Membrane Interaction Characteristics of the RTX Toxins and the Cholesterol-Dependence of Their Cytolytic/Cytotoxic Activity
Abstract
:1. RTX Toxins
2. Interaction of the RTX Toxins with the Target Cell Membrane
2.1. Binding through Cell-Specific Receptors
2.2. Binding in the Absence of a Specific Proteinaceous Receptor
3. Cholesterol Dependence of the Cytolytic/Cytotoxic Activity of RTX Toxins
3.1. Binding of the RTX Toxins to Membrane Cholesterol
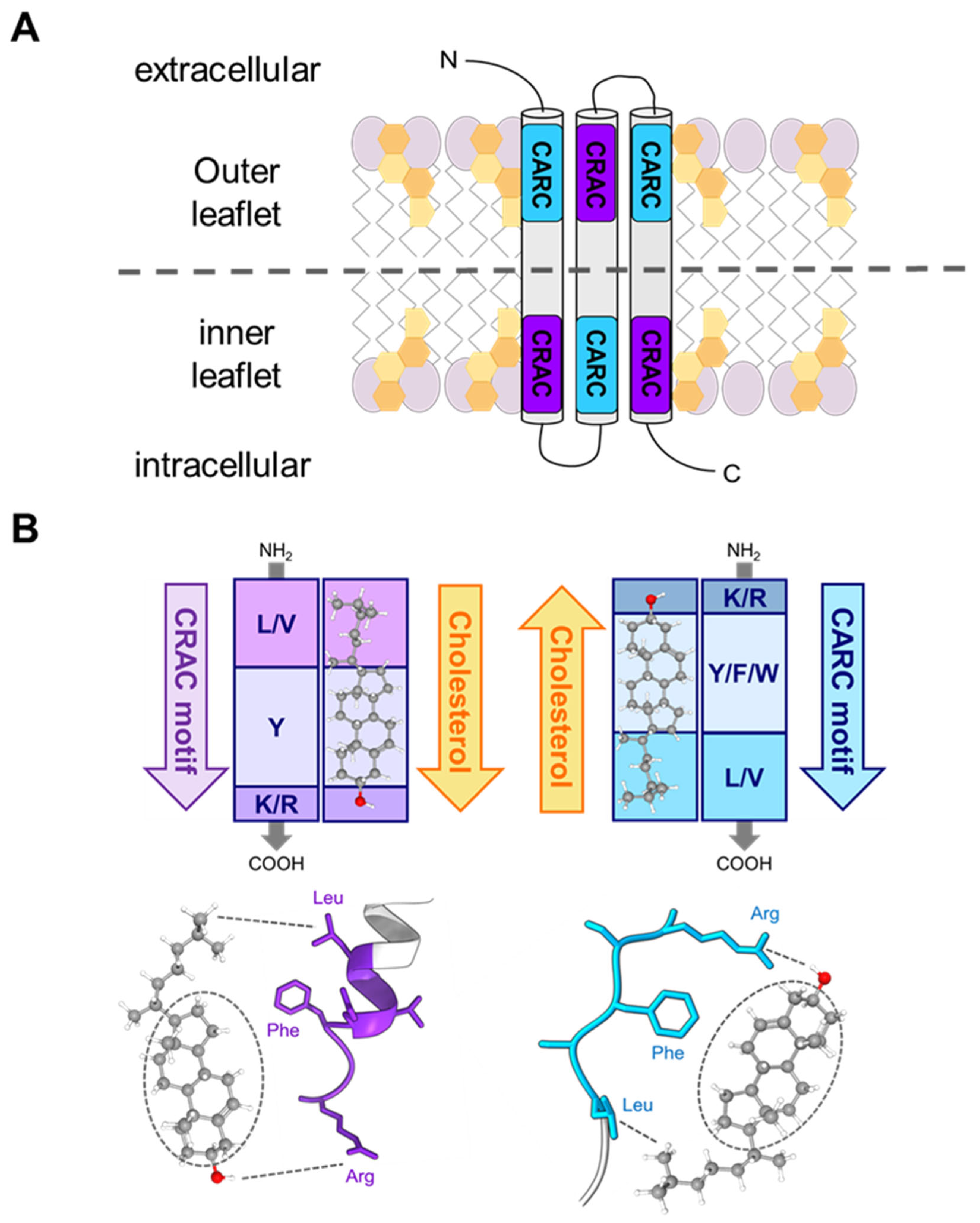
3.2. CRAC/CARC Motifs in the RTX Toxins as Possible Molecular Determinants of the Interaction with Membrane Cholesterol
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Welch, R.A. Pore-Forming Cytolysins of Gram-Negative Bacteria. Mol. Microbiol. 1991, 5, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Linhartová, I.; Bumba, L.; Mašín, J.; Basler, M.; Osička, R.; Kamanová, J.; Procházková, K.; Adkins, I.; Hejnová-Holubová, J.; Sadílková, L.; et al. RTX Proteins: A Highly Diverse Family Secreted by a Common Mechanism. FEMS Microbiol. Rev. 2010, 34, 1076–1112. [Google Scholar] [CrossRef] [PubMed]
- Welch, R.A. RTX Toxin Structure and Function: A Story of Numerous Anomalies and Few Analogies in Toxin Biology. Curr. Top. Microbiol. Immunol. 2001, 257, 85–111. [Google Scholar] [CrossRef] [PubMed]
- Coote, J.G. Structural and Functional Relationships among the RTX Toxin Determinants of Gram-Negative Bacteria. FEMS Microbiol. Rev. 1992, 8, 137–161. [Google Scholar] [CrossRef] [PubMed]
- Stanley, P.; Packman, L.C.; Koronakis, V.; Hughes, C. Fatty Acylation of Two Internal Lysine Residues Required for the Toxic Activity of Escherichia coli Hemolysin. Science 1994, 266, 1992–1996. [Google Scholar] [CrossRef] [PubMed]
- Balashova, N.V.; Shah, C.; Patel, J.K.; Megalla, S.; Kachlany, S.C. Aggregatibacter actinomycetemcomitans LtxC Is Required for Leukotoxin Activity and Initial Interaction between Toxin and Host Cells. Gene 2009, 443, 42–47. [Google Scholar] [CrossRef]
- Stanley, P.; Koronakis, V.; Hughes, C. Acylation of Escherichia coli Hemolysin: A Unique Protein Lipidation Mechanism Underlying Toxin Function. Microbiol. Mol. Biol. Rev. 1998, 62, 309–333. [Google Scholar] [CrossRef]
- Barry, E.M.; Weiss, A.A.; Ehrmann, I.E.; Gray, M.C.; Hewlett, E.L.; Goodwin, M.S. Bordetella pertussis Adenylate Cyclase Toxin and Hemolytic Activities Require a Second Gene, cyaC, for Activation. J. Bacteriol. 1991, 173, 720–726. [Google Scholar] [CrossRef]
- Osickova, A.; Khaliq, H.; Masin, J.; Jurnecka, D.; Sukova, A.; Fiser, R.; Holubova, J.; Stanek, O.; Sebo, P.; Osicka, R. Acyltransferase-Mediated Selection of the Length of the Fatty Acyl Chain and of the Acylation Site Governs Activation of Bacterial RTX Toxins. J. Biol. Chem. 2020, 295, 9268–9280. [Google Scholar] [CrossRef]
- Greene, N.P.; Crow, A.; Hughes, C.; Koronakis, V. Structure of a Bacterial Toxin-Activating Acyltransferase. Proc. Natl. Acad. Sci. USA 2015, 112, E3058–E3066. [Google Scholar] [CrossRef]
- Holland, I.B.; Schmitt, L.; Young, J. Type 1 Protein Secretion in Bacteria, the ABC-Transporter Dependent Pathway (Review). Mol. Membr. Biol. 2005, 22, 29–39. [Google Scholar] [CrossRef]
- Holland, I.B.; Peherstorfer, S.; Kanonenberg, K.; Lenders, M.; Reimann, S.; Schmitt, L. Type I Protein Secretion-Deceptively Simple yet with a Wide Range of Mechanistic Variability across the Family. EcoSal Plus 2016, 7, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Gygi, D.; Nicolet, J.; Frey, J.; Cross, M.; Koronakis, V.; Hughes, C. Isolation of the Actinobacillus pleuropneumoniae Haemolysin Gene and the Activation and Secretion of the Prohaemolysin by the HlyC, HlyB and HlyD Proteins of Escherichia coli. Mol. Microbiol. 1990, 4, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Wandersman, C.; Delepelaire, P. TolC, an Escherichia coli Outer Membrane Protein Required for Hemolysin Secretion. Proc. Natl. Acad. Sci. USA 1990, 87, 4776–4780. [Google Scholar] [CrossRef] [PubMed]
- Glaser, P.; Sakamoto, H.; Bellalou, J.; Ullmann, A.; Danchin, A. Secretion of Cyclolysin, the Calmodulin-Sensitive Adenylate Cyclase-Haemolysin Bifunctional Protein of Bordetella pertussis. EMBO J. 1988, 7, 3997–4004. [Google Scholar] [CrossRef] [PubMed]
- Basler, M.; Knapp, O.; Masin, J.; Fiser, R.; Maier, E.; Benz, R.; Sebo, P.; Osicka, R. Segments Crucial for Membrane Translocation and Pore-Forming Activity of Bordetella Adenylate Cyclase Toxin. J. Biol. Chem. 2007, 282, 12419–12429. [Google Scholar] [CrossRef] [PubMed]
- Roderova, J.; Osickova, A.; Sukova, A.; Mikusova, G.; Fiser, R.; Sebo, P.; Osicka, R.; Masin, J. Residues 529 to 549 Participate in Membrane Penetration and Pore-Forming Activity of the Bordetella Adenylate Cyclase Toxin. Sci. Rep. 2019, 9, 5758. [Google Scholar] [CrossRef] [PubMed]
- Osičková, A.; Osička, R.; Maier, E.; Benz, R.; Šebo, P. An Amphipathic α-Helix Including Glutamates 509 and 516 Is Crucial for Membrane Translocation of Adenylate Cyclase Toxin and Modulates Formation and Cation Selectivity of Its Membrane Channels. J. Biol. Chem. 1999, 274, 37644–37650. [Google Scholar] [CrossRef] [PubMed]
