GnRH Induces Citrullination of the Cytoskeleton in Murine Gonadotrope Cells
Abstract
1. Introduction
2. Results
2.1. GnRHa Stimulation of LβT2 Cells Rapidly Induces Protein Citrullination
2.2. Cytoskeletal Proteins Are Temporally Citrullinated following GnRHa Stimulation of LβT2 Cells
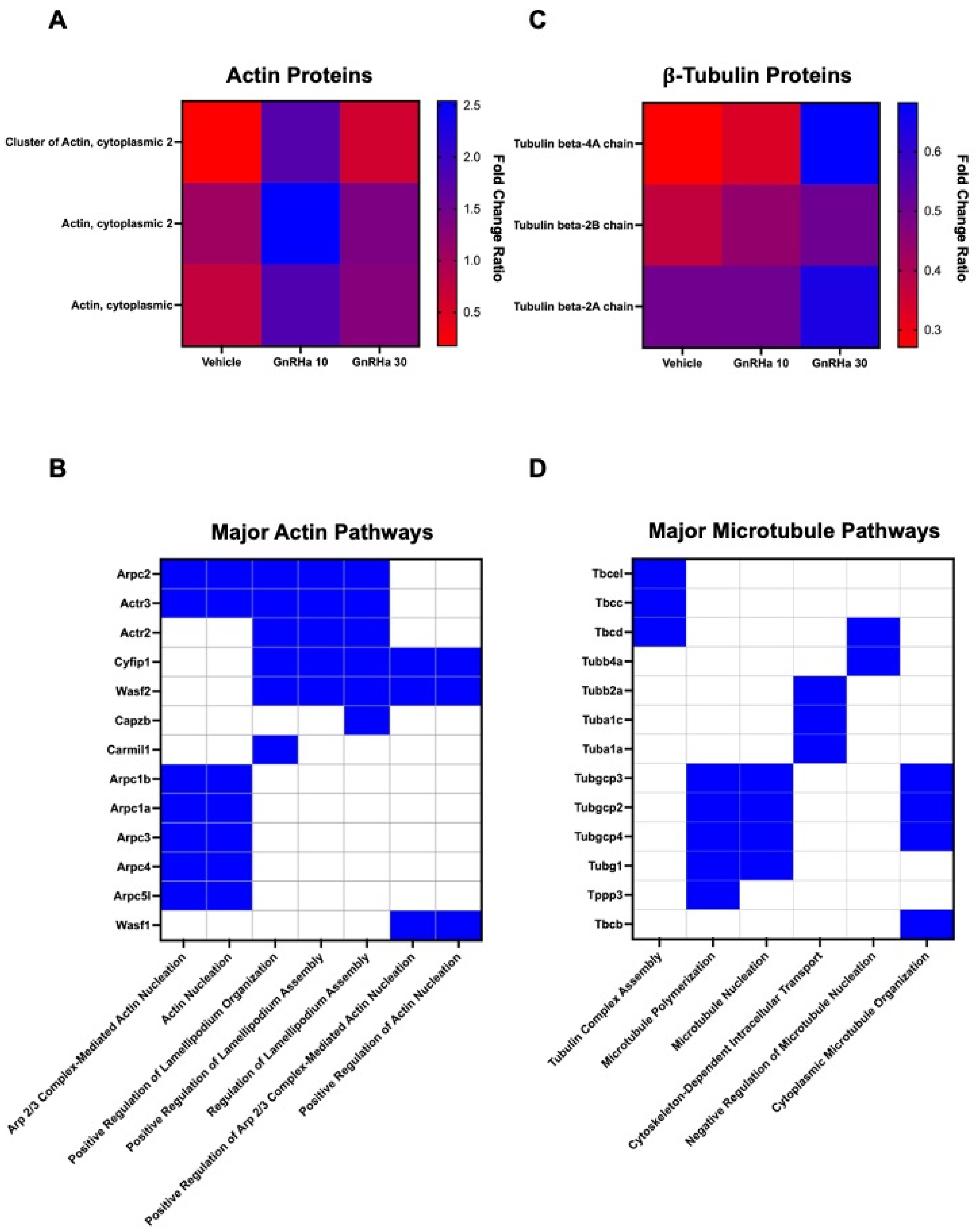
2.3. GnRHa Stimulates the Citrullination of β-Actin to Alter Gonadotrope Cytoskeletal Architecture
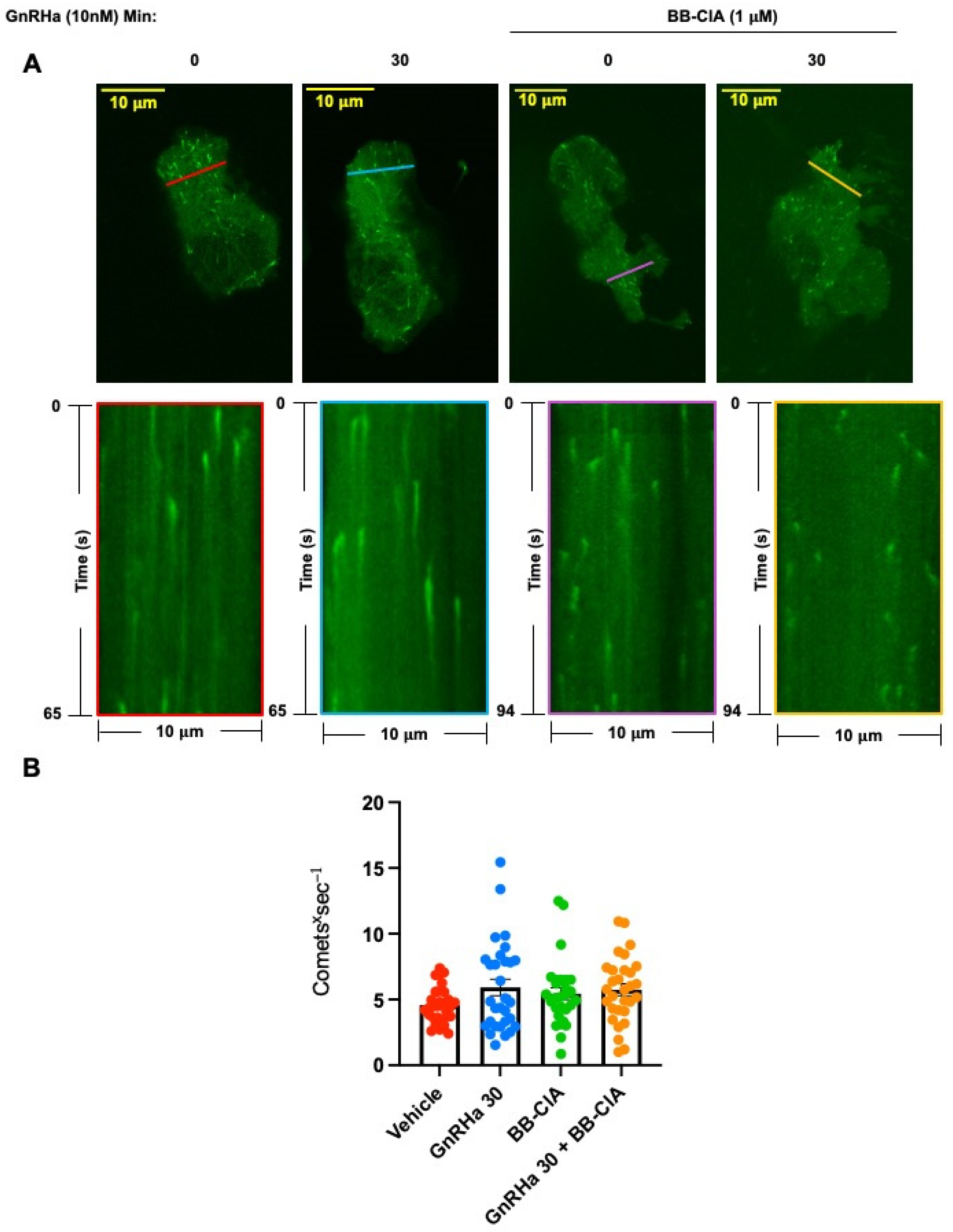
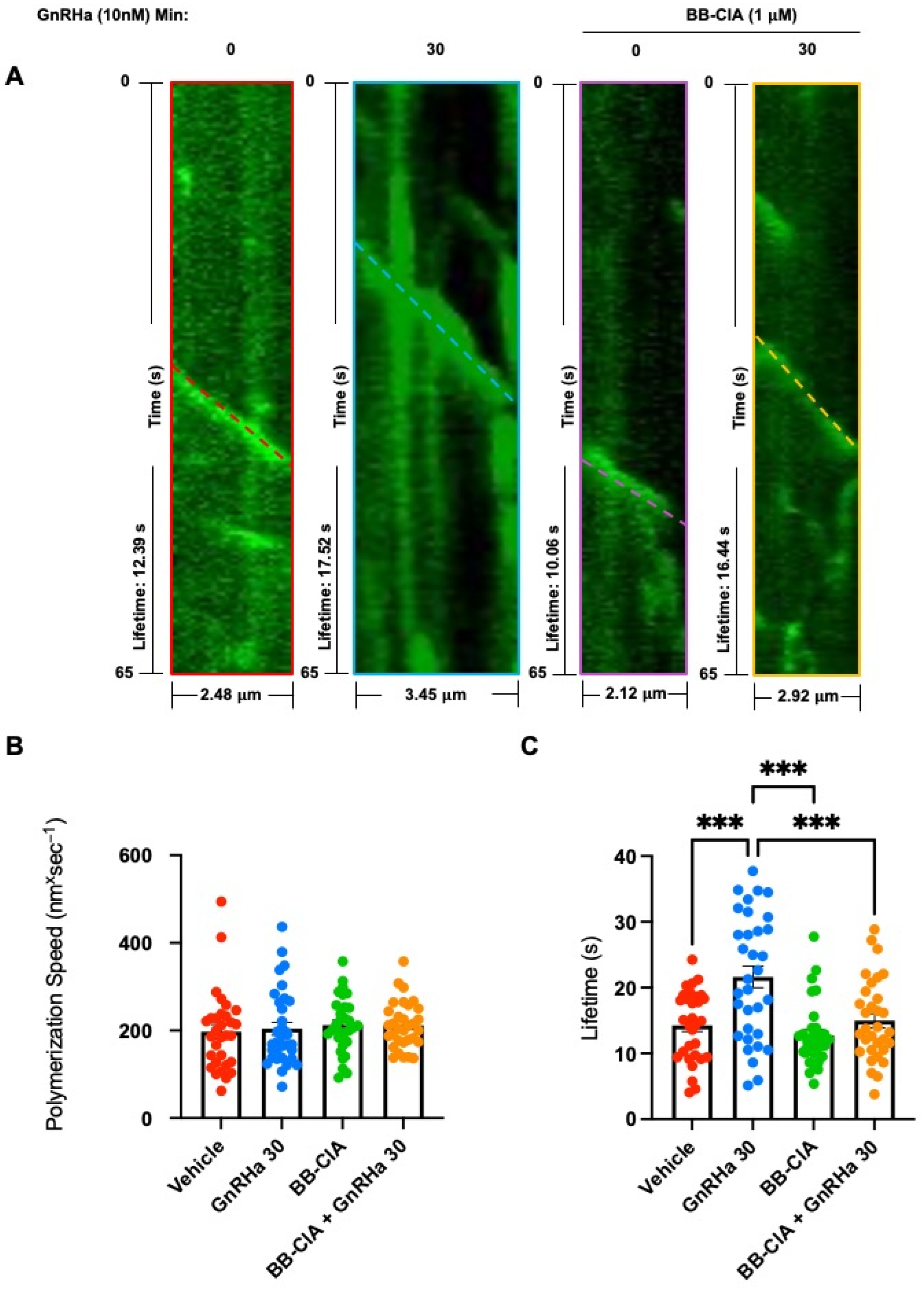
2.4. GnRHa-Induced Citrullination of β-Tubulin Increases MT Lifetimes but Has No Effect on Polymerization Speed or Nucleation Rate
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Citrullinated Protein Purification Using Biotin-Phenylglyoxal (PG) Immunoprecipitation
4.4. Filter-Aided Sample Preparation and Orbitrap Eclipse Analysis
Proteomics Data Analysis
4.5. Western Blots
4.6. Mouse Pituitary Cultures
4.7. LβT2 and Primary Gonadotrope Immunofluorescence
4.8. EB1-GFP Live Cell Imaging
4.8.1. Measurement of EB1-GFP Flux
4.8.2. Measurement of MT Polymerization Speeds and Lifetimes
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vossenaar, E.R.; Zendman, A.J.; van Venrooij, W.J.; Pruijn, G.J. PAD, a growing family of citrullinating enzymes: Genes, features and involvement in disease. Bioessays 2003, 25, 1106–1118. [Google Scholar] [CrossRef]
- Terakawa, H.; Takahara, H.; Sugawara, K. Three types of mouse peptidylarginine deiminase: Characterization and tissue distribution. J. Biochem. 1991, 110, 661–666. [Google Scholar] [CrossRef]
- Watanabe, K.; Akiyama, K.; Hikichi, K.; Ohtsuka, R.; Okuyama, A.; Senshu, T. Combined biochemical and immunochemical comparison of peptidylarginine deiminases present in various tissues. Biochim. Biophys. Acta 1988, 966, 375–383. [Google Scholar] [CrossRef]
- Williams, J.P.C.; Walport, L.J. PADI6: What we know about the elusive fifth member of the peptidyl arginine deiminase family. Philos. Trans. R. Soc. B 2023, 378, 20220242. [Google Scholar] [CrossRef]
