Circulating microRNAs in Cancer: A 5-Year Update with a Focus on Breast and Lung Cancers
Abstract
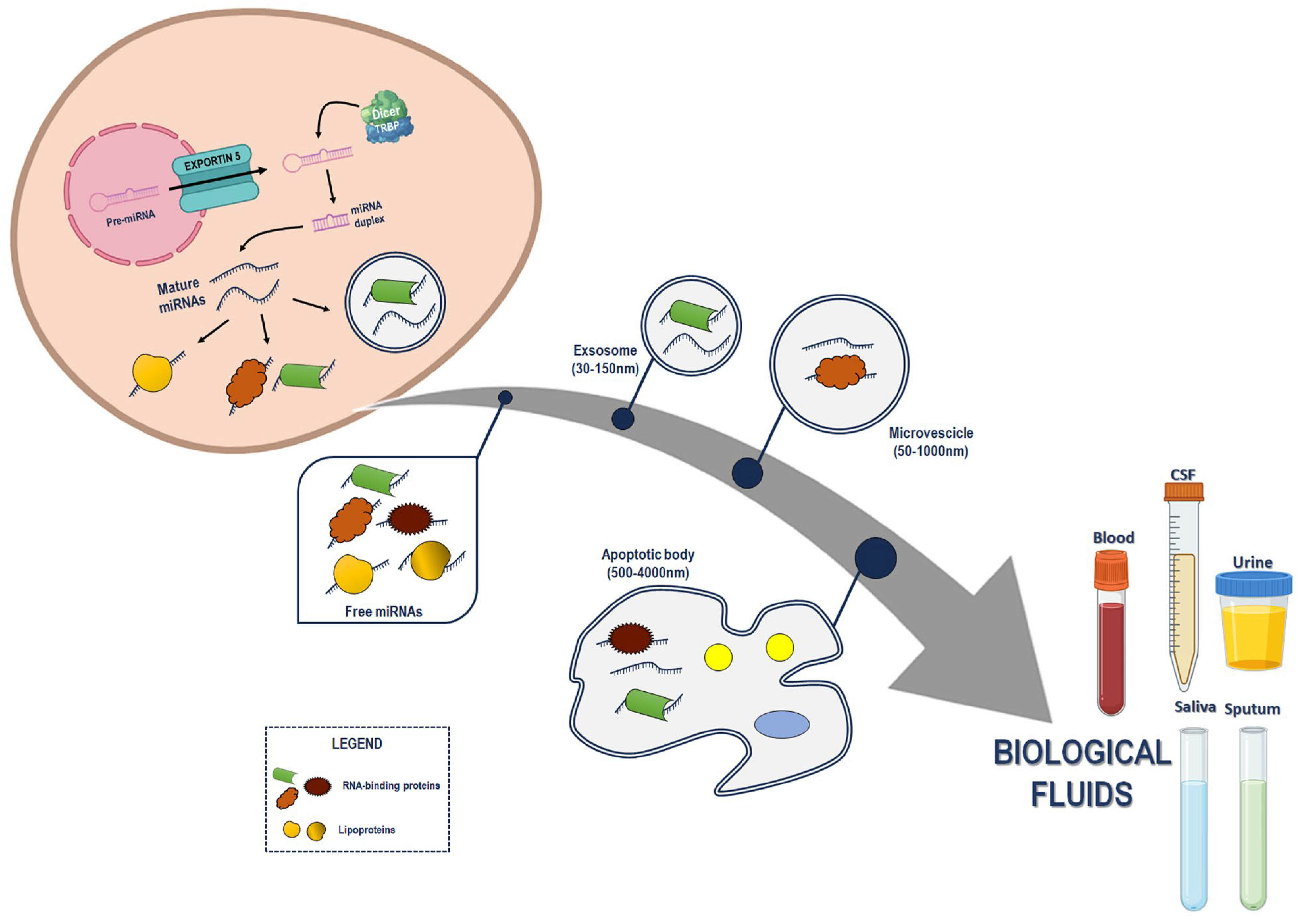
1. Introduction
2. C-miRNAs in Cancer: An Update of the Past Five Years’ Literature
3. BC and C-miRNAs
3.1. C-miRNAs and BC Diagnosis
3.2. C-miRNAs and Genetic and Environmental Factors in BC
3.3. C-miRNAs and BC Treatments
3.4. C-miRNAs and BC Recurrence
3.5. Novel Detection Systems and Approaches for C-miRNA Detection in BC
4. LC and C-miRNAs
4.1. C-miRNAs and LC Diagnosis
4.2. C-miRNAs, LC and EVs
4.3. C-miRNAs and LC Treatments
4.4. Novel Detection Systems and Approaches for C-miRNA Detection in LC
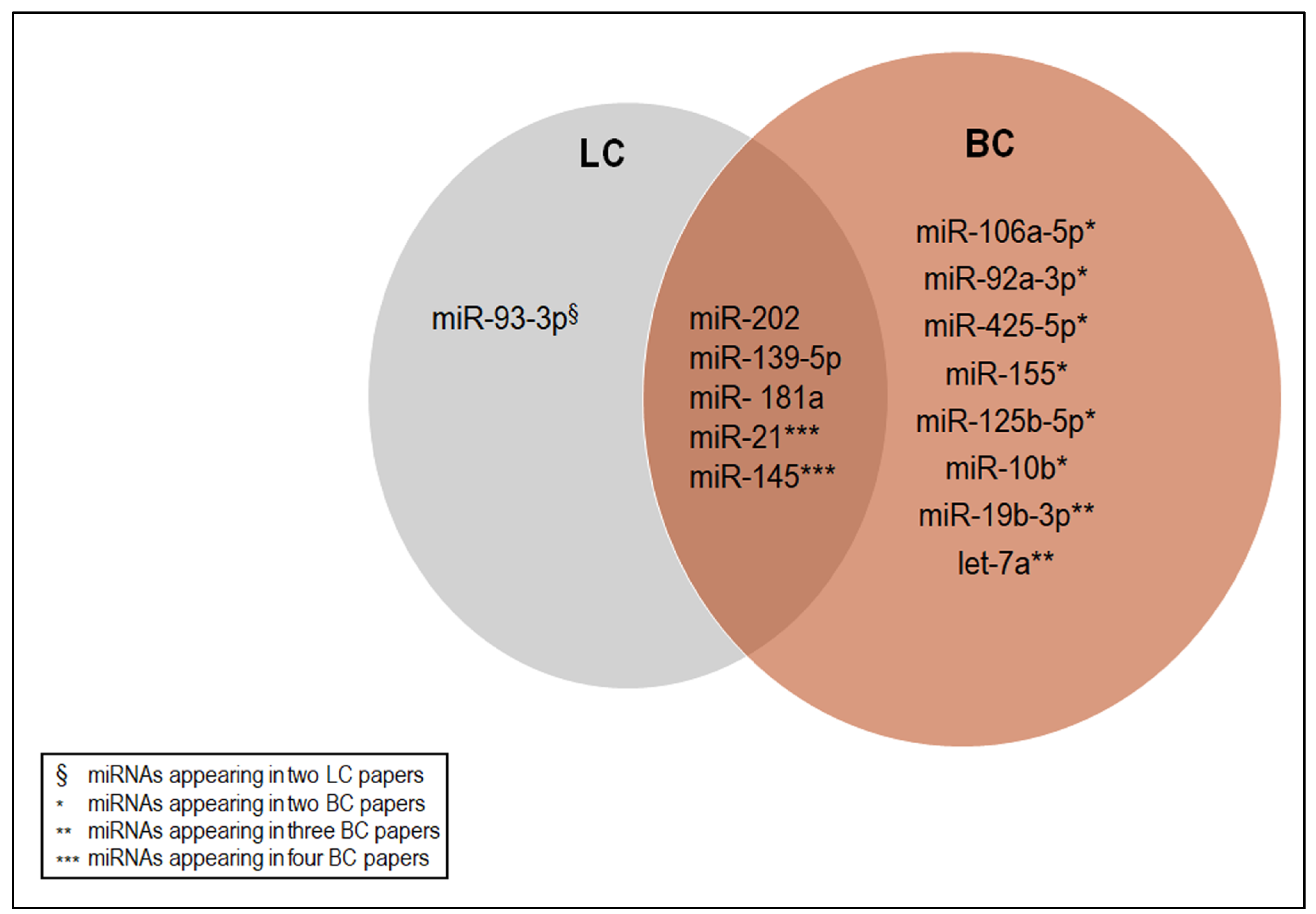
5. The C-miRNAs Shared among the Selected Studies
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Conte, R.; Finicelli, M.; Borrone, A.; Margarucci, S.; Peluso, G.; Calarco, A.; Bosetti, M. MMP-2 Silencing through siRNA Loaded Positively-Charged Nanoparticles (AcPEI-NPs) Counteracts Chondrocyte De-Differentiation. Polymers 2023, 15, 1172. [Google Scholar] [CrossRef] [PubMed]
- Precazzini, F.; Detassis, S.; Imperatori, A.S.; Denti, M.A.; Campomenosi, P. Measurements Methods for the Development of MicroRNA-Based Tests for Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1176. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Lampignano, R.; Kloten, V.; Krahn, T.; Schlange, T. Integrating circulating miRNA analysis in the clinical management of lung cancer: Present or future? Mol. Asp. Med. 2020, 72, 100844. [Google Scholar] [CrossRef]
- Valihrach, L.; Androvic, P.; Kubista, M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol. Asp. Med. 2020, 72, 100825. [Google Scholar] [CrossRef] [PubMed]
- Suarez, B.; Sole, C.; Marquez, M.; Nanetti, F.; Lawrie, C.H. Circulating MicroRNAs as Cancer Biomarkers in Liquid Biopsies. Adv. Exp. Med. Biol. 2022, 1385, 23–73. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B. Free circulating nucleic acids in plasma and serum (CNAPS)—Useful for the detection of lung cancer patients? Cancer Biomark. 2010, 6, 211–219. [Google Scholar] [CrossRef]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenet. 2018, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Capri, M.; Bonafe, M.; Morsiani, C.; Jung, H.J.; Spazzafumo, L.; Vina, J.; Suh, Y. Circulating miRNAs and miRNA shuttles as biomarkers: Perspective trajectories of healthy and unhealthy aging. Mech. Ageing Dev. 2017, 165, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ding, H.; Zhang, Y.; Wang, Y.; Zhu, W.; Li, P. Circulating MicroRNAs: Biogenesis and Clinical Significance in Acute Myocardial Infarction. Front. Physiol. 2020, 11, 1088. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell communication: microRNAs as hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef] [PubMed]
- Ruivo, C.F.; Adem, B.; Silva, M.; Melo, S.A. The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res. 2017, 77, 6480–6488. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Godoy, P.M.; Bhakta, N.R.; Barczak, A.J.; Cakmak, H.; Fisher, S.; MacKenzie, T.C.; Patel, T.; Price, R.W.; Smith, J.F.; Woodruff, P.G.; et al. Large Differences in Small RNA Composition Between Human Biofluids. Cell Rep. 2018, 25, 1346–1358. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Mattiuzzi, C.; Lippi, G. Current Cancer Epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Zheng, R.; Zhang, S.; Wang, S.; Chen, R.; Sun, K.; Zeng, H.; Zhou, J.; Wei, W. Global patterns of breast cancer incidence and mortality: A population-based cancer registry data analysis from 2000 to 2020. Cancer Commun. 2021, 41, 1183–1194. [Google Scholar] [CrossRef]
- Anwar, S.L.; Tanjung, D.S.; Fitria, M.S.; Kartika, A.I.; Sari, D.N.I.; Rakhmina, D.; Wardana, T.; Astuti, I.; Haryana, S.M.; Aryandono, T. Dynamic Changes of Circulating Mir-155 Expression and the Potential Application as a Non-Invasive Biomarker in Breast Cancer. Asian Pac. J. Cancer Prev. 2020, 21, 491–497. [Google Scholar] [CrossRef]