- Valeva, A.; Walev, I.; Kemmer, H.; Weis, S.; Siegel, I.; Boukhallouk, F.; Wassenaar, T.M.; Chavakis, T.; Bhakdi, S. Binding of Escherichia coli Hemolysin and Activation of the Target Cells Is Not Receptor-Dependent. J. Biol. Chem. 2005, 280, 36657–36663. [Google Scholar] [CrossRef]
- Benz, R.; Schmid, A.; Wagner, W.; Goebel, W. Pore Formation by the Escherichia coli Hemolysin: Evidence for an Association-Dissociation Equilibrium of the Pore-Forming Aggregates. Infect. Immun. 1989, 57, 887–895. [Google Scholar] [CrossRef]
- Bhakdi, S.; Mackman, N.; Nicaud, J.M.; Holland, I.B. Escherichia coli Hemolysin May Damage Target Cell Membranes by Generating Transmembrane Pores. Infect. Immun. 1986, 52, 63–69. [Google Scholar] [CrossRef]
- Bárcena-Uribarri, I.; Benz, R.; Winterhalter, M.; Zakharian, E.; Balashova, N. Pore Forming Activity of the Potent RTX-Toxin Produced by Pediatric Pathogen Kingella kingae: Characterization and Comparison to Other RTX-Family Members. Biochim. Biophys. Acta 2015, 1848, 1536–1544. [Google Scholar] [CrossRef]
- Ludwig, A.; Schmid, A.; Benz, R.; Goebel, W. Mutations Affecting Pore Formation by Haemolysin from Escherichia coli. Mol. Gen. Genet. 1991, 226, 198–208. [Google Scholar] [CrossRef]
- Baumann, U.; Wu, S.; Flaherty, K.M.; McKay, D.B. Three-Dimensional Structure of the Alkaline Protease of Pseudomonas Aeruginosa: A Two-Domain Protein with a Calcium Binding Parallel Beta Roll Motif. EMBO J. 1993, 12, 3357–3364. [Google Scholar] [CrossRef]
- Bauche, C.; Chenal, A.; Knapp, O.; Bodenreider, C.; Benz, R.; Chaffotte, A.; Ladant, D. Structural and Functional Characterization of an Essential RTX Subdomain of Bordetella pertussis Adenylate Cyclase Toxin. J. Biol. Chem. 2006, 281, 16914–16926. [Google Scholar] [CrossRef]
- Chenal, A.; Karst, J.C.; Sotomayor Pérez, A.C.; Wozniak, A.K.; Baron, B.; England, P.; Ladant, D. Calcium-Induced Folding and Stabilization of the Intrinsically Disordered RTX Domain of the CyaA Toxin. Biophys. J. 2010, 99, 3744–3753. [Google Scholar] [CrossRef]
- Nicaud, J.-M.; Mackman, N.; Gray, L.; Holland, I.B. The C-Terminal, 23 kDa Peptide of E. coli Haemolysin 2001 Contains All the Information Necessary for Its Secretion by the Haemolysin (Hly) Export Machinery. FEBS Lett. 1986, 204, 331–335. [Google Scholar] [CrossRef]
- Mackman, N.; Baker, K.; Gray, L.; Haigh, R.; Nicaud, J.M.; Holland, I.B. Release of a Chimeric Protein into the Medium from Escherichia coli Using the C-Terminal Secretion Signal of Haemolysin. EMBO J. 1987, 6, 2835–2841. [Google Scholar] [CrossRef] [PubMed]
- Koronakis, V.; Koronakis, E.; Hughes, C. Isolation and Analysis of the C-Terminal Signal Directing Export of Escherichia coli Hemolysin Protein across Both Bacterial Membranes. EMBO J. 1989, 8, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.B.; Walker, C.R.B.; Guo, L.; Pellett, S.; Shabanowitz, J.; Hunt, D.F.; Hewlett, E.L.; Ludwig, A.; Goebel, W.; Welch, R.A.; et al. Escherichia coli α-Hemolysin (HlyA) Is Heterogeneously Acylated in Vivo with 14-, 15-, and 17-Carbon Fatty Acids. J. Biol. Chem. 2000, 275, 36698–36702. [Google Scholar] [CrossRef] [PubMed]
- Fong, K.P.; Tang, H.-Y.; Brown, A.C.; Kieba, I.R.; Speicher, D.W.; Boesze-Battaglia, K.; Lally, E.T. Aggregatibacter actinomycetemcomitans Leukotoxin Is Post-Translationally Modified by Addition of Either Saturated or Hydroxylated Fatty Acyl Chains. Mol. Oral Microbiol. 2011, 26, 262–276. [Google Scholar] [CrossRef]
- Basar, T.; Havlíček, V.; Bezoušková, S.; Hackett, M.; Šebo, P. Acylation of Lysine 983 Is Sufficient for Toxin Activity of Bordetella pertussis Adenylate Cyclase: Substitutions of Alanine 140 Modulate Acylation Site Selectivity of The Toxin Acyltransferase CyaC. J. Biol. Chem. 2001, 276, 348–354. [Google Scholar] [CrossRef]
- Nicaud, J.-M.; Mackman, N.; Gray, L.; Holland, I.B. Characterisation of HlyC and Mechanism of Activation and Secretion of Haemolysin from E. coli 2001. FEBS Lett. 1985, 187, 339–344. [Google Scholar] [CrossRef]
- Masin, J.; Osicka, R.; Bumba, L.; Sebo, P. Bordetella Adenylate Cyclase Toxin: A Unique Combination of a Pore-Forming Moiety with a Cell-Invading Adenylate Cyclase Enzyme. Pathog. Dis. 2015, 73, ftv075. [Google Scholar] [CrossRef]
- Döbereiner, A.; Schmid, A.; Ludwig, A.; Goebel, W.; Benz, R. The Effects of Calcium and Other Polyvalent Cations on Channel Formation by Escherichia coli Alpha-Hemolysin in Red Blood Cells and Lipid Bilayer Membranes. Eur. J. Biochem. 1996, 240, 454–460. [Google Scholar] [CrossRef]
- Soloaga, A.; Ostolaza, H.; Goñi, F.M.; De La Cruz, F. Purification of Escherichia coli Pro-Haemolysin, and a Comparison with the Properties of Mature α-Haemolysin. Eur. J. Biochem. 1996, 238, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Masín, J.; Konopásek, I.; Svobodová, J.; Sebo, P. Different Structural Requirements for Adenylate Cyclase Toxin Interactions with Erythrocyte and Liposome Membranes. Biochim. Biophys. Acta 2004, 1660, 144–154. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, D.P.; Cannella, S.E.; Voegele, A.; Raoux-Barbot, D.; Davi, M.; Douché, T.; Matondo, M.; Brier, S.; Ladant, D.; Chenal, A.A. Post-translational Acylation Controls the Folding and Functions of the CyaA RTX Toxin. FASEB J. 2019, 33, 10065–10076. [Google Scholar] [CrossRef] [PubMed]