- Raijmakers, R.; Zendman, A.J.; Egberts, W.V.; Vossenaar, E.R.; Raats, J.; Soede-Huijbregts, C.; Rutjes, F.P.; van Veelen, P.A.; Drijfhout, J.W.; Pruijn, G.J. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J. Mol. Biol. 2007, 367, 1118–1129. [Google Scholar] [CrossRef]
- Christensen, A.O.; Li, G.; Young, C.H.; Snow, B.; Khan, S.A.; DeVore, S.B.; Edwards, S.; Bouma, G.J.; Navratil, A.M.; Cherrington, B.D.; et al. Peptidylarginine deiminase (PAD) enzymes and Citrullinated proteins in female reproductive physiology and associated diseases. Biol. Reprod. 2022, 107, 1395–1410. [Google Scholar] [CrossRef]
- Takahara, H.; Tsuchida, M.; Kusubata, M.; Akutsu, K.; Tagami, S.; Sugawara, K. Peptidylarginine deiminase of the mouse. Distribution, properties, and immunocytochemical localization. J. Biol. Chem. 1989, 264, 13361–13368. [Google Scholar] [CrossRef]
- Takahara, H.; Kusubata, M.; Tsuchida, M.; Kohsaka, T.; Tagami, S.; Sugawara, K. Expression of peptidylarginine deiminase in the uterine epithelial cells of mouse is dependent on estrogen. J. Biol. Chem. 1992, 267, 520–525. [Google Scholar] [CrossRef]
- Khan, S.A.; Edwards, B.S.; Muth, A.; Thompson, P.R.; Cherrington, B.D.; Navratil, A.M. GnRH Stimulates Peptidylarginine Deiminase Catalyzed Histone Citrullination in Gonadotrope Cells. Mol. Endocrinol. 2016, 30, 1081–1091. [Google Scholar] [CrossRef]
- Jiang, Z.; Cui, Y.; Wang, L.; Zhao, Y.; Yan, S.; Chang, X. Investigating citrullinated proteins in tumour cell lines. World J. Surg. Oncol. 2013, 11, 260. [Google Scholar] [CrossRef]
- Kholia, S.; Jorfi, S.; Thompson, P.R.; Causey, C.P.; Nicholas, A.P.; Inal, J.M.; Lange, S. A novel role for peptidylarginine deiminases in microvesicle release reveals therapeutic potential of PAD inhibition in sensitizing prostate cancer cells to chemotherapy. J. Extracell. Vesicles 2015, 4, 26192. [Google Scholar] [CrossRef]
- Chang, X.; Zhao, Y.; Wang, Y.; Chen, Y.; Yan, X. Screening citrullinated proteins in synovial tissues of rheumatoid arthritis using 2-dimensional western blotting. J. Rheumatol. 2013, 40, 219–227. [Google Scholar] [CrossRef]