- Uyisenga, J.P.; Debit, A.; Poulet, C.; Freres, P.; Poncin, A.; Thiry, J.; Mutesa, L.; Jerusalem, G.; Bours, V.; Josse, C. Differences in plasma microRNA content impair microRNA-based signature for breast cancer diagnosis in cohorts recruited from heterogeneous environmental sites. Sci. Rep. 2021, 11, 11698. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Ali, O.S.; Shabayek, M.I.; Seleem, M.M.; Abdelaziz, H.G.; Makhlouf, D.O. MicroRNAs 182 and 375 Sera Expression as Prognostic Biochemical Markers in Breast Cancer. Clin. Breast Cancer 2018, 18, e1373–e1379. [Google Scholar] [CrossRef]
- McAnena, P.; Tanriverdi, K.; Curran, C.; Gilligan, K.; Freedman, J.E.; Brown, J.A.L.; Kerin, M.J. Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer 2019, 19, 436. [Google Scholar] [CrossRef]
- Zou, X.; Xia, T.; Li, M.; Wang, T.; Liu, P.; Zhou, X.; Huang, Z.; Zhu, W. MicroRNA profiling in serum: Potential signatures for breast cancer diagnosis. Cancer Biomark. 2021, 30, 41–53. [Google Scholar] [CrossRef]
- Ozawa, P.M.M.; Vieira, E.; Lemos, D.S.; Souza, I.L.M.; Zanata, S.M.; Pankievicz, V.C.; Tuleski, T.R.; Souza, E.M.; Wowk, P.F.; Urban, C.A.; et al. Identification of miRNAs Enriched in Extracellular Vesicles Derived from Serum Samples of Breast Cancer Patients. Biomolecules 2020, 10, 150. [Google Scholar] [CrossRef]
- Itani, M.M.; Nassar, F.J.; Tfayli, A.H.; Talhouk, R.S.; Chamandi, G.K.; Itani, A.R.S.; Makoukji, J.; Boustany, R.N.; Hou, L.; Zgheib, N.K.; et al. A Signature of Four Circulating microRNAs as Potential Biomarkers for Diagnosing Early-Stage Breast Cancer. Int. J. Mol. Sci. 2021, 22, 6121. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, S.; Hwang, D.; Kim, S.I.; Lee, H. Diagnostic Value of Circulating miR-202 in Early-Stage Breast Cancer in South Korea. Medicina 2020, 56, 340. [Google Scholar] [CrossRef]
- Zou, R.; Loke, S.Y.; Tang, Y.C.; Too, H.P.; Zhou, L.; Lee, A.S.G.; Hartman, M. Development and validation of a circulating microRNA panel for the early detection of breast cancer. Br. J. Cancer 2022, 126, 472–481. [Google Scholar] [CrossRef]
- Ibrahim, A.M.; Said, M.M.; Hilal, A.M.; Medhat, A.M.; Abd Elsalam, I.M. Candidate circulating microRNAs as potential diagnostic and predictive biomarkers for the monitoring of locally advanced breast cancer patients. Tumour Biol. 2020, 42, 1010428320963811. [Google Scholar] [CrossRef] [PubMed]
- Khalighfard, S.; Alizadeh, A.M.; Irani, S.; Omranipour, R. Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci. Rep. 2018, 8, 17981. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Casey, M.C.; McGuire, A.; Waldron, R.M.; Paganga, M.; Holian, E.; Newell, J.; Heneghan, H.M.; McDermott, A.M.; Keane, M.M.; et al. Evaluating the Role of Circulating MicroRNAs to Aid Therapeutic Decision Making for Neoadjuvant Chemotherapy in Breast Cancer: A Prospective, Multicenter Clinical Trial. Ann. Surg. 2022, 276, 905–912. [Google Scholar] [CrossRef]
- Davey, M.G.; McGuire, A.; Casey, M.C.; Waldron, R.M.; Paganga, M.; Holian, E.; Newell, J.; Heneghan, H.M.; McDermott, A.M.; Keane, M.M.; et al. Evaluating the Role of Circulating MicroRNAs in Predicting Long-Term Survival Outcomes in Breast Cancer: A Prospective, Multicenter Clinical Trial. J. Am. Coll. Surg. 2023, 236, 317–327. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Li, C.; Xiang, Q.; Xu, L.; Liu, Q.; Pang, X.; Zhang, W.; Zhang, H.; Zhang, S.; et al. Circulating microRNAs as indicators in the prediction of neoadjuvant chemotherapy response in luminal B breast cancer. Thorac. Cancer 2021, 12, 3396–3406. [Google Scholar] [CrossRef]
- Zhu, W.; Liu, M.; Fan, Y.; Ma, F.; Xu, N.; Xu, B. Dynamics of circulating microRNAs as a novel indicator of clinical response to neoadjuvant chemotherapy in breast cancer. Cancer Med. 2018, 7, 4420–4433. [Google Scholar] [CrossRef]
- Di Cosimo, S.; Appierto, V.; Pizzamiglio, S.; Silvestri, M.; Baselga, J.; Piccart, M.; Huober, J.; Izquierdo, M.; de la Pena, L.; Hilbers, F.S.; et al. Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy. Int. J. Mol. Sci. 2020, 21, 1386. [Google Scholar] [CrossRef]