- Ostolaza, H.; Soloaga, A.; Goñi, F.M. The Binding of Divalent Cations to Escherichia coli Alpha-Haemolysin. Eur. J. Biochem. 1995, 228, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Karst, J.C.; Ntsogo Enguéné, V.Y.; Cannella, S.E.; Subrini, O.; Hessel, A.; Debard, S.; Ladant, D.; Chenal, A. Calcium, Acylation, and Molecular Confinement Favor Folding of Bordetella pertussis Adenylate Cyclase CyaA Toxin into a Monomeric and Cytotoxic Form. J. Biol. Chem. 2014, 289, 30702–30716. [Google Scholar] [CrossRef] [PubMed]
- Blenner, M.A.; Shur, O.; Szilvay, G.R.; Cropek, D.M.; Banta, S. Calcium-Induced Folding of a Beta Roll Motif Requires C-Terminal Entropic Stabilization. J. Mol. Biol. 2010, 400, 244–256. [Google Scholar] [CrossRef]
- Chenal, A.; Guijarro, J.I.; Raynal, B.; Delepierre, M.; Ladant, D. RTX Calcium Binding Motifs Are Intrinsically Disordered in the Absence of Calcium: Implication for Protein Secretion. J. Biol. Chem. 2009, 284, 1781–1789. [Google Scholar] [CrossRef]
- Rose, T.; Sebo, P.; Bellalou, J.; Ladant, D. Interaction of Calcium with Bordetella pertussis Adenylate Cyclase Toxin. Characterization of Multiple Calcium-Binding Sites and Calcium-Induced Conformational Changes. J. Biol. Chem. 1995, 270, 26370–26376. [Google Scholar] [CrossRef]
- Rhodes, C.R.; Gray, M.C.; Watson, J.M.; Muratore, T.L.; Kim, S.B.; Hewlett, E.L.; Grisham, C.M. Structural Consequences of Divalent Metal Binding by the Adenylyl Cyclase Toxin of Bordetella pertussis. Arch. Biochem. Biophys. 2001, 395, 169–176. [Google Scholar] [CrossRef]
- Benz, R. Channel Formation by RTX-Toxins of Pathogenic Bacteria: Basis of Their Biological Activity. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 526–537. [Google Scholar] [CrossRef]
- Hyland, C.; Vuillard, L.; Hughes, C.; Koronakis, V. Membrane Interaction of Escherichia coli Hemolysin: Flotation and Insertion-Dependent Labeling by Phospholipid Vesicles. J. Bacteriol. 2001, 183, 5364–5370. [Google Scholar] [CrossRef]
- Powthongchin, B.; Angsuthanasombat, C. Effects on Haemolytic Activity of Single Proline Substitutions in the Bordetella pertussis CyaA Pore-Forming Fragment. Arch. Microbiol. 2009, 191, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Guermonprez, P.; Khelef, N.; Blouin, E.; Rieu, P.; Ricciardi-Castagnoli, P.; Guiso, N.; Ladant, D.; Leclerc, C. The Adenylate Cyclase Toxin of Bordetella pertussis Binds to Target Cells via the αMβ2 Integrin (Cd11b/Cd18). J. Exp. Med. 2001, 193, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Ristow, L.C.; Tran, V.; Schwartz, K.J.; Pankratz, L.; Mehle, A.; Sauer, J.-D.; Welch, R.A. The Extracellular Domain of the Β2 Integrin β Subunit (CD18) Is Sufficient for Escherichia coli Hemolysin and Aggregatibacter actinomycetemcomitans Leukotoxin Cytotoxic Activity. mBio 2019, 10, e01459-19. [Google Scholar] [CrossRef] [PubMed]
- Lally, E.T.; Kieba, I.R.; Sato, A.; Green, C.L.; Rosenbloom, J.; Korostoff, J.; Wang, J.F.; Shenker, B.J.; Ortlepp, S.; Robinson, M.K.; et al. RTX Toxins Recognize a Beta2 Integrin on the Surface of Human Target Cells. J. Biol. Chem. 1997, 272, 30463–30469. [Google Scholar] [CrossRef] [PubMed]
- Vanden Bergh, P.G.A.C.; Zecchinon, L.L.M.; Fett, T.; Desmecht, D. Porcine CD18 Mediates Actinobacillus pleuropneumoniae ApxIII Species-Specific Toxicity. Vet. Res. 2009, 40, 33. [Google Scholar] [CrossRef] [PubMed]
- Confer, D.L.; Eaton, J.W. Phagocyte Impotence Caused by an Invasive Bacterial Adenylate Cyclase. Science 1982, 217, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Gueirard, P.; Le Blay, K.; Le Coustumier, A.; Chaby, R.; Guiso, N. Variation in Bordetella bronchiseptica Lipopolysaccharide during Human Infection. FEMS Microbiol. Lett. 1998, 162, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Kamanova, J.; Kofronova, O.; Masin, J.; Genth, H.; Vojtova, J.; Linhartova, I.; Benada, O.; Just, I.; Sebo, P. Adenylate Cyclase Toxin Subverts Phagocyte Function by RhoA Inhibition and Unproductive Ruffling. J. Immunol. 2008, 181, 5587–5597. [Google Scholar] [CrossRef]
- Khelef, N.; Guiso, N. Induction of Macrophage Apoptosis by Bordetella pertussis Adenylate Cyclase-Hemolysin. FEMS Microbiol. Lett. 1995, 134, 27–32. [Google Scholar] [CrossRef]
- Pearson, R.D.; Symes, P.; Conboy, M.; Weiss, A.A.; Hewlett, E.L. Inhibition of Monocyte Oxidative Responses by Bordetella pertussis Adenylate Cyclase Toxin. J. Immunol. 1987, 139, 2749–2754. [Google Scholar] [CrossRef]
- Weingart, C.L.; Weiss, A.A. Bordetella pertussis Virulence Factors Affect Phagocytosis by Human Neutrophils. Infect. Immun. 2000, 68, 1735–1739. [Google Scholar] [CrossRef]
- Dhakal, B.K.; Mulvey, M.A. The UPEC Pore-Forming Toxin α-Hemolysin Triggers Proteolysis of Host Proteins to Disrupt Cell Adhesion, Inflammatory, and Survival Pathways. Cell Host Microbe 2012, 11, 58–69. [Google Scholar] [CrossRef]
