Quantitative Proteomics Reveal That CB2R Agonist JWH-133 Downregulates NF-κB Activation, Oxidative Stress, and Lysosomal Exocytosis from HIV-Infected Macrophages
Abstract
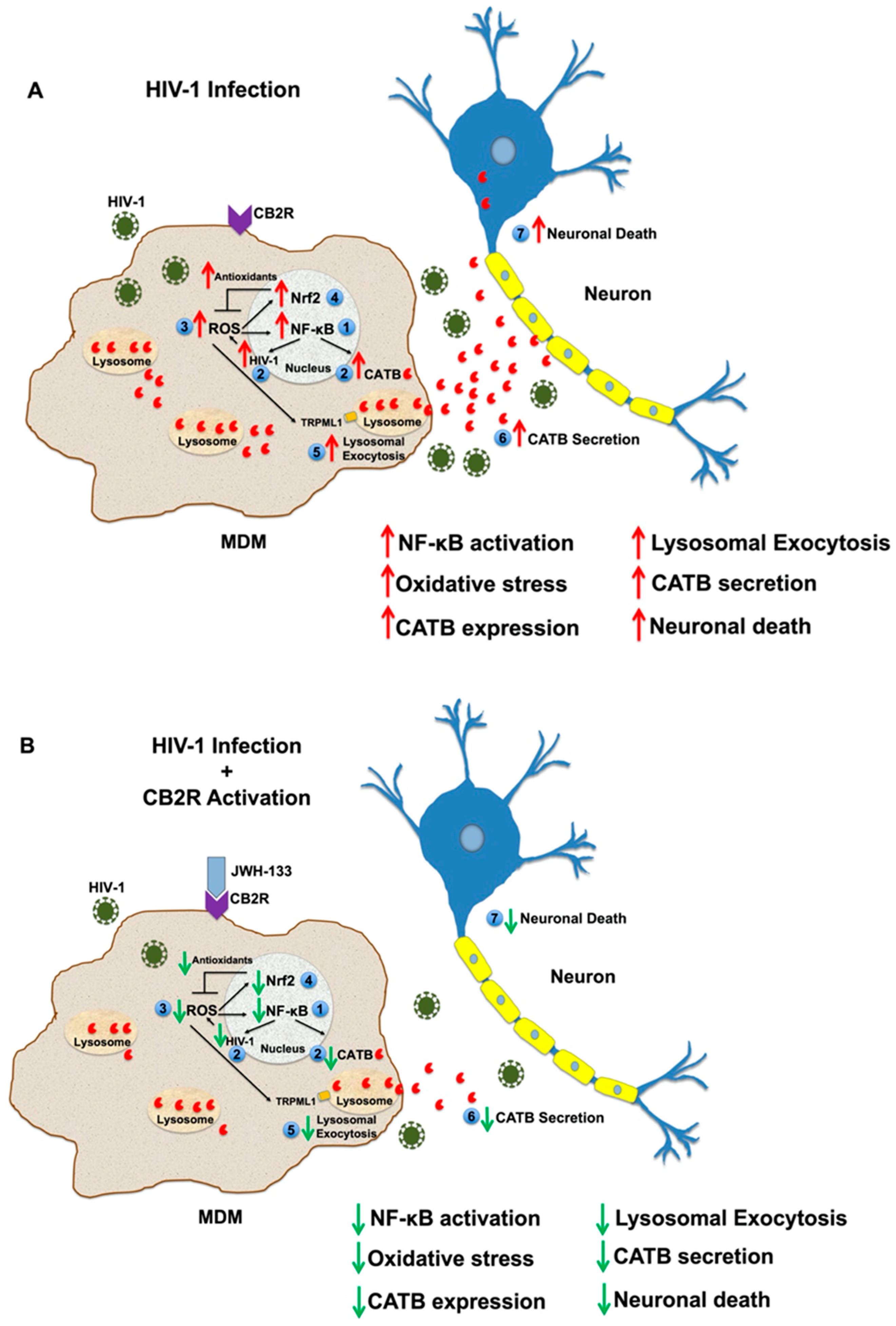
:1. Introduction
2. Results
2.1. JWH-133 Inhibits HIV-Induced Activation of Nrf2-Mediated Oxidative Stress Response and NF-κB Signaling
2.2. JWH-133 Decreases the Expression of Proteins Associated with Lysosomal Exocytosis
2.3. JWH-133 Decreases the Expression of CATB and Associated Proteins
3. Discussion
4. Materials and Methods
4.1. Isolation of Macrophages, HIV-1ADA Infection, and Treatments with JWH-133 Agonist
4.2. Total CATB ELISA
4.3. Preparation of MDM Lysates for TMT Labeling
4.4. TMT Labeling
4.5. Mass Spectrometry Analyses
4.6. Protein Identification and Quantitative Analyses
4.7. Bioinformatics and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saylor, D.; Dickens, A.M.; Sacktor, N.; Haughey, N.; Slusher, B.; Pletnikov, M.; Mankowski, J.L.; Brown, A.; Volsky, D.J.; McArthur, J.C. HIV-Associated Neurocognitive Disorder—Pathogenesis and Prospects for Treatment. Nat. Rev. Neurol. 2016, 12, 234–248. [Google Scholar] [CrossRef]
- Williams, D.; Veenstra, M.; Gaskill, P.; Morgello, S.; Calderon, T.; Berman, J. Monocytes Mediate HIV Neuropathogenesis: Mechanisms That Contribute to HIV Associated Neurocognitive Disorders. Curr. HIV Res. 2014, 12, 85–96. [Google Scholar] [CrossRef]
- Hong, S.; Banks, W.A. Role of the Immune System in HIV-Associated Neuroinflammation and Neurocognitive Implications. Brain Behav. Immun. 2015, 45, 1–12. [Google Scholar] [CrossRef]
- Rappaport, J.; Volsky, D.J. Role of the Macrophage in HIV-Associated Neurocognitive Disorders and Other Comorbidities in Patients on Effective Antiretroviral Treatment. J. Neurovirol. 2015, 21, 235. [Google Scholar] [CrossRef]
- Subra, C.; Trautmann, L. Role of T Lymphocytes in HIV Neuropathogenesis. Curr. HIV/AIDS Rep. 2019, 16, 236. [Google Scholar] [CrossRef]
- Wahl, A.; Al-Harthi, L. HIV Infection of Non-Classical Cells in the Brain. Retrovirology 2023, 20, 1. [Google Scholar] [CrossRef]
- Rodriguez-Franco, E.J.; Cantres-Rosario, Y.M.; Plaud-Valentin, M.; Romeu, R.; Rodríguez, Y.; Skolasky, R.; Meléndez, V.; Cadilla, C.L.; Melendez, L.M. Dysregulation of Macrophage-Secreted Cathepsin B Contributes to HIV-1-Linked Neuronal Apoptosis. PLoS ONE 2012, 7, e36571. [Google Scholar] [CrossRef]
- Cantres-Rosario, Y.; Plaud-Valentín, M.; Gerena, Y.; Skolasky, R.L.; Wojna, V.; Meléndez, L.M. Cathepsin B and Cystatin B in HIV-Seropositive Women Are Associated with Infection and HIV-1-Associated Neurocognitive Disorders. AIDS 2013, 27, 347–356. [Google Scholar] [CrossRef]