- Papadaki, C.; Stratigos, M.; Markakis, G.; Spiliotaki, M.; Mastrostamatis, G.; Nikolaou, C.; Mavroudis, D.; Agelaki, S. Circulating microRNAs in the early prediction of disease recurrence in primary breast cancer. Breast Cancer Res. 2018, 20, 72. [Google Scholar] [CrossRef]
- Colleoni, M.; Sun, Z.; Price, K.N.; Karlsson, P.; Forbes, J.F.; Thurlimann, B.; Gianni, L.; Castiglione, M.; Gelber, R.D.; Coates, A.S.; et al. Annual Hazard Rates of Recurrence for Breast Cancer During 24 Years of Follow-Up: Results From the International Breast Cancer Study Group Trials I to V. J. Clin. Oncol. 2016, 34, 927–935. [Google Scholar] [CrossRef]
- Fischer, C.; Deutsch, T.M.; Feisst, M.; Rippinger, N.; Riedel, F.; Hartkopf, A.D.; Brucker, S.Y.; Domschke, C.; Fremd, C.; Michel, L.; et al. Circulating miR-200 family as predictive markers during systemic therapy of metastatic breast cancer. Arch. Gynecol. Obstet. 2022, 306, 875–885. [Google Scholar] [CrossRef]
- Fischer, C.; Turchinovich, A.; Feisst, M.; Riedel, F.; Hassdenteufel, K.; Scharli, P.; Hartkopf, A.D.; Brucker, S.Y.; Michel, L.; Burwinkel, B.; et al. Circulating miR-200 Family and CTCs in Metastatic Breast Cancer before, during, and after a New Line of Systemic Treatment. Int. J. Mol. Sci. 2022, 23, 9535. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, M.; Mokhtari, S.; Abidi, H.; Alipoor, B.; Nazer Mozaffari, M.A.; Sadeghi, H.; Mahmoudi, R.; Nikseresht, M. Multi-Drug Resistance against Second-Line Medication and MicroRNA Plasma Level in Metastatic Breast Cancer Patients. Iran J. Med. Sci. 2023, 48, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef]
- Squillaro, T.; Finicelli, M.; Alessio, N.; Del Gaudio, S.; Di Bernardo, G.; Melone, M.A.B.; Peluso, G.; Galderisi, U. A rapid, safe, and quantitative in vitro assay for measurement of uracil-DNA glycosylase activity. J. Mol. Med. 2019, 97, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Forte, A.; Finicelli, M.; Grossi, M.; Vicchio, M.; Alessio, N.; Sante, P.; De Feo, M.; Cotrufo, M.; Berrino, L.; Rossi, F.; et al. DNA damage and repair in a model of rat vascular injury. Clin. Sci. 2010, 118, 473–485. [Google Scholar] [CrossRef][Green Version]
- Li, Z.; Xiao, H.; Li, J.; Yang, Z.; Jiang, J.; Ji, J.; Peng, C.; He, Y. Graphene Oxide-Based Highly Sensitive Assay of Circulating MicroRNAs for Early Prediction of the Response to Neoadjuvant Chemotherapy in Breast Cancer. Anal. Chem. 2022, 94, 16254–16264. [Google Scholar] [CrossRef]
- Fan, T.; Mao, Y.; Sun, Q.; Liu, F.; Lin, J.S.; Liu, Y.; Cui, J.; Jiang, Y. Branched rolling circle amplification method for measuring serum circulating microRNA levels for early breast cancer detection. Cancer Sci. 2018, 109, 2897–2906. [Google Scholar] [CrossRef]
- Sadeghi, S.; Rahaie, M. Design and Fabrication of a DNA-copper Nanocluster-based Biosensor for Multiple Detections of Circulating miRNAs in Early Screening of Breast Cancer. J. Fluoresc. 2022, 32, 2297–2307. [Google Scholar] [CrossRef]
- Agahi, M.; Rahaie, M. A novel DNA tweezers-based nanobiosensor for multiple detections of circulating exosomal microRNAs in breast cancer. Anal. Biochem. 2022, 651, 114697. [Google Scholar] [CrossRef]
- Garrido-Cano, I.; Pla, L.; Santiago-Felipe, S.; Simon, S.; Ortega, B.; Bermejo, B.; Lluch, A.; Cejalvo, J.M.; Eroles, P.; Martinez-Manez, R. Nanoporous Anodic Alumina-Based Sensor for miR-99a-5p Detection as an Effective Early Breast Cancer Diagnostic Tool. ACS Sens. 2021, 6, 1022–1029. [Google Scholar] [CrossRef]
- Kim, J.; Park, J.; Ki, J.; Rho, H.W.; Huh, Y.M.; Kim, E.; Son, H.Y.; Haam, S. Simultaneous dual-targeted monitoring of breast cancer circulating miRNA via surface-enhanced Raman spectroscopy. Biosens. Bioelectron. 2022, 207, 114143. [Google Scholar] [CrossRef]
- Wong, C.L.; Loke, S.Y.; Lim, H.Q.; Balasundaram, G.; Chan, P.; Chong, B.K.; Tan, E.Y.; Lee, A.S.G.; Olivo, M. Circulating microRNA breast cancer biomarker detection in patient sera with surface plasmon resonance imaging biosensor. J. Biophotonics 2021, 14, e202100153. [Google Scholar] [CrossRef]