- Wiles, T.J.; Kulesus, R.R.; Mulvey, M.A. Origins and Virulence Mechanisms of Uropathogenic Escherichia coli. Exp. Mol. Pathol. 2008, 85, 11–19. [Google Scholar] [CrossRef]
- El-Azami-El-Idrissi, M.; Bauche, C.; Loucka, J.; Osicka, R.; Sebo, P.; Ladant, D.; Leclerc, C. Interaction of Bordetella pertussis Adenylate Cyclase with CD11b/CD18: Role of Toxin Acylation and Identification of the Main Integrin Interaction Domain. J. Biol. Chem. 2003, 278, 38514–38521. [Google Scholar] [CrossRef] [PubMed]
- Osicka, R.; Osickova, A.; Hasan, S.; Bumba, L.; Cerny, J.; Sebo, P. Bordetella Adenylate Cyclase Toxin Is a Unique Ligand of the Integrin Complement Receptor 3. eLife 2014, 4, e10766. [Google Scholar] [CrossRef]
- Goldsmith, J.A.; DiVenere, A.M.; Maynard, J.A.; McLellan, J.S. Structural Basis for Non-Canonical Integrin Engagement by Bordetella Adenylate Cyclase Toxin. Cell Rep. 2022, 40, 111196. [Google Scholar] [CrossRef] [PubMed]
- Nygren, P.; Balashova, N.; Brown, A.C.; Kieba, I.; Dhingra, A.; Boesze-Battaglia, K.; Lally, E.T. Aggregatibacter actinomycetemcomitans Leukotoxin Causes Activation of Lymphocyte Function-Associated Antigen 1. Cell. Microbiol. 2019, 21, e12967. [Google Scholar] [CrossRef] [PubMed]
- Ristow, L.C.; Welch, R.A. RTX Toxins Ambush Immunity’s First Cellular Responders. Toxins 2019, 11, 720. [Google Scholar] [CrossRef] [PubMed]
- Masin, J.; Osickova, A.; Jurnecka, D.; Klimova, N.; Khaliq, H.; Sebo, P.; Osicka, R. Retargeting from the CR3 to the LFA-1 Receptor Uncovers the Adenylyl Cyclase Enzyme–Translocating Segment of Bordetella Adenylate Cyclase Toxin. J. Biol. Chem. 2020, 295, 9349–9365. [Google Scholar] [CrossRef]
- Martín, C.; Requero, M.-A.; Masin, J.; Konopasek, I.; Goñi, F.M.; Sebo, P.; Ostolaza, H. Membrane Restructuring by Bordetella pertussis Adenylate Cyclase Toxin, a Member of the RTX Toxin Family. J. Bacteriol. 2004, 186, 3760–3765. [Google Scholar] [CrossRef]
- Ostolaza, H.; Bartolomé, B.; Serra, J.L.; de la Cruz, F.; Goñi, F.M. α-Haemolysin from E. coli Purification and Self-Aggregation Properties. FEBS Lett. 1991, 280, 195–198. [Google Scholar] [CrossRef]
- Bakás, L.; Veiga, M.P.; Soloaga, A.; Ostolaza, H.; Goñi, F.M. Calcium-Dependent Conformation of E. coli α-Haemolysin. Implications for the Mechanism of Membrane Insertion and Lysis. Biochim. Biophys. Acta (BBA)-Biomembr. 1998, 1368, 225–234. [Google Scholar] [CrossRef]
- Brown, A.C.; Balashova, N.V.; Epand, R.M.; Epand, R.F.; Bragin, A.; Kachlany, S.C.; Walters, M.J.; Du, Y.; Boesze-Battaglia, K.; Lally, E.T. Aggregatibacter actinomycetemcomitans Leukotoxin Utilizes a Cholesterol Recognition/Amino Acid Consensus Site for Membrane Association. J. Biol. Chem. 2013, 288, 23607–23621. [Google Scholar] [CrossRef]
- Keane, W.F.; Welch, R.; Gekker, G.; Peterson, P.K. Mechanism of Escherichia coli Alpha-Hemolysin-Induced Injury to Isolated Renal Tubular Cells. Am. J. Pathol. 1987, 126, 350–357. [Google Scholar]
- Suttorp, N.; Flöer, B.; Schnittler, H.; Seeger, W.; Bhakdi, S. Effects of Escherichia coli Hemolysin on Endothelial Cell Function. Infect. Immun. 1990, 58, 3796–3801. [Google Scholar] [CrossRef]
- Gadeberg, O.V.; Orskov, I. In Vitro Cytotoxic Effect of Alpha-Hemolytic Escherichia coli on Human Blood Granulocytes. Infect. Immun. 1984, 45, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Bhakdi, S.; Muhly, M.; Korom, S.; Hugo, F. Release of Interleukin-1 Beta Associated with Potent Cytocidal Action of Staphylococcal Alpha-Toxin on Human Monocytes. Infect. Immun. 1989, 57, 3512–3519. [Google Scholar] [CrossRef] [PubMed]
- Bhakdi, S.; Muhly, M.; Korom, S.; Schmidt, G. Effects of Escherichia coli Hemolysin on Human Monocytes. Cytocidal Action and Stimulation of Interleukin 1 Release. J. Clin. Investig. 1990, 85, 1746–1753. [Google Scholar] [CrossRef] [PubMed]
- Mobley, H.L.; Green, D.M.; Trifillis, A.L.; Johnson, D.E.; Chippendale, G.R.; Lockatell, C.V.; Jones, B.D.; Warren, J.W. Pyelonephritogenic Escherichia coli and Killing of Cultured Human Renal Proximal Tubular Epithelial Cells: Role of Hemolysin in Some Strains. Infect. Immun. 1990, 58, 1281–1289. [Google Scholar] [CrossRef]
- Crosby, J.A.; Kachlany, S.C. TdeA, a TolC-like Protein Required for Toxin and Drug Export in Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene 2007, 388, 83–92. [Google Scholar] [CrossRef]
- Balashova, N.V.; Crosby, J.A.; Al Ghofaily, L.; Kachlany, S.C. Leukotoxin Confers Beta-Hemolytic Activity to Actinobacillus actinomycetemcomitans. Infect. Immun. 2006, 74, 2015–2021. [Google Scholar] [CrossRef]
- Murphy, G.L.; Whitworth, L.C.; Clinkenbeard, K.D.; Clinkenbeard, P.A. Hemolytic Activity of the Pasteurella Haemolytica Leukotoxin. Infect. Immun. 1995, 63, 3209–3212. [Google Scholar] [CrossRef] [PubMed]
- Osickova, A.; Balashova, N.; Masin, J.; Sulc, M.; Roderova, J.; Wald, T.; Brown, A.C.; Koufos, E.; Chang, E.H.; Giannakakis, A.; et al. Cytotoxic Activity of Kingella kingae RtxA Toxin Depends on Post-Translational Acylation of Lysine Residues and Cholesterol Binding. Emerg. Microbes Infect. 2018, 7, 1–15. [Google Scholar] [CrossRef]