- Cantres-Rosario, Y.M.; Hernandez, N.; Negron, K.; Perez-Laspiur, J.; Leszyk, J.; Shaffer, S.A.; Meléndez, L.M. Interacting Partners of Macrophage-Secreted Cathepsin B Contribute to HIV-Induced Neuronal Apoptosis. AIDS 2015, 29, 2081–2092. [Google Scholar] [CrossRef]
- Rosario-Rodríguez, L.J.; Colón, K.; Borges-Vélez, G.; Negrón, K.; Meléndez, L.M. Dimethyl Fumarate Prevents HIV-Induced Lysosomal Dysfunction and Cathepsin B Release from Macrophages. J. Neuroimmune Pharmacol. 2018, 13, 345–354. [Google Scholar] [CrossRef]
- López, O.V.; Gorantla, S.; Segarra, A.C.; Andino Norat, M.C.; Álvarez, M.; Skolasky, R.L.; Meléndez, L.M. Sigma-1 Receptor Antagonist (BD1047) Decreases Cathepsin B Secretion in HIV-Infected Macrophages Exposed to Cocaine. J. Neuroimmune Pharmacol. 2019, 14, 226–240. [Google Scholar] [CrossRef]
- Zenón, F.; Segarra, A.C.; Gonzalez, M.; Meléndez, L.M. Cocaine Potentiates Cathepsin B Secretion and Neuronal Apoptosis from HIV-Infected Macrophages. J. Neuroimmune Pharmacol. 2014, 9, 703–715. [Google Scholar] [CrossRef]
- Zenón-Meléndez, C.N.; Carrasquillo Carrión, K.; Cantres Rosario, Y.; Roche Lima, A.; Meléndez, L.M. Inhibition of Cathepsin B and SAPC Secreted by HIV-Infected Macrophages Reverses Common and Unique Apoptosis Pathways. J. Proteome Res. 2022, 21, 301–312. [Google Scholar] [CrossRef]
- Rosario-Rodríguez, L.J.; Gerena, Y.; García-Requena, L.A.; Cartagena-Isern, L.J.; Cuadrado-Ruiz, J.C.; Borges-Vélez, G.; Meléndez, L.M. Cannabinoid Receptor Type 2 Agonist JWH-133 Decreases Cathepsin B Secretion and Neurotoxicity from HIV-Infected Macrophages. Sci. Rep. 2022, 12, 233. [Google Scholar] [CrossRef]
- Cantres-Rosario, Y.M.; Ortiz-Rodríguez, S.C.; Santos-Figueroa, A.G.; Plaud, M.; Negron, K.; Cotto, B.; Langford, D.; Melendez, L.M. HIV Infection Induces Extracellular Cathepsin B Uptake and Damage to Neurons. Sci. Rep. 2019, 9, 8006. [Google Scholar] [CrossRef] [PubMed]
- Hamer, I.; Delaive, E.; Dieu, M.; Abdel-Sater, F.; Mercy, L.; Jadot, M.; Arnould, T. Up-Regulation of Cathepsin B Expression and Enhanced Secretion in Mitochondrial DNA-Depleted Osteosarcoma Cells. Biol. Cell 2009, 101, 31–43. [Google Scholar] [CrossRef]