- Duan, X.; Qiao, S.; Li, D.; Li, S.; Zheng, Z.; Wang, Q.; Zhu, X. Circulating miRNAs in Serum as Biomarkers for Early Diagnosis of Non-small Cell Lung Cancer. Front. Genet. 2021, 12, 673926. [Google Scholar] [CrossRef] [PubMed]
- Wiemer, E.A.C. Prognostic Circulating MicroRNA Biomarkers in Early-Stage Non-Small Cell Lung Cancer: A Role for miR-150. Clin. Pharmacol. Ther. 2018, 103, 968–970. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, A.; Seam, R.K.; Gupta, M.; Rana, M.K.; Prakash, H.; Vasquez, K.M.; Jain, A. Circulating microRNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 2020, 111, 826–839. [Google Scholar] [CrossRef]
- Singh, A.; Kant, R.; Saluja, T.S.; Tripathi, T.; Srivastava, K.; Naithani, M.; Gupta, A.; Mirza, A.A.; Prakash, V.; Singh, S.K. Differential diagnosis of non-small cell lung carcinoma by circulating microRNA. J. Cancer Res. Ther. 2020, 16, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Fehlmann, T.; Kahraman, M.; Ludwig, N.; Backes, C.; Galata, V.; Keller, V.; Geffers, L.; Mercaldo, N.; Hornung, D.; Weis, T.; et al. Evaluating the Use of Circulating MicroRNA Profiles for Lung Cancer Detection in Symptomatic Patients. JAMA Oncol. 2020, 6, 714–723. [Google Scholar] [CrossRef]
- Abdollahi, A.; Rahmati, S.; Ghaderi, B.; Sigari, N.; Nikkhoo, B.; Sharifi, K.; Abdi, M. A combined panel of circulating microRNA as a diagnostic tool for detection of the non-small cell lung cancer. QJM Int. J. Med. 2019, 112, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Du, L.; Zou, R.; Shi, L.; Zhang, N.; Jin, J.; Xu, C.; Zhang, F.; Zhu, C.; Wu, J.; et al. Development of a serum miRNA panel for detection of early stage non-small cell lung cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 25036–25042. [Google Scholar] [CrossRef]
- Zhang, Y.; Roth, J.A.; Yu, H.; Ye, Y.; Xie, K.; Zhao, H.; Chang, D.W.; Huang, M.; Li, H.; Qu, J.; et al. A 5-microRNA signature identified from serum microRNA profiling predicts survival in patients with advanced stage non-small cell lung cancer. Carcinogenesis 2019, 40, 643–650. [Google Scholar] [CrossRef]
- Zhao, J.; Qiao, C.R.; Ding, Z.; Sheng, Y.L.; Li, X.N.; Yang, Y.; Zhu, D.Y.; Zhang, C.Y.; Liu, D.L.; Wu, K.; et al. A novel pathway in NSCLC cells: miR-191, targeting NFIA, is induced by chronic hypoxia, and promotes cell proliferation and migration. Mol. Med. Rep. 2017, 15, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Vadla, G.P.; Daghat, B.; Patterson, N.; Ahmad, V.; Perez, G.; Garcia, A.; Manjunath, Y.; Kaifi, J.T.; Li, G.; Chabu, C.Y. Combining plasma extracellular vesicle Let-7b-5p, miR-184 and circulating miR-22-3p levels for NSCLC diagnosis and drug resistance prediction. Sci. Rep. 2022, 12, 6693. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, H. Serum exosomal miR-378 upregulation is associated with poor prognosis in non-small-cell lung cancer patients. J. Clin. Lab. Anal. 2020, 34, e23237. [Google Scholar] [CrossRef]
- Zappasodi, R.; Merghoub, T.; Wolchok, J.D. Emerging Concepts for Immune Checkpoint Blockade-Based Combination Therapies. Cancer Cell 2018, 33, 581–598. [Google Scholar] [CrossRef] [PubMed]
- Kleinovink, J.W.; Marijt, K.A.; Schoonderwoerd, M.J.A.; van Hall, T.; Ossendorp, F.; Fransen, M.F. PD-L1 expression on malignant cells is no prerequisite for checkpoint therapy. Oncoimmunology 2017, 6, e1294299. [Google Scholar] [CrossRef]
- Shukuya, T.; Ghai, V.; Amann, J.M.; Okimoto, T.; Shilo, K.; Kim, T.K.; Wang, K.; Carbone, D.P. Circulating MicroRNAs and Extracellular Vesicle-Containing MicroRNAs as Response Biomarkers of Anti-programmed Cell Death Protein 1 or Programmed Death-Ligand 1 Therapy in NSCLC. J. Thorac. Oncol. 2020, 15, 1773–1781. [Google Scholar] [CrossRef]
- Fan, J.; Yin, Z.; Xu, J.; Wu, F.; Huang, Q.; Yang, L.; Jin, Y.; Yang, G. Circulating microRNAs predict the response to anti-PD-1 therapy in non-small cell lung cancer. Genomics 2020, 112, 2063–2071. [Google Scholar] [CrossRef]