- Kehl-Fie, T.E.; St. Geme, J.W. Identification and Characterization of an RTX Toxin in the Emerging Pathogen Kingella kingae. J. Bacteriol. 2007, 189, 430–436. [Google Scholar] [CrossRef]
- Ostolaza, H.; Bartolomé, B.; de Zárate, I.O.; de la Cruz, F.; Goñi, F.M. Release of Lipid Vesicle Contents by the Bacterial Protein Toxin α-Haemolysin. Biochim. Biophys. Acta (BBA)-Biomembr. 1993, 1147, 81–88. [Google Scholar] [CrossRef]
- Brown, A.C.; Boesze-Battaglia, K.; Du, Y.; Stefano, F.P.; Kieba, I.R.; Epand, R.F.; Kakalis, L.; Yeagle, P.L.; Epand, R.M.; Lally, E.T. Aggregatibacter actinomycetemcomitans Leukotoxin Cytotoxicity Occurs through Bilayer Destabilization. Cell. Microbiol. 2012, 14, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Fiser, R.; Konopásek, I. Different Modes of Membrane Permeabilization by Two RTX Toxins: HlyA from Escherichia coli and CyaA from Bordetella pertussis. Biochim. Biophys. Acta 2009, 1788, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Bakás, L.; Ostolaza, H.; Vaz, W.L.; Goñi, F.M. Reversible Adsorption and Nonreversible Insertion of Escherichia coli Alpha-Hemolysin into Lipid Bilayers. Biophys. J. 1996, 71, 1869–1876. [Google Scholar] [CrossRef] [PubMed]
- Ostolaza, H.; Bakás, L.; Goñi, F.M. Balance of Electrostatic and Hydrophobic Interactions in the Lysis of Model Membranes by E. coliα-Haemolysin. J. Membr. Biol. 1997, 158, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Iwase, M.; Lally, E.T.; Berthold, P.; Korchak, H.M.; Taichman, N.S. Effects of Cations and Osmotic Protectants on Cytolytic Activity of Actinobacillus actinomycetemcomitans Leukotoxin. Infect. Immun. 1990, 58, 1782–1788. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Magraner, L.; Viguera, A.R.; García-Pacios, M.; Garcillán, M.P.; Arrondo, J.-L.R.; de la Cruz, F.; Goñi, F.M.; Ostolaza, H. The Calcium-Binding C-Terminal Domain of Escherichia coli α-Hemolysin Is a Major Determinant in the Surface-Active Properties of the Protein. J. Biol. Chem. 2007, 282, 11827–11835. [Google Scholar] [CrossRef]
- Forman, M.S.; Nishikubo, J.B.; Han, R.K.; Le, A.; Balashova, N.V.; Kachlany, S.C. Gangliosides Block Aggregatibacter actinomycetemcomitans Leukotoxin (LtxA)-Mediated Hemolysis. Toxins 2010, 2, 2824–2836. [Google Scholar] [CrossRef] [PubMed]
- Munksgaard, P.S.; Skals, M.; Reinholdt, J.; Poulsen, K.; Jensen, M.R.; Yang, C.; Leipziger, J.; Vorup-Jensen, T.; Praetorius, H.A. Sialic Acid Residues Are Essential for Cell Lysis Mediated by Leukotoxin from Aggregatibacter actinomycetemcomitans. Infect. Immun. 2014, 82, 2219–2228. [Google Scholar] [CrossRef] [PubMed]
- Morova, J.; Osicka, R.; Masin, J.; Sebo, P. RTX Cytotoxins Recognize Beta2 Integrin Receptors through N-Linked Oligosaccharides. Proc. Natl. Acad. Sci. USA 2008, 105, 5355–5360. [Google Scholar] [CrossRef]
- Gable, P.; Eaton, J.; Confer, D. Intoxication of Human Phagocytes by Bordetella Adenylate-Cyclase Toxin-Implication of a Ganglioside Receptor. Clin. Res. 1985, 33, A844. [Google Scholar]
- Gordon, V.M.; Young, W.W.; Lechler, S.M.; Gray, M.C.; Leppla, S.H.; Hewlett, E.L. Adenylate Cyclase Toxins from Bacillus anthracis and Bordetella pertussis. J. Biol. Chem. 1989, 264, 14792–14796. [Google Scholar] [CrossRef]
- Mrówczyńska, L.; Bobrowska-Hägerstrand, M.; Lindqvist, C.; Hägerstrand, H. Bordetella Adenylate Cyclase Toxin Can Bind Ganglioside GM1. BIO 2011, 1, 67–71. [Google Scholar] [CrossRef]
- Hasan, S.; Osickova, A.; Bumba, L.; Novák, P.; Sebo, P.; Osicka, R. Interaction of Bordetella Adenylate Cyclase Toxin with Complement Receptor 3 Involves Multivalent Glycan Binding. FEBS Lett. 2015, 589, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Rahman, W.U.; Osickova, A.; Klimova, N.; Lora, J.; Balashova, N.; Osicka, R. Binding of Kingella kingae RtxA Toxin Depends on Cell Surface Oligosaccharides, but Not on Β2 Integrins. Int. J. Mol. Sci. 2020, 21, 9092. [Google Scholar] [CrossRef]
- Fong, K.P.; Pacheco, C.M.F.; Otis, L.L.; Baranwal, S.; Kieba, I.R.; Harrison, G.; Hersh, E.V.; Boesze-Battaglia, K.; Lally, E.T. Actinobacillus actinomycetemcomitans Leukotoxin Requires Lipid Microdomains for Target Cell Cytotoxicity. Cell Microbiol. 2006, 8, 1753–1767. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.C.; Koufos, E.; Balashova, N.V.; Boesze-Battaglia, K.; Lally, E.T. Inhibition of LtxA Toxicity by Blocking Cholesterol Binding with Peptides. Mol. Oral Microbiol. 2016, 31, 94–105. [Google Scholar] [CrossRef]
- Bumba, L.; Masin, J.; Fiser, R.; Sebo, P. Bordetella Adenylate Cyclase Toxin Mobilizes Its Beta2 Integrin Receptor into Lipid Rafts to Accomplish Translocation across Target Cell Membrane in Two Steps. PLoS Pathog. 2010, 6, e1000901. [Google Scholar] [CrossRef] [PubMed]
- Herlax, V.; Maté, S.; Rimoldi, O.; Bakás, L. Relevance of Fatty Acid Covalently Bound to Escherichia coli α-Hemolysin and Membrane Microdomains in the Oligomerization Process. J. Biol. Chem. 2009, 284, 25199–25210. [Google Scholar] [CrossRef]