- Chae, H.J.; Ha, K.C.; Lee, G.Y.; Yang, S.K.; Yun, K.J.; Kim, E.C.; Kim, S.H.; Chae, S.W.; Kim, H.R. Interleukin-6 and Cyclic AMP Stimulate Release of Cathepsin B in Human Osteoblasts. Immunopharmacol. Immunotoxicol. 2007, 29, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Fiore, L.S.; Richards, D.L.; Yang, Y.; Liu, J.; Wang, C.; Plattner, R. Abl and Arg Mediate Cysteine Cathepsin Secretion to Facilitate Melanoma Invasion and Metastasis. Sci. Signal 2018, 11, 518. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Rivera, L.; Perez--Laspiur, J.; Colón, K.; Meléndez, L.M. Inhibition of Interferon Response by Cystatin B: Implication in HIV Replication of Macrophage Reservoirs. J. Neurovirol. 2012, 18, 20. [Google Scholar] [CrossRef]
- Rivera, L.E.; Colón, K.; Cantres-Rosario, Y.M.; Zenón, F.M.; Meléndez, L.M. Macrophage Derived Cystatin B/Cathepsin B in HIV Replication and Neuropathogenesis. Curr. HIV Res. 2014, 12, 111. [Google Scholar] [CrossRef] [PubMed]
- Rivera, L.E.; Kraiselburd, E.; Meléndez, L.M. Cystatin B and HIV Regulate the STAT-1 Signaling Circuit in HIV Infected and INF-β Treated Human Macrophages. J. Neurovirol. 2016, 22, 666. [Google Scholar] [CrossRef]
- Luciano-Montalvo, C.; Meléndez, L.M. Cystatin B Associates with Signal Transducer and Activator of Transcription 1 in Monocyte-Derived and Placental Macrophages. Placenta 2009, 30, 464. [Google Scholar] [CrossRef]
- Vomund, S.; Schäfer, A.; Parnham, M.J.; Brüne, B.; von Knethen, A. Nrf2, the Master Regulator of Anti-Oxidative Responses. Int. J. Mol. Sci. 2017, 18, 2772. [Google Scholar] [CrossRef]
- Shaw, P.; Chattopadhyay, A. Nrf2–ARE Signaling in Cellular Protection: Mechanism of Action and the Regulatory Mechanisms. J. Cell. Physiol. 2020, 235, 3119–3130. [Google Scholar] [CrossRef]
- Ngo, V.; Duennwald, M.L. Nrf2 and Oxidative Stress: A General Overview of Mechanisms and Implications in Human Disease. Antioxidants 2022, 11, 2345. [Google Scholar] [CrossRef] [PubMed]
- Gill, A.J.; Kovacsics, C.E.; Vance, P.J.; Collman, R.G.; Kolson, D.L. Induction of Heme Oxygenase-1 Deficiency and Associated Glutamate-Mediated Neurotoxicity Is a Highly Conserved HIV Phenotype of Chronic Macrophage Infection That Is Resistant to Antiretroviral Therapy. J. Virol. 2015, 89, 10656. [Google Scholar] [CrossRef]
- Staitieh, B.S.; Ding, L.; Neveu, W.A.; Spearman, P.; Guidot, D.M.; Fan, X. HIV-1 Decreases Nrf2/ARE Activity and Phagocytic Function in Alveolar Macrophages. J. Leukoc. Biol. 2017, 102, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Murray, S.C.; Staitieh, B.S.; Spearman, P.; Guidot, D.M. HIV Impairs Alveolar Macrophage Function via MicroRNA-144-Induced Suppression of Nrf2. Am. J. Med. Sci. 2021, 361, 90–97. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Ivanova, O.N.; Kochetkov, S.N.; Starodubova, E.S.; Bartosch, B.; Isaguliants, M.G. Oxidative Stress during HIV Infection: Mechanisms and Consequences. Oxid. Med. Cell. Longev. 2016, 2016, 8910396. [Google Scholar] [CrossRef]