- Halvorsen, A.R.; Sandhu, V.; Sprauten, M.; Flote, V.G.; Kure, E.H.; Brustugun, O.T.; Helland, A. Circulating microRNAs associated with prolonged overall survival in lung cancer patients treated with nivolumab. Acta Oncol. 2018, 57, 1225–1231. [Google Scholar] [CrossRef]
- Monastirioti, A.; Papadaki, C.; Rounis, K.; Kalapanida, D.; Mavroudis, D.; Agelaki, S. A Prognostic Role for Circulating microRNAs Involved in Macrophage Polarization in Advanced Non-Small Cell Lung Cancer. Cells 2021, 10, 1988. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Yu, W.; Chen, C.; Guo, S.; Tian, X.; Miao, Y.; Ma, L.; Zhang, X.; Yu, Y.; Huang, L.; et al. A Versatile Electrochemical Biosensor for the Detection of Circulating MicroRNA toward Non-Small Cell Lung Cancer Diagnosis. Small 2022, 18, e2200784. [Google Scholar] [CrossRef]
- Zhuang, P.; Zhang, H.; Welchko, R.M.; Thompson, R.C.; Xu, S.; Turner, D.L. Combined microRNA and mRNA detection in mammalian retinas by in situ hybridization chain reaction. Sci. Rep. 2020, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Lin, M.; Wang, P.; Pei, H.; Yan, J.; Shi, J.; Huang, Q.; He, D.; Fan, C.; Zuo, X. Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor. Anal. Chem. 2014, 86, 2124–2130. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Yuan, C.; Liu, Y.; Li, Z.; Xia, K.; Li, M.; Xie, F.; Chen, Q.; Chen, M.; Fu, W.; et al. A novel quantification platform for point-of-care testing of circulating MicroRNAs based on allosteric spherical nanoprobe. J. Nanobiotechnol. 2020, 18, 158. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, L.; Yang, L.; Ni, W.; Li, Y.; Wu, Y. Tandem reassembly of split luciferase-DNA chimeras for bioluminescent detection of attomolar circulating microRNAs using a smartphone. Biosens. Bioelectron. 2021, 173, 112824. [Google Scholar] [CrossRef]
- Yao, Q.; Wang, Y.; Wang, J.; Chen, S.; Liu, H.; Jiang, Z.; Zhang, X.; Liu, S.; Yuan, Q.; Zhou, X. An Ultrasensitive Diagnostic Biochip Based on Biomimetic Periodic Nanostructure-Assisted Rolling Circle Amplification. ACS Nano 2018, 12, 6777–6783. [Google Scholar] [CrossRef] [PubMed]
- Detassis, S.; Grasso, M.; Tabraue-Chavez, M.; Marin-Romero, A.; Lopez-Longarela, B.; Ilyine, H.; Ress, C.; Ceriani, S.; Erspan, M.; Maglione, A.; et al. New Platform for the Direct Profiling of microRNAs in Biofluids. Anal. Chem. 2019, 91, 5874–5880. [Google Scholar] [CrossRef]
- Muller, V.; Gade, S.; Steinbach, B.; Loibl, S.; von Minckwitz, G.; Untch, M.; Schwedler, K.; Lubbe, K.; Schem, C.; Fasching, P.A.; et al. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: A translational research project within the Geparquinto trial. Breast Cancer Res. Treat. 2014, 147, 61–68. [Google Scholar] [CrossRef]
- Wei, J.; Gao, W.; Zhu, C.J.; Liu, Y.Q.; Mei, Z.; Cheng, T.; Shu, Y.Q. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin. J. Cancer 2011, 30, 407–414. [Google Scholar] [CrossRef]
- Liu, B.; Su, F.; Lv, X.; Zhang, W.; Shang, X.; Zhang, Y.; Zhang, J. Serum microRNA-21 predicted treatment outcome and survival in HER2-positive breast cancer patients receiving neoadjuvant chemotherapy combined with trastuzumab. Cancer Chemother. Pharmacol. 2019, 84, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Aleckovic, M.; Kang, Y. Regulation of cancer metastasis by cell-free miRNAs. Biochim. Biophys. Acta 2015, 1855, 24–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, L.; Sun, S.; Wu, J.; Wang, Q. Quantitative measurement of serum microRNA-21 expression in relation to breast cancer metastasis in Chinese females. Ann. Lab. Med. 2015, 35, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Fang, M.; Li, S.; Yan, Y.; Zhong, Y.; Du, B. miR-21 is involved in transforming growth factor beta1-induced chemoresistance and invasion by targeting PTEN in breast cancer. Oncol. Lett. 2017, 14, 6929–6936. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, J.; Zhan, Y.; Feng, J.; Luo, J.; Fan, S. MicroRNAs associated with therapy of non-small cell lung cancer. Int. J. Biol. Sci. 2018, 14, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Kang, X.; Xia, X.; Wo, L.; Gu, X.; Hu, Y.; Xie, X.; Chang, H.; Lou, L.; Shen, X. miR-145 suppresses breast cancer cell migration by targeting FSCN-1 and inhibiting epithelial-mesenchymal transition. Am. J. Transl. Res. 2016, 8, 3106–3114. [Google Scholar] [PubMed]