- Vazquez, R.F.; Maté, S.M.; Bakás, L.S.; Fernández, M.M.; Malchiodi, E.L.; Herlax, V.S. Novel Evidence for the Specific Interaction between Cholesterol and α-Haemolysin of Escherichia coli. Biochem. J. 2014, 458, 481–489. [Google Scholar] [CrossRef]
- González Bullón, D.; Uribe, K.B.; Amuategi, J.; Martín, C.; Ostolaza, H. Cholesterol Stimulates the Lytic Activity of Adenylate Cyclase Toxin on Lipid Membranes by Promoting Toxin Oligomerization and Formation of Pores with a Greater Effective Size. FEBS J. 2021, 288, 6795–6814. [Google Scholar] [CrossRef]
- Fantini, J.; Garmy, N.; Mahfoud, R.; Yahi, N. Lipid Rafts: Structure, Function and Role in HIV, Alzheimer’s and Prion Diseases. Expert. Rev. Mol. Med. 2002, 4, 1–22. [Google Scholar] [CrossRef]
- Chang, H.M.; Reitstetter, R.; Mason, R.P.; Gruener, R. Attenuation of Channel Kinetics and Conductance by Cholesterol: An Interpretation Using Structural Stress as a Unifying Concept. J. Membr. Biol. 1995, 143, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Hanson, M.A.; Cherezov, V.; Griffith, M.T.; Roth, C.B.; Jaakola, V.-P.; Chien, E.Y.T.; Velasquez, J.; Kuhn, P.; Stevens, R.C. A Specific Cholesterol Binding Site Is Established by the 2.8 A Structure of the Human Beta2-Adrenergic Receptor. Structure 2008, 16, 897–905. [Google Scholar] [CrossRef]
- Rosenhouse-Dantsker, A.; Noskov, S.; Durdagi, S.; Logothetis, D.E.; Levitan, I. Identification of Novel Cholesterol-Binding Regions in Kir2 Channels. J. Biol. Chem. 2013, 288, 31154–31164. [Google Scholar] [CrossRef]
- Fantini, J.; Di Scala, C.; Baier, C.J.; Barrantes, F.J. Molecular Mechanisms of Protein-Cholesterol Interactions in Plasma Membranes: Functional Distinction between Topological (Tilted) and Consensus (CARC/CRAC) Domains. Chem. Phys. Lipids 2016, 199, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Baier, C.J.; Fantini, J.; Barrantes, F.J. Disclosure of Cholesterol Recognition Motifs in Transmembrane Domains of the Human Nicotinic Acetylcholine Receptor. Sci. Rep. 2011, 1, 69. [Google Scholar] [CrossRef]
- Amuategi, J.; Alonso, R.; Ostolaza, H. Four Cholesterol-Recognition Motifs in the Pore-Forming and Translocation Domains of Adenylate Cyclase Toxin Are Essential for Invasion of Eukaryotic Cells and Lysis of Erythrocytes. Int. J. Mol. Sci. 2022, 23, 8703. [Google Scholar] [CrossRef] [PubMed]
- Krueger, E.; Brown, A.C. Aggregatibacter actinomycetemcomitans Leukotoxin: From Mechanism to Targeted Anti-Toxin Therapeutics. Mol. Oral Microbiol. 2020, 35, 85–105. [Google Scholar] [CrossRef]
- Cortajarena, A.L.; Goñi, F.M.; Ostolaza, H. Glycophorin as a Receptor for Escherichia coli α-Hemolysin in Erythrocytes. J. Biol. Chem. 2001, 276, 12513–12519. [Google Scholar] [CrossRef]
- Fantini, J.; Barrantes, F.J. Sphingolipid/Cholesterol Regulation of Neurotransmitter Receptor Conformation and Function. Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 2345–2361. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Chattopadhyay, A. Transbilayer Organization of Membrane Cholesterol at Low Concentrations: Implications in Health and Disease. Biochim. Biophys. Acta (BBA)-Biomembr. 2011, 1808, 19–25. [Google Scholar] [CrossRef]
- Mattjus, P.; Slotte, J.P. Does Cholesterol Discriminate between Sphingomyelin and Phosphatidylcholine in Mixed Monolayers Containing Both Phospholipids? Chem. Phys. Lipids 1996, 81, 69–80. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Anderson, T.G.; McConnell, H.M. Condensed Complexes, Rafts, and the Chemical Activity of Cholesterol in Membranes. Proc. Natl. Acad. Sci. USA 2000, 97, 12422–12427. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.A.; London, E. Structure and Origin of Ordered Lipid Domains in Biological Membranes. J. Membr. Biol. 1998, 164, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Ikonen, E. Functional Rafts in Cell Membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.G.W.; Jacobson, K. A Role for Lipid Shells in Targeting Proteins to Caveolae, Rafts, and Other Lipid Domains. Science 2002, 296, 1821–1825. [Google Scholar] [CrossRef] [PubMed]
- Infante, R.E.; Abi-Mosleh, L.; Radhakrishnan, A.; Dale, J.D.; Brown, M.S.; Goldstein, J.L. Purified NPC1 Protein. I. Binding of Cholesterol and Oxysterols to a 1278-Amino Acid Membrane Protein. J. Biol Chem 2008, 283, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Rose, I.A.; Hanson, K.R.; Wilkinson, K.D.; Wimmer, M.J. A Suggestion for Naming Faces of Ring Compounds. Proc. Natl. Acad. Sci. USA 1980, 77, 2439–2441. [Google Scholar] [CrossRef]
- Ramstedt, B.; Slotte, J.P. Interaction of Cholesterol with Sphingomyelins and Acyl-Chain-Matched Phosphatidylcholines: A Comparative Study of the Effect of the Chain Length. Biophys. J. 1999, 76, 908–915. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Garmy, N. Cholesterol Accelerates the Binding of Alzheimer’s β-Amyloid Peptide to Ganglioside GM1 through a Universal Hydrogen-Bond-Dependent Sterol Tuning of Glycolipid Conformation. Front. Physiol. 2013, 4, 120. [Google Scholar] [CrossRef]