- Senshu, T.; Kan, S.; Ogawa, H.; Manabe, M.; Asaga, H. Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochem. Biophys. Res. Commun. 1996, 225, 712–719. [Google Scholar] [CrossRef]
- Inagaki, M.; Takahara, H.; Nishi, Y.; Sugawara, K.; Sato, C. Ca2+-dependent deimination-induced disassembly of intermediate filaments involves specific modification of the amino-terminal head domain. J. Biol. Chem 1989, 264, 18119–18127. [Google Scholar] [CrossRef]
- Chang, X.; Jian, X.; Yan, X. Expression and citrullination of keratin in synovial tissue of rheumatoid arthritis. Rheumatol. Int. 2009, 29, 1337–1342. [Google Scholar] [CrossRef]
- Tilvawala, R.; Nguyen, S.H.; Maurais, A.J.; Nemmara, V.V.; Nagar, M.; Salinger, A.J.; Nagpal, S.; Weerapana, E.; Thompson, P.R. The Rheumatoid Arthritis-Associated Citrullinome. Cell Chem. Biol. 2018, 25, 691–704.e696. [Google Scholar] [CrossRef]
- Izumi, T.; Kasai, K.; Gomi, H. Secretory vesicle docking to the plasma membrane: Molecular mechanism and functional significance. Diabetes Obes. Metab. 2007, 9 (Suppl. S2), 109–117. [Google Scholar] [CrossRef]
- Porat-Shliom, N.; Milberg, O.; Masedunskas, A.; Weigert, R. Multiple roles for the actin cytoskeleton during regulated exocytosis. Cell Mol. Life Sci. 2013, 70, 2099–2121. [Google Scholar] [CrossRef]
- Wloga, D.; Joachimiak, E.; Fabczak, H. Tubulin Post-Translational Modifications and Microtubule Dynamics. Int. J. Mol. Sci. 2017, 18, 2207. [Google Scholar] [CrossRef]
- Gadadhar, S.; Bodakuntla, S.; Natarajan, K.; Janke, C. The tubulin code at a glance. J. Cell Sci. 2017, 130, 1347–1353. [Google Scholar] [CrossRef]
- Yu, I.; Garnham, C.P.; Roll-Mecak, A. Writing and Reading the Tubulin Code. J. Biol. Chem. 2015, 290, 17163–17172. [Google Scholar] [CrossRef]
- Navratil, A.M.; Dozier, M.G.; Whitesell, J.D.; Clay, C.M.; Roberson, M.S. Role of cortactin in dynamic actin remodeling events in gonadotrope cells. Endocrinology 2014, 155, 548–557. [Google Scholar] [CrossRef]