- Zhang, Y.; Lin, Q. MicroRNA-145 inhibits migration and invasion by down-regulating FSCN1 in lung cancer. Int. J. Clin. Exp. Med. 2015, 8, 8794–8802. [Google Scholar]
- Ahmed, E.A.; Rajendran, P.; Scherthan, H. The microRNA-202 as a Diagnostic Biomarker and a Potential Tumor Suppressor. Int. J. Mol. Sci. 2022, 23, 5870. [Google Scholar] [CrossRef]
- Bustos, M.A.; Yokoe, T.; Shoji, Y.; Kobayashi, Y.; Mizuno, S.; Murakami, T.; Zhang, X.; Sekhar, S.C.; Kim, S.; Ryu, S.; et al. MiR-181a targets STING to drive PARP inhibitor resistance in BRCA- mutated triple-negative breast cancer and ovarian cancer. Cell Biosci. 2023, 13, 200. [Google Scholar] [CrossRef]
- Niu, J.; Xue, A.; Chi, Y.; Xue, J.; Wang, W.; Zhao, Z.; Fan, M.; Yang, C.H.; Shao, Z.M.; Pfeffer, L.M.; et al. Induction of miRNA-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer. Oncogene 2016, 35, 1302–1313. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, T.; Du, Y.; Hu, X.; Xia, W. LncRNA LUCAT1/miR-181a-5p axis promotes proliferation and invasion of breast cancer via targeting KLF6 and KLF15. BMC Mol. Cell Biol. 2020, 21, 69. [Google Scholar] [CrossRef] [PubMed]
- Simiene, J.; Dabkeviciene, D.; Stanciute, D.; Prokarenkaite, R.; Jablonskiene, V.; Askinis, R.; Normantaite, K.; Cicenas, S.; Suziedelis, K. Potential of miR-181a-5p and miR-630 as clinical biomarkers in NSCLC. BMC Cancer 2023, 23, 857. [Google Scholar] [CrossRef] [PubMed]
- Pirlog, R.; Chiroi, P.; Rusu, I.; Jurj, A.M.; Budisan, L.; Pop-Bica, C.; Braicu, C.; Crisan, D.; Sabourin, J.C.; Berindan-Neagoe, I. Cellular and Molecular Profiling of Tumor Microenvironment and Early-Stage Lung Cancer. Int. J. Mol. Sci. 2022, 23, 5346. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, Z. MicroRNA-139-5p suppresses non-small cell lung cancer progression by targeting ATAD2. Pathol. Res. Pract. 2023, 249, 154719. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.Q.; Luo, J.H.; Yao, W.X. The regulation of miR-139-5p on the biological characteristics of breast cancer cells by targeting COL11A1. Math. Biosci. Eng. 2019, 17, 1428–1441. [Google Scholar] [CrossRef]
- Gao, B.; Li, R.; Song, X.; Hu, S.; Yang, F. miR-139-5p and miR-451a as a Diagnostic Biomarker in LUSC. Pharmgenomics Pers. Med. 2023, 16, 313–323. [Google Scholar] [CrossRef]
- Sun, H.; Dai, J.; Chen, M.; Chen, Q.; Xie, Q.; Zhang, W.; Li, G.; Yan, M. miR-139-5p Was Identified as Biomarker of Different Molecular Subtypes of Breast Carcinoma. Front. Oncol. 2022, 12, 857714. [Google Scholar] [CrossRef]
- Hong, H.C.; Chuang, C.H.; Huang, W.C.; Weng, S.L.; Chen, C.H.; Chang, K.H.; Liao, K.W.; Huang, H.D. A panel of eight microRNAs is a good predictive parameter for triple-negative breast cancer relapse. Theranostics 2020, 10, 8771–8789. [Google Scholar] [CrossRef]

| Breast Cancer | ||||
|---|---|---|---|---|
| miRNA | Source | Population | Function | References |
| ↑ miR-182; ↑ miR-375. | Serum | Pre- and post-menopausal BC patients and patients with benign tumors | Positive association with estrogen and progesterone receptors | [27] |
| ↑ miR-331; ↓ miR-195. | Plasma/ Whole blood | Patients with metastatic BC compared to those suffering from local disease or those who are healthy subjects | Identification of metastatic disease | [28] |
| ↑ miR-106a-3p; ↑ miR-106a-5p; ↑ miR-20b-5p; ↑ miR-92a-2-5p. | Plasma | BC patients and healthy controls | Upregulation in BC and prognostic significance | [26] |
| ↑ miR-106a-5p; ↑ miR-19b-3p; ↑ miR-20b-5p; ↑ miR-92a-3p. | Serum | BC patients and healthy controls | Upregulation in BC and prognostic significance | [26] |