- Li, H.; Papadopoulos, V. Peripheral-Type Benzodiazepine Receptor Function in Cholesterol Transport. Identification of a Putative Cholesterol Recognition/Interaction Amino Acid Sequence and Consensus Pattern. Endocrinology 1998, 139, 4991–4997. [Google Scholar] [CrossRef]
- Jamin, N.; Neumann, J.-M.; Ostuni, M.A.; Vu, T.K.N.; Yao, Z.-X.; Murail, S.; Robert, J.-C.; Giatzakis, C.; Papadopoulos, V.; Lacapère, J.-J. Characterization of the Cholesterol Recognition Amino Acid Consensus Sequence of the Peripheral-Type Benzodiazepine Receptor. Mol. Endocrinol. 2005, 19, 588–594. [Google Scholar] [CrossRef]
- Ulmschneider, M.B.; Sansom, M.S. Amino Acid Distributions in Integral Membrane Protein Structures. Biochim. Biophys. Acta 2001, 1512, 1–14. [Google Scholar] [CrossRef]
- Fantini, J.; Barrantes, F.J. How Cholesterol Interacts with Membrane Proteins: An Exploration of Cholesterol-Binding Sites Including CRAC, CARC, and Tilted Domains. Front. Physiol. 2013, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Di Scala, C.; Evans, L.S.; Williamson, P.T.F.; Barrantes, F.J. A Mirror Code for Protein-Cholesterol Interactions in the Two Leaflets of Biological Membranes. Sci. Rep. 2016, 6, 21907. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Epand, R.M.; Barrantes, F.J. Cholesterol-Recognition Motifs in Membrane Proteins. Adv. Exp. Med. Biol. 2019, 1135, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Azzaz, F.; Chahinian, H.; Yahi, N.; Di Scala, C.; Baier, C.J.; Barrantes, F.J.; Fantini, J. Chapter 7—Cholesterol-Recognizing Amino Acid Consensus Motifs in Transmembrane Proteins: Comparative Analysis of in Silico Studies and Structural Data. In Cholesterol; Bukiya, A.N., Dopico, A.M., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 127–145. ISBN 978-0-323-85857-1. [Google Scholar]
- Masin, J.; Roderova, J.; Osickova, A.; Novak, P.; Bumba, L.; Fiser, R.; Sebo, P.; Osicka, R. The Conserved Tyrosine Residue 940 Plays a Key Structural Role in Membrane Interaction of Bordetella Adenylate Cyclase Toxin. Sci. Rep. 2017, 7, 9330. [Google Scholar] [CrossRef] [PubMed]
- Koufos, E.; Chang, E.H.; Rasti, E.S.; Krueger, E.; Brown, A.C. Use of a Cholesterol Recognition Amino Acid Consensus Peptide to Inhibit Binding of a Bacterial Toxin to Cholesterol. Biochemistry 2016, 55, 4787–4797. [Google Scholar] [CrossRef] [PubMed]
- Cané, L.; Guzmán, F.; Balatti, G.; Daza Millone, M.A.; Pucci Molineris, M.; Maté, S.; Martini, M.F.; Herlax, V. Biophysical Analysis to Assess the Interaction of CRAC and CARC Motif Peptides of Alpha Hemolysin of Escherichia coli with Membranes. Biochemistry 2023, 62, 1994–2011. [Google Scholar] [CrossRef] [PubMed]

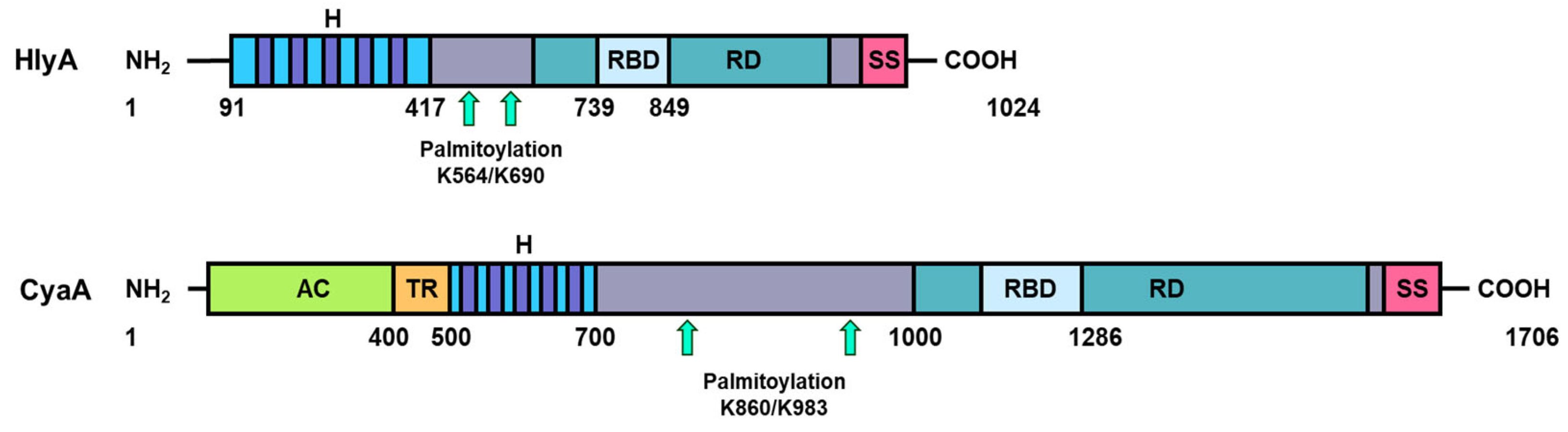
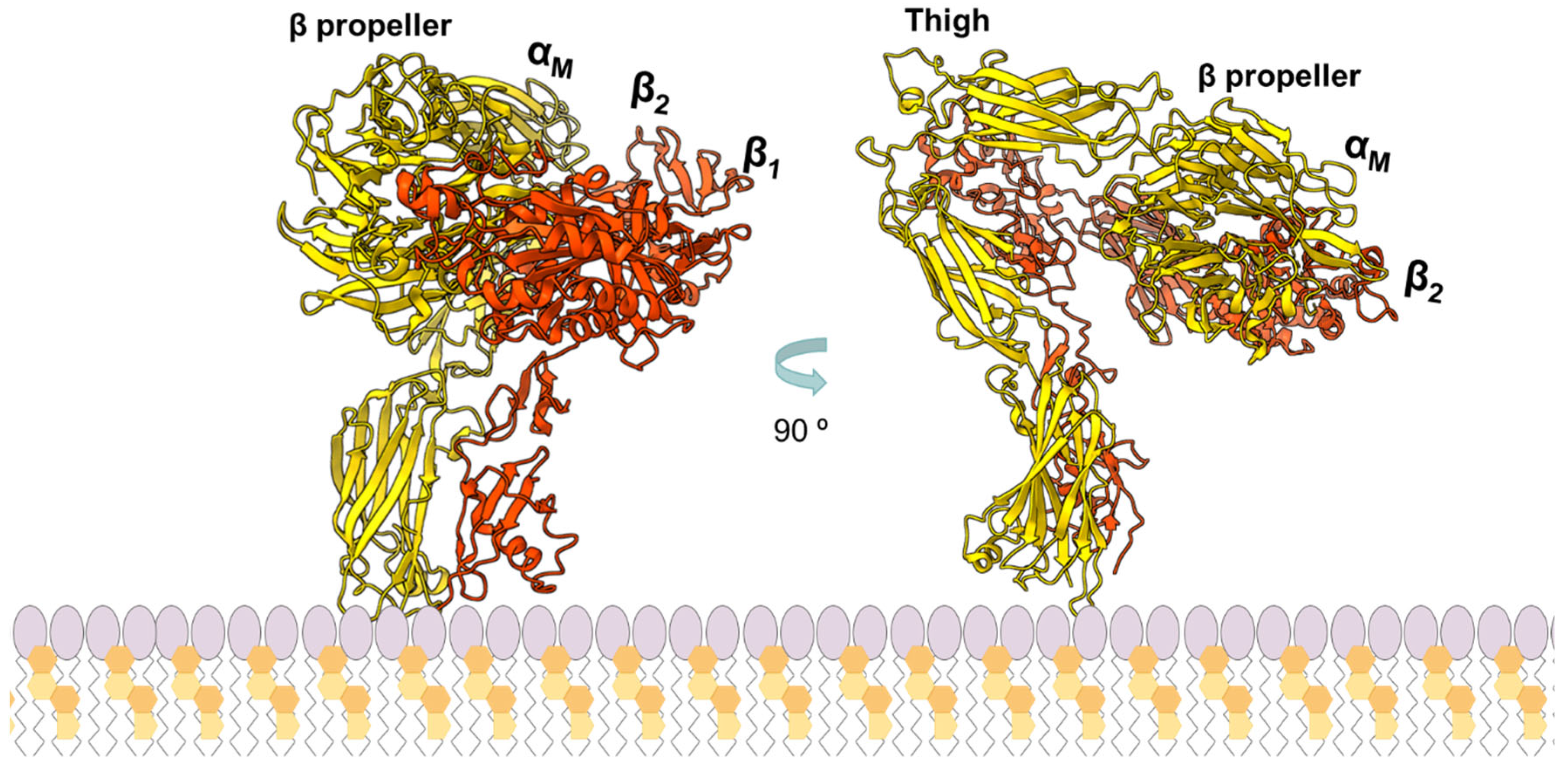
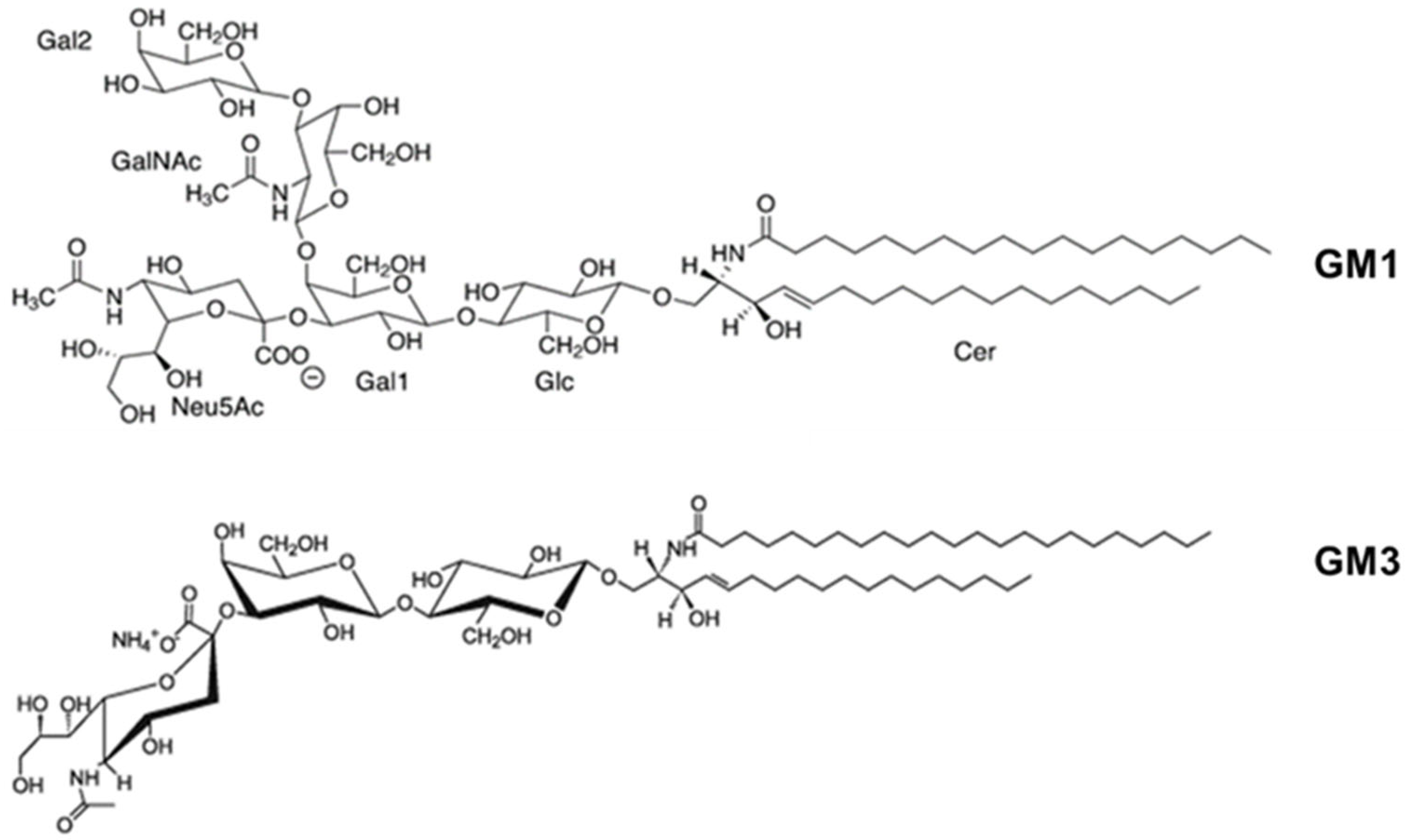
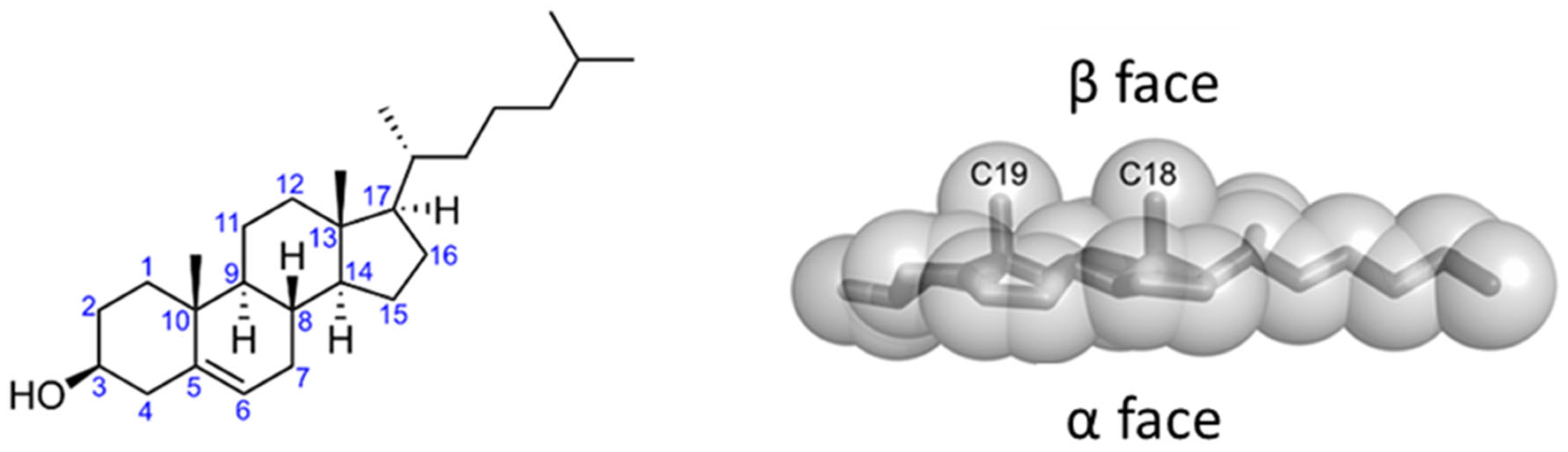
| RTX Toxin | Lipid Composition | Affinity (M) | Measurement Technique | Reference |
|---|---|---|---|---|
| A. Actinomycetemcomitans LtxA | POPC/CHOL (3:2 molar ratio) | KD = 10−12 | SPR | Brown and colleagues [69] |
| CHOL | KD = 2.31 × 10−10 | DSC | Krueger and Brown [106] | |
| K. kingae RtxA | POPC/CHOL (3:1 molar ratio) | KD = 1.71 × 10−10 | SPR | Osickova and colleagues [79] |
| UPEC HlyA | DOPC/CHOL (4:1 molar ratio) | KD = 1.6 × 10−5 | SPR | Vazquez and colleagues [100] |
| RTX Toxin | Bacterium | Residues | CRAC Motifs | CARC Motifs | Total No. CRAC/CARC Motifs |
|---|---|---|---|---|---|
| EhxA | Enterohemorrhagic (EHEC) Escherichia coli | 997 | 16 | 15 | 31 |
| LktA | Mannheimia haemolytica | 953 | 15 | 13 | 28 |
| PlLktA | M. varigena | 953 | 21 | 16 | 37 |
| PaxA | Pasteurella aerogenes | 1049 | 19 | 21 | 40 |
| MmxA | Morganella morganii | 1024 | 13 | 15 | 28 |
| HlyA | UPEC E. coli | 1023 | 13 | 13 | 26 |
| CyaA | Bordetella pertussis | 1706 | 22 | 18 | 40 |
| LtxA | Aggregatibacter actinomycetemcomitans | 1055 | 24 | 20 | 44 |
| ApxIA | Actinobacillus pleuropneumoniae | 1022 | 18 | 16 | 34 |
| ApxIIA | A. pleuropneumoniae | 956 | 16 | 16 | 32 |
| ApxIIIA | A. pleuropneumoniae | 1052 | 22 | 19 | 41 |
| MbxA | Moraxella bovis | 927 | 25 | 18 | 43 |
| RtxA | Kingella kingae | 956 | 12 | 14 | 26 |
| Motif | RTX Toxin | Bacterium | Residues | Sequence |
|---|---|---|---|---|
| CARC | EhxA | EHEC E. coli | 434–443 | RHAAFLEDSL |
| 400–407 | KQAMFEHV | |||
| 328–340 | RFKKLNYEGDALL | |||
| 271–278 | KAVSQYIL | |||
| 102–110 | KLLQKYQKV | |||
| LktA | M. haemolytica | 405–412 | KQAMFEHV | |
| 333–345 | RFKKLGYDGDNLL | |||
| 276–287 | KAVSSYILAQRV | |||
| 259–270 | KVGAGFELANQV | |||
| PlLktA | M. varigena | 405–412 | KQAMFEHV | |
| 333–345 | RFKKLGYDGDDLL | |||
| 276–287 | KAVSSYILAQRV | |||
| 258–270 | RKVGAGFELVNQV | |||
| 221–231 | KNFSGFSKAGL | |||
| 199–207 | KINQFGSKL | |||
| PaxA | P. aerogenes | 455–464 | RHKAFLEDSL | |
| 349–361 | RFKKLGYEGDKLL | |||
| 344–353 | REFAERFKKL | |||
| 292–303 | KAVSSYILAQRL | |||
| 275–286 | KVAAGFELSNQV | |||
| 115–124 | RGLTLFAPQL | |||
| MmxA | M. morganii | 458–469 | KLLSKYSEEYSV | |
| 414–421 | KQAMFEHV | |||
| 342–354 | RFKKFGYEGDSLL | |||
| 285–296 | KAVSQYILAQRV | |||
| 105–114 | RGIAIFAPQL | |||