- Khar, A.; Kunert-Radek, J.; Jutisz, M. Involvement of microtubule and microfilament system in the GnRH-induced release of gonadotropins by rat anterior pituitary cells in culture. FEBS Lett. 1979, 104, 410–414. [Google Scholar] [CrossRef]
- Adams, T.E.; Nett, T.M. Interaction of GnRH with anterior pituitary. III. Role of divalent cations, microtubules and microfilaments in the GnRH activated gonadotroph. Biol. Reprod. 1979, 21, 1073–1086. [Google Scholar] [CrossRef]
- Clay, C.M.; Cherrington, B.D.; Navratil, A.M. Plasticity of Anterior Pituitary Gonadotrope Cells Facilitates the Pre-Ovulatory LH Surge. Front. Endocrinol. 2020, 11, 616053. [Google Scholar] [CrossRef]
- Lewallen, D.M.; Bicker, K.L.; Subramanian, V.; Clancy, K.W.; Slade, D.J.; Martell, J.; Dreyton, C.J.; Sokolove, J.; Weerapana, E.; Thompson, P.R. Chemical Proteomic Platform To Identify Citrullinated Proteins. ACS Chem. Biol. 2015, 10, 2520–2528. [Google Scholar] [CrossRef]
- Li, G.; Young, C.H.; Snow, B.; Christensen, A.O.; Demoruelle, M.K.; Nemmara, V.V.; Thompson, P.R.; Rothfuss, H.M.; Cherrington, B.D. Identification and Characterization of the Lactating Mouse Mammary Gland Citrullinome. Int. J. Mol. Sci. 2020, 21, 2634. [Google Scholar] [CrossRef]
- Bliss, S.P.; Navratil, A.M.; Xie, J.; Roberson, M.S. GnRH signaling, the gonadotrope and endocrine control of fertility. Front. Neuroendocrinol. 2010, 31, 322–340. [Google Scholar] [CrossRef]
- Roberson, M.S.; Misra-Press, A.; Laurance, M.E.; Stork, P.J.; Maurer, R.A. A role for mitogen-activated protein kinase in mediating activation of the glycoprotein hormone alpha-subunit promoter by gonadotropin-releasing hormone. Mol. Cell. Biol. 1995, 15, 3531–3539. [Google Scholar] [CrossRef]
- Kanasaki, H.; Bedecarrats, G.Y.; Kam, K.Y.; Xu, S.; Kaiser, U.B. Gonadotropin-releasing hormone pulse frequency-dependent activation of extracellular signal-regulated kinase pathways in perifused LβT2 cells. Endocrinology 2005, 146, 5503–5513. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Keely, P.J. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J. Cell Sci. 2011, 124, 1195–1205. [Google Scholar] [CrossRef]
