.43 | 82.03 | −0.625 | Cytoplasm |
| AsHSP90-7 | AVESA.00010b.r2.5DG0939350.1 | 707 | 80,750.8 | 4.67 | 40.95 | 82.48 | −0.586 | Chloroplast |
| AsHSP90-8 | AVESA.00010b.r2.4CG1254200.1 | 713 | 81,379.5 | 4.67 | 41.36 | 82.61 | −0.577 | Cytoplasm mitochondrion |
| AsHSP90-9 | AVESA.00010b.r2.6AG1070350.1 | 713 | 81,411.4 | 4.62 | 41.45 | 82.47 | −0.577 | Chloroplast |
| AsHSP90-10 | AVESA.00010b.r2.5CG0924830.1 | 781 | 88,373.9 | 4.6 | 47.64 | 79.14 | −0.531 | Nucleus |
| AsHSP90-11 | AVESA.00010b.r2.7DG1384250.1 | 781 | 88,429.9 | 4.58 | 46.62 | 80.01 | −0.528 | Chloroplast |
| AsHSP90-12 | AVESA.00010b.r2.7AG1224100.1 | 781 | 88,432 | 4.6 | 46.45 | 79.51 | −0.534 | Chloroplast |
| AsHSP90-13 | AVESA.00010b.r2.5AG0822120.1 | 787 | 88,667.1 | 5.05 | 44.63 | 76.24 | −0.565 | Cytoplasm |
| AsHSP90-14 | AVESA.00010b.r2.5CG0884240.1 | 784 | 88,810.7 | 4.7 | 44.1 | 78.37 | −0.544 | Cytoplasm |
| AsHSP90-15 | AVESA.00010b.r2.5DG0960220.1 | 785 | 89,044.9 | 4.65 | 44.92 | 78.14 | −0.553 | Endoplasmic reticulum |
| AsHSP90-16 | AVESA.00010b.r2.5AG0849600.1 | 786 | 89,235.1 | 4.69 | 44.7 | 78.04 | −0.553 | Cytoplasm |
| AsHSP90-17 | AVESA.00010b.r2.5CG0914170.1 | 809 | 91,165.9 | 5.09 | 44.57 | 78.74 | −0.539 | Chloroplast |
| AsHSP90-18 | AVESA.00010b.r2.7DG1344440.1 | 806 | 92,426.4 | 4.65 | 37.35 | 79.6 | −0.703 | Cytoplasm |
| AsHSP90-19 | AVESA.00010b.r2.2AG0260460.1 | 806 | 92,568.6 | 4.63 | 37.28 | 80.57 | −0.69 | Chloroplast |
| AsHSP90-20 | AVESA.00010b.r2.6CG1147820.1 | 806 | 92,623.6 | 4.6 | 37.68 | 80.09 | −0.699 | Endoplasmic reticulum |
| Paralogous HSP90 Pairs | chr. Location | Duplication Type | AsHSP90 Group | Ka | Ks | Ka_Ks |
|---|---|---|---|---|---|---|
| AsHSP90-8 | chr4C | Segmental | Group 4 | 0.0036 | 0.0908 | 0.0396 |
| AsHSP90-9 | chr6A | Group 4 | ||||
| AsHSP90-7 | chr5D | Segmental | Group 4 | 0.0018 | 0.1265 | 0.0143 |
| AsHSP90-8 | chr4C | Group 4 | ||||
| AsHSP90-7 | chr5D | Segmental | Group 4 | 0.0042 | 0.1343 | 0.0316 |
| AsHSP90-9 | chr6A | Group 4 | ||||
| AsHSP90-12 | chr7A | Segmental | Group 3 | 0.0016 | 0.0234 | 0.0706 |
| AsHSP90-11 | chr7D | Group 3 | ||||
| AsHSP90-12 | chr7A | Segmental | Group 3 | 0.0080 | 0.0928 | 0.0862 |
| AsHSP90-10 | chr5C | Group 3 | ||||
| AsHSP90-12 | chr7A | Segmental | Group 3 | 0.0638 | 0.9539 | 0.0669 |
| AsHSP90-15 | chr5D | Group 3 | ||||
| AsHSP90-12 | chr7A | Segmental | Group 3 | 0.0656 | 0.9452 | 0.0694 |
| AsHSP90-14 | chr5C | Group 3 | ||||
| AsHSP90-11 | chr7D | Segmental | Group 3 | 0.0097 | 0.0798 | 0.1211 |
| AsHSP90-10 | chr5C | Group 3 | ||||
| AsHSP90-11 | chr7D | Segmental | Group 3 | 0.0647 | 0.9356 | 0.0691 |
| AsHSP90-15 | chr5D | Group 3 | ||||
| AsHSP90-11 | chr7D | Segmental | Group 3 | 0.0665 | 0.9406 | 0.0707 |
| AsHSP90-14 | chr5C | Group 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, J.; Liu, S.; Wu, J.; Liu, T.; Liu, B.; Xiong, Y.; Zhao, J.; You, M.; Lei, X.; Ma, X. Genome-Wide Analysis of the Oat (Avena sativa) HSP90 Gene Family Reveals Its Identification, Evolution, and Response to Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 2305. https://doi.org/10.3390/ijms25042305
Peng J, Liu S, Wu J, Liu T, Liu B, Xiong Y, Zhao J, You M, Lei X, Ma X. Genome-Wide Analysis of the Oat (Avena sativa) HSP90 Gene Family Reveals Its Identification, Evolution, and Response to Abiotic Stress. International Journal of Molecular Sciences. 2024; 25(4):2305. https://doi.org/10.3390/ijms25042305
Chicago/Turabian StylePeng, Jinghan, Siyu Liu, Jiqiang Wu, Tianqi Liu, Boyang Liu, Yi Xiong, Junming Zhao, Minghong You, Xiong Lei, and Xiao Ma. 2024. "Genome-Wide Analysis of the Oat (Avena sativa) HSP90 Gene Family Reveals Its Identification, Evolution, and Response to Abiotic Stress" International Journal of Molecular Sciences 25, no. 4: 2305. https://doi.org/10.3390/ijms25042305