| HlyA | UPEC E. coli | 457–468 | KILSQYNKEYSV | |
| 413–420 | KQAMFEHV | |||
| 341–353 | RFKKLGYDGDSLL | |||
| 105–114 | RGVTIFAPQL | |||
| CyaA | B. pertussis | 527–534 | RWAGGFGV | |
| 413–420 | RSFSLGEV | |||
| 399–410 | RQDSGYDSLDGV | |||
| LtxA | A. actinomycetemcomitans | 456–461 | KLFNEL | |
| 446–455 | RHSAFLEDSL | |||
| 340–352 | RFKKFGYNGDSLL | |||
| 326–334 | KQFDRARML | |||
| 218–227 | KHFGSFGDKL | |||
| ApxIA | A. pleuropneumoniae | 461–469 | KEYSVERVV | |
| 410–417 | KQAIFERV | |||
| 338–350 | RFKKFGYEGDSLL | |||
| 281–292 | KAVSQYIIAQRV | |||
| ApxIIA | A. pleuropneumoniae | 410–417 | KQAMFEHV | |
| 338–350 | RFQKLGYDGDRLL | |||
| 333–342 | KSYSERFQKL | |||
| 281–292 | KAVSSYILAQRV | |||
| ApxIIIA | A. pleuropneumoniae | 352–360 | KLGYDGDKL | |
| 349–361 | RFKKLGYDGDKLL | |||
| 292–303 | KAVSSYILAQRL | |||
| 275–286 | KVAAGFELSNQV | |||
| 115–124 | RGLTLFAPQL | |||
| MbxA | M. bovis | 422–431 | RYAAYLANNL | |
| 417–427 | KGYDSRYAAYL | |||
| 387–394 | KQAMFESV | |||
| 317–327 | RKFGYDGDHLL | |||
| 258–269 | KAISSYVLAQRV | |||
| 240–252 | KKVAAGFELSNQV | |||
| 202–213 | KLQNLNFSKTNL | |||
| RtxA | K. kingae | 444–453 | RHAHYLERNL | |
| 409–416 | KQAMFESV | |||
| 339–349 | KKFGYDGDSLL | |||
| 280–287 | KAISSYVL | |||
| 262–274 | KKVAAGFELSNQV |
| Motif | RTX Toxin | Bacterium | Residues | Sequence |
|---|---|---|---|---|
| CRAC | EhxA | EHEC E. coli | 407–414 | VADKFAAR |
| 339–345 | LLAAFHK | |||
| 322–331 | LESYSERFKK | |||
| 310–320 | LAIADKFERAK | |||
| 307–315 | LSFLAIADK | |||
| 273–281 | VSQYILAQR | |||
| LktA | M. haemolytica | 401–405 | LQYSK | |
| 344–350 | LLAEYQR | |||
| 327–336 | LESYAERFKK | |||
| 312–320 | LAFAGIADK | |||
| 278–286 | VSSYILAQR | |||
| PlLktA | M. varigena | 401–405 | LQYSK | |
| 344–350 | LLAQYQR | |||
| 327–336 | LESYAERFKK | |||
| 312–320 | LAFAGIADK | |||
| 278–286 | VSSYILAQR | |||
| 220–228 | LKNFSGFSK | |||
| PaxA | P. aerogenes | 417–421 | LEFSK | |
| 353–359 | LGYEGDK | |||
| 331–341 | LRVADNFNRSK | |||
| 328–332 | LSFLR | |||
| 294–302 | VSSYILAQR | |||
| 259–268 | VTASFTLADK | |||
| 124–133 | LDKFLQQHSK | |||
| 117–126 | LTLFAPQLDK | |||
| MmxA | M. morganii | 453–462 | LEDNFKLLSK | |
| 324–334 | LAVADKFKRAR | |||
| 321–329 | LSFLAVADK | |||
| 287–295 | VSQYILAQR | |||
| HlyA | UPEC E. coli | 459–464 | LSQYNK | |
| 352–358 | LLAAFHK | |||
| 323–334 | LSIADKFKRANK | |||
| 320–328 | LSFLSIADK | |||
| 198–210 | LNNVNSFSQQLNK | |||
| 117–127 | LLQKYQKAGNK | |||
| CyaA | B. pertussis | 732–741 | LGGPQAYFEK | |
| 721–728 | LANDYARK | |||
| 653–661 | LLAQLYRDK | |||
| 626–638 | LVQQSHYADQLDK | |||
| 518–527 | VSGFFRGSSR | |||
| 481–487 | LMTQFGR | |||
| LtxA | A. actinomycetemcomitans | 455–464 | LKLFNELREK | |
| 351–357 | LLGQFYK | |||
| 334–343 | LEEYSKRFKK | |||
| 322–332 | LGIAKQFDRAR | |||
| 319–326 | LSFLGIAK | |||
| 217–228 | VKHFGSFGDKLK | |||
| 214–226 | LGQVKHFGSFGDK | |||
| 200–209 | VDTFSKQLNK | |||
| ApxIA | A. pleuropneumoniae | 455–461 | LLSQYNK | |
| 349–355 | LLASFYR | |||
| 332–341 | LEQYSERFKK | |||
| 320–330 | LNVADKFERAK | |||
| 317–325 | LSFLNVADK | |||
| 283–291 | VSQYIIAQR | |||
| 247–257 | VVSASFILSNK | |||
| 199–208 | VDAFAEQLGK | |||
| 106–117 | LFAPQFDKLLNK | |||
| ApxIIA | A. pleuropneumoniae | 449–461 | LQDNMKFLINLNK | |
| 406–410 | LEYSK | |||
| 349–355 | LLADFHR | |||
| 342–348 | LGYDGDR | |||
| 331–341 | LIKSYSERFQK | |||
| 314–325 | VTPLSFLNVADK | |||
| 283–291 | VSSYILAQR | |||
| 195–207 | VQTVDAFAEQISK | |||
| 101–106 | LGFTDR | |||
| ApxIIIA | A. pleuropneumoniae | 417–421 | LEFSK | |
| 353–359 | LGYDGDK | |||
| 331–341 | LRVADNFNRSK | |||
| 328–332 | LAFLR | |||
| 294–302 | VSSYILAQR | |||
| 259–268 | VTASFALANK | |||
| 124–133 | LDQFLQKHSK | |||
| MbxA | M. bovis | 427–439 | LANNLKFLSELNK | |
| 326–332 | LLAEYQR | |||
| 309–318 | LDEFAKQFRK | |||
| 294–302 | LAFMNAADK | |||
| 203–210 | LQNLNFSK | |||
| RtxA | K. kingae | 348–354 | LLAEYQR | |
| 330–340 | LIDEFAKQFKK | |||
| 316–324 | LAFMNAADK | |||
| 224–232 | LQNLPNFGK | |||
| 194–206 | VQSIEAFSEQLGR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ostolaza, H.; Amuategi, J. Membrane Interaction Characteristics of the RTX Toxins and the Cholesterol-Dependence of Their Cytolytic/Cytotoxic Activity. Int. J. Mol. Sci. 2024, 25, 3131. https://doi.org/10.3390/ijms25063131
Ostolaza H, Amuategi J. Membrane Interaction Characteristics of the RTX Toxins and the Cholesterol-Dependence of Their Cytolytic/Cytotoxic Activity. International Journal of Molecular Sciences. 2024; 25(6):3131. https://doi.org/10.3390/ijms25063131
Chicago/Turabian StyleOstolaza, Helena, and Jone Amuategi. 2024. "Membrane Interaction Characteristics of the RTX Toxins and the Cholesterol-Dependence of Their Cytolytic/Cytotoxic Activity" International Journal of Molecular Sciences 25, no. 6: 3131. https://doi.org/10.3390/ijms25063131
APA StyleOstolaza, H., & Amuategi, J. (2024). Membrane Interaction Characteristics of the RTX Toxins and the Cholesterol-Dependence of Their Cytolytic/Cytotoxic Activity. International Journal of Molecular Sciences, 25(6), 3131. https://doi.org/10.3390/ijms25063131