- Heng, Y.W.; Koh, C.G. Actin cytoskeleton dynamics and the cell division cycle. Int. J. Biochem. Cell. Biol. 2010, 42, 1622–1633. [Google Scholar] [CrossRef]
- McNally, F.J. Modulation of microtubule dynamics during the cell cycle. Curr. Opin. Cell. Biol. 1996, 8, 23–29. [Google Scholar] [CrossRef]
- Navratil, A.M.; Knoll, J.G.; Whitesell, J.D.; Tobet, S.A.; Clay, C.M. Neuroendocrine plasticity in the anterior pituitary: Gonadotropin-releasing hormone-mediated movement in vitro and in vivo. Endocrinology 2007, 148, 1736–1744. [Google Scholar] [CrossRef]
- Matov, A.; Applegate, K.; Kumar, P.; Thoma, C.; Krek, W.; Danuser, G.; Wittmann, T. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods 2010, 7, 761–768. [Google Scholar] [CrossRef]
- Nehlig, A.; Molina, A.; Rodrigues-Ferreira, S.; Honore, S.; Nahmias, C. Regulation of end-binding protein EB1 in the control of microtubule dynamics. Cell. Mol. Life Sci. 2017, 74, 2381–2393. [Google Scholar] [CrossRef]
- Jiang, Y.Y.; Lechtreck, K.; Gaertig, J. Total internal reflection fluorescence microscopy of intraflagellar transport in Tetrahymena thermophila. Methods Cell. Biol. 2015, 127, 445–456. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Hamilton, R.S.; Pauli, A.; Davis, I.; Nasmyth, K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell. Biol. 2010, 12, 185–192. [Google Scholar] [CrossRef]
- Kim, C.; Choi, H.; Jung, E.S.; Lee, W.; Oh, S.; Jeon, N.L.; Mook-Jung, I. HDAC6 inhibitor blocks amyloid beta-induced impairment of mitochondrial transport in hippocampal neurons. PLoS ONE 2012, 7, e42983. [Google Scholar] [CrossRef]
- Akiyama, K.; Nagata, S.; Tanaka, S.; Inoue, K.; Watanabe, K.; Senshu, T. Search for functional significance of peptidylarginine deiminase in rat pituitaries: Variation during pregnancy and ultrastructural localization in prolactin cells. Cell. Biol. Int. 1993, 17, 487–494. [Google Scholar] [CrossRef]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Marshall, K.A.; et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009, 37, D885–D890. [Google Scholar] [CrossRef]