| ↑ let-7b-5p; ↑ miR-106a-5p; ↑ miR-19a-3p; ↑ miR-19b-3p; ↑ miR-20a-5p; miR-223-3p; ↑ miR-25-3p; ↑ miR-425-5p; ↑ miR-451a; ↑ miR-92a-3p; ↑miR-93-5p; ↑ miR-16-5p. | Serum | BC patients and healthy controls | Distinguishing BC in different stages from controls | [29] |
| ↕ miR-142-5p; ↕ miR-150-5p; ↕ miR-320a; ↕ miR-4433b-5p. | Serum | EVs derived for the sera of CT subjects, Luminal A BC patients and triple negative BC patients | Distinguishing BC patients from CT subjects | [30] |
| ↑ miR-202; ↑ miR-21; ↑ miR-155; ↑ miR-23a; ↑ miR-130a; ↑ miR-145; ↑ miR-425-5p; ↑ miR-139-5p. | Plasma | BC patients from South Korea and Lebanese BC patients | Differential expression of circulating miRNAs in different ethnic groups | [31,32] |
| ↑ miR-133a-3p; ↑ miR-497-5p; ↑ mir-24-3p; ↑ miR-125b-5p; ↓ miR-377-3p; ↓ miR-374c-5p; ↓ miR-324-5p; ↓miR-19b-3p. | Serum | Caucasian and Asian BC patients and healthy controls | Distinguishing between BC patients and healthy individuals | [33] |
| ↑ miR-155. | Plasma | BC patients at diagnosis and after treatment | Potential role in diagnosis and therapeutic monitoring | [24] |
| ↑ miR-21; ↑ miR-181a; ↑ miR-10b; ↓ miR-145; ↓ let-7a. | Plasma | Locally advanced BC patients at diagnosis, during treatment, and after tumor restriction | Differential expression of miRNAs in BC patients with respect to HCs. Expression levels of the miRNAs returned to control values once the treatment finished. | [34] |
| ↑ miR-21; ↑ miR-55; ↑miR-10b; ↓ let-7a. | Plasma | Non-metastatic Luminal A patients undergoing the common treatments, such as surgery, chemotherapy, and radiotherapy | Treatments reversed the expression patterns of miRNAs | [35] |
| ↑ Let-7a; ↓ miR-145. | Whole blood | Luminal B and HER2+ BCs | Predicting the response to NAC in BC patients | [36] |
| ↑ miR-145. | Whole blood | Patients undergoing neoadjuvant chemotherapy across eight Irish centers. | Improved recurrence-free survival | [37] |
| ↕ miR-718; ↕ miR-4516; ↕ miR-210; ↕ miR-125b-5p. | Plasma | Luminal B-HER2-negative patients undergoing NAC | Association with chemosensitivity in Luminal B-HER2-negative patients undergoing NAC | [38] |
| ↕ miR-222, ↕ miR-20a; ↕ mir-451. | Plasma | HR+/HER2+ BC patients | Association with the chemosensitivity in a cohort of HR+/HER2+ BC patients | [39] |
| ↑ miR-148a-3p; ↑miR-374a-5p. | Plasma | HER2-positive BC patients receiving trastuzumab-based neoadjuvant therapy (NeoALLTTO trial) | Prognostic significance in identifying patients likely to respond to therapy | [40] |
| ↓ miR-200a; ↓ miR-200b; ↓ miR-141. | Plasma | BC patients receiving a complete cycle of systemic therapy | These values returned at the basal level upon the progression of disease, suggesting the potential effectiveness of these miRNAs in the management of metastatic BC | [43,44] |
| ↑ miR-21; ↑ miR-23b; ↑ miR-200c; ↓mir-190. | Plasma | BC patients before adjuvant chemotherapy | Efficacy as biomarkers for BC recurrence | [41] |
| ↑ miR-199a; ↓ miR-633b. | Plasma | Metastatic BC patients and healthy subjects | Correlation with chemoresistance in metastatic BC subjects | [45] |
| Lung Cancer | ||||
|---|---|---|---|---|
| miRNA | Source | Population | Function | Reference |
| ↓ miR-590-5p. | Plasma | 80 NSCLC patients compared to healthy controls | Association between low levels of miR-590-5p and poor prognosis, in terms of median survival | [58] |
| ↑ mir-2114; ↑mir-449c; ↑mir-2115. | Plasma | Lung adenocarcinoma and squamous cell carcinoma cases, compared with those from healthy individuals | Increased expression levels of mir-2114 and mir-449c in AC and mir-2115 in SCC; potential diagnostic significance | [59] |
| ↕ miR-1285-3p; ↕ miR-205-5p; ↕ miR-1260a; ↕ miR-1260b; ↕ miR-3152-3p; ↕ miR-378b; ↕ miR-17-3p; ↕ miR-1202; ↕ miR-139-5p; ↕ miR-16-2-3p; ↕ miR-18a-3p; ↕ miR-23b-3p; ↕ miR-3907; ↕ miR-551b-3p; ↕ miR-93-3p. | Whole blood | LC patients (both NSCLC and SCLC), patients with non-tumor lung diseases, patients with no pulmonary diseases, and unaffected control participants. | miRNA signature was used to discriminate between LC-diagnosed subject and all other individuals | [60] |