- Filant, J.; Spencer, T.E. Uterine glands: Biological roles in conceptus implantation, uterine receptivity and decidualization. Int. J. Dev. Biol. 2014, 58, 107–116. [Google Scholar] [CrossRef]
- Badillo-Soto, M.A.; Rodriguez-Rodriguez, M.; Perez-Perez, M.E.; Daza-Benitez, L.; Bollain, Y.G.J.J.; Carrillo-Jimenez, M.A.; Avalos-Diaz, E.; Herrera-Esparza, R. Potential protein targets of the peptidylarginine deiminase 2 and peptidylarginine deiminase 4 enzymes in rheumatoid synovial tissue and its possible meaning. Eur. J. Rheumatol. 2016, 3, 44–49. [Google Scholar] [CrossRef]
- van Beers, J.J.; Schwarte, C.M.; Stammen-Vogelzangs, J.; Oosterink, E.; Bozic, B.; Pruijn, G.J. The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein E, myeloid nuclear differentiation antigen, and beta-actin. Arthritis Rheum. 2013, 65, 69–80. [Google Scholar] [CrossRef]
- Gallart-Palau, X.; Serra, A.; Lee, B.S.T.; Guo, X.; Sze, S.K. Brain ureido degenerative protein modifications are associated with neuroinflammation and proteinopathy in Alzheimer’s disease with cerebrovascular disease. J. Neuroinflammation 2017, 14, 175. [Google Scholar] [CrossRef]
- Xu, M.; Du, R.; Xing, W.; Chen, X.; Wan, J.; Wang, S.; Xiong, L.; Nandakumar, K.S.; Holmdahl, R.; Geng, H. Platelets derived citrullinated proteins and microparticles are potential autoantibodies ACPA targets in RA patients. Front. Immunol. 2023, 14, 1084283. [Google Scholar] [CrossRef]
- Fert-Bober, J.; Giles, J.T.; Holewinski, R.J.; Kirk, J.A.; Uhrigshardt, H.; Crowgey, E.L.; Andrade, F.; Bingham, C.O., 3rd; Park, J.K.; Halushka, M.K.; et al. Citrullination of myofilament proteins in heart failure. Cardiovasc. Res. 2015, 108, 232–242. [Google Scholar] [CrossRef]
- Janke, C.; Montagnac, G. Causes and Consequences of Microtubule Acetylation. Curr. Biol. 2017, 27, R1287–R1292. [Google Scholar] [CrossRef]