| ↕ let-7g-3p; ↕ miR-1202; ↕ miR-1285-3p; ↕ miR-17- 3p; ↕ miR-193a-5p; ↕ miR-205-5p; ↕ miR-21-3p; ↕ miR-3610; ↕ miR-4282; ↕ miR-4286; ↕ miR-452-3p; ↕ miR- 516a-3p; ↕ miR-572; ↕ miR-625-5p. | miRNA signature was used to discriminate between LC and non-tumor lung diseases | |||
| ↕ miR-1260a; ↕ miR-1260b; ↕ miR-1285-3p; ↕ miR-17-3p; ↕ miR-205-5p; ↕ miR-3152-3p; ↕ miR-374b-5p; ↕ miR-378b; ↕miR-564. | miRNA signature was used to discriminate between early-stage LC patients vs. individuals without LC | |||
| ↑ miR-21; ↓ miR-638; ↓ miR148; ↓ miR-152. | Whole blood | NSCLC patients and non-cancerous subjects | Diagnostic efficacy in distinguishing between LC patients and non-cancerous subjects | [61] |
| ↓ let-7a-5p; ↓ miR375; ↑ miR-1-3p; ↑miR-1291; ↑ miR-241-3p. | Serum | NSCLC patients and matched controls, including smokers and nonsmokers, male and female | miRNA signature was used for the identification of early-stage NSCLC | [62] |
| ↑ miR-191. | Serum | NSCLC patients and controls | Upregulation of miRNA in cancerous vs. non-cancerous tissues and its role in sustaining the proliferation and migration of LC cells in hypoxic conditions | [63,64] |
| ↕ let-7-5p; ↕ miR-184; ↕ miR-22-3p. | Plasma | NSCLC patients and high-risk subjects | Let-7-5p, miR-184 from Evs, and miR-22-3p from c-miRNAs were able to discriminate between the two groups. | [65] |
| ↑ miR-378. | Serum | NSCLC patients, subjects with a non-malignant disease, and healthy controls | The expression levels of exosomal miR-378 increased in cancerous sera compared to heathy sera; this increase was also associated with lymph node metastasis and the TNM stage | [66] |
| ↓ miR-200c-3p; ↓ miR-21-5p; ↓ miR-28-5p. | Plasma | Small cohort of advanced NSCLC patients treated with a single-agent anti–PD-1 or an anti–PD-L1 antibody | Expression levels of miRNAs significantly decreased in responders when compared to non-responder NSCLC patients | [69] |
| ↑ miR-93; ↑ miR-138-5p; ↑ miR-200; ↑ miR-27a; ↑ miR-424; ↑ miR-34a; ↑ miR-28; ↑ miR-106b; ↑ miR-193a-3p; ↑ miR-181a. | Serum | NSCLC patients undergoing immunotherapy and divided into responder and non-responder subjects | 10-fold increase in the expression levels of miRNAs from pre- to post-treatment; the highly expressed signature was further associated with the improvement of progression-free survival | [70] |
| ↕ miR-215-5p; ↕ miR-411-3p; ↕ miR- 493-5p; ↕ miR-494-3p; ↕ miR-495-3p; ↕ miR-548j-5p; ↕ miR-93-3p. | Serum | NSCLC patients treated with nivolumab | Association with OS after treatment with the immune check point inhibitor nivolumab | [71] |
| ↑ miR-202. | Plasma | NSCLC patients treated with first-line platinum-based chemotherapy. | Correlation with disease progression in NCSLC patients and a prognostic significance for shorter progression-free survival and OS | [72] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siniscalco, D.; Galderisi, U.; Peluso, G.; Finicelli, M. Circulating microRNAs in Cancer: A 5-Year Update with a Focus on Breast and Lung Cancers. Int. J. Mol. Sci. 2024, 25, 3140. https://doi.org/10.3390/ijms25063140
Siniscalco D, Galderisi U, Peluso G, Finicelli M. Circulating microRNAs in Cancer: A 5-Year Update with a Focus on Breast and Lung Cancers. International Journal of Molecular Sciences. 2024; 25(6):3140. https://doi.org/10.3390/ijms25063140
Chicago/Turabian StyleSiniscalco, Dario, Umberto Galderisi, Gianfranco Peluso, and Mauro Finicelli. 2024. "Circulating microRNAs in Cancer: A 5-Year Update with a Focus on Breast and Lung Cancers" International Journal of Molecular Sciences 25, no. 6: 3140. https://doi.org/10.3390/ijms25063140
APA StyleSiniscalco, D., Galderisi, U., Peluso, G., & Finicelli, M. (2024). Circulating microRNAs in Cancer: A 5-Year Update with a Focus on Breast and Lung Cancers. International Journal of Molecular Sciences, 25(6), 3140. https://doi.org/10.3390/ijms25063140