- Reed, N.A.; Cai, D.; Blasius, T.L.; Jih, G.T.; Meyhofer, E.; Gaertig, J.; Verhey, K.J. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 2006, 16, 2166–2172. [Google Scholar] [CrossRef]
- Portran, D.; Schaedel, L.; Xu, Z.; Thery, M.; Nachury, M.V. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat. Cell. Biol. 2017, 19, 391–398. [Google Scholar] [CrossRef]
- Bhuwania, R.; Castro-Castro, A.; Linder, S. Microtubule acetylation regulates dynamics of KIF1C-powered vesicles and contact of microtubule plus ends with podosomes. Eur. J. Cell. Biol. 2014, 93, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quigley, E.B.; DeVore, S.B.; Khan, S.A.; Geisterfer, Z.M.; Rothfuss, H.M.; Sequoia, A.O.; Thompson, P.R.; Gatlin, J.C.; Cherrington, B.D.; Navratil, A.M. GnRH Induces Citrullination of the Cytoskeleton in Murine Gonadotrope Cells. Int. J. Mol. Sci. 2024, 25, 3181. https://doi.org/10.3390/ijms25063181
Quigley EB, DeVore SB, Khan SA, Geisterfer ZM, Rothfuss HM, Sequoia AO, Thompson PR, Gatlin JC, Cherrington BD, Navratil AM. GnRH Induces Citrullination of the Cytoskeleton in Murine Gonadotrope Cells. International Journal of Molecular Sciences. 2024; 25(6):3181. https://doi.org/10.3390/ijms25063181
Chicago/Turabian StyleQuigley, Elizabeth B., Stanley B. DeVore, Shaihla A. Khan, Zachary M. Geisterfer, Heather M. Rothfuss, Ari O. Sequoia, Paul R. Thompson, Jesse C. Gatlin, Brian D. Cherrington, and Amy M. Navratil. 2024. "GnRH Induces Citrullination of the Cytoskeleton in Murine Gonadotrope Cells" International Journal of Molecular Sciences 25, no. 6: 3181. https://doi.org/10.3390/ijms25063181
APA StyleQuigley, E. B., DeVore, S. B., Khan, S. A., Geisterfer, Z. M., Rothfuss, H. M., Sequoia, A. O., Thompson, P. R., Gatlin, J. C., Cherrington, B. D., & Navratil, A. M. (2024). GnRH Induces Citrullination of the Cytoskeleton in Murine Gonadotrope Cells. International Journal of Molecular Sciences, 25(6), 3181. https://doi.org/10.3390/ijms25063181