- Ronaldson, P.T.; Bendayan, R. HIV-1 Viral Envelope Glycoprotein Gp120 Produces Oxidative Stress and Regulates the Functional Expression of Multidrug Resistance Protein-1 (Mrp1) in Glial Cells. J. Neurochem. 2008, 106, 1298–1313. [Google Scholar] [CrossRef] [PubMed]
- Isaguliants, M.; Smirnova, O.; Ivanov, A.V.; Kilpelainen, A.; Kuzmenko, Y.; Petkov, S.; Latanova, A.; Krotova, O.; Engström, G.; Karpov, V.; et al. Oxidative Stress Induced by HIV-1 Reverse Transcriptase Modulates the Enzyme’s Performance in Gene Immunization. Hum. Vaccines Immunother. 2013, 9, 2111–2119. [Google Scholar] [CrossRef]
- Fan, X.; Staitieh, B.S.; Jensen, J.S.; Mould, K.J.; Greenberg, J.A.; Joshi, P.C.; Koval, M.; Guidot, D.M. Activating the Nrf2-Mediated Antioxidant Response Element Restores Barrier Function in the Alveolar Epithelium of HIV-1 Transgenic Rats. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2013, 305, L267. [Google Scholar] [CrossRef]
- Mastrantonio, R.; Cervelli, M.; Pietropaoli, S.; Mariottini, P.; Colasanti, M.; Persichini, T. HIV-Tat Induces the Nrf2/ARE Pathway through NMDA Receptor-Elicited Spermine Oxidase Activation in Human Neuroblastoma Cells. PLoS ONE 2016, 11, e0149802. [Google Scholar] [CrossRef]
- Gill, A.J.; Kolson, D.L. Dimethyl Fumarate Modulation of Immune and Antioxidant Responses: Application to HIV Therapy. Crit. Rev. Immunol. 2013, 33, 307–359. [Google Scholar] [CrossRef]
- Ravi, S.; Peña, K.A.; Chu, C.T.; Kiselyov, K. Biphasic Regulation of Lysosomal Exocytosis by Oxidative Stress. Cell Calcium 2016, 60, 356–362. [Google Scholar] [CrossRef]
- He, X.; Li, X.; Tian, W.; Li, C.; Li, P.; Zhao, J.; Yang, S.; Li, S. The Role of Redox-Mediated Lysosomal Dysfunction and Therapeutic Strategies. Biomed. Pharmacother. 2023, 165, 115121. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gulbins, E.; Zhang, Y. Oxidative Stress Triggers Ca2+-Dependent Lysosome Trafficking and Activation of Acid Sphingomyelinase. Cell. Physiol. Biochem. 2012, 30, 815. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gu, Y.; Wen, R.; Shen, F.; Tian, H.L.; Yang, G.Y.; Zhang, Z. Lysosome Exocytosis Is Involved in Astrocyte ATP Release after Oxidative Stress Induced by H2O2. Neurosci. Lett. 2019, 705, 251–258. [Google Scholar] [CrossRef]
- Fan, Y.; He, J.J. HIV-1 Tat Promotes Lysosomal Exocytosis in Astrocytes and Contributes to Astrocyte-Mediated Tat Neurotoxicity. J. Biol. Chem. 2016, 291, 22830–22840. [Google Scholar] [CrossRef] [PubMed]
- Datta, G.; Miller, N.M.; Afghah, Z.; Geiger, J.D.; Chen, X. Hiv-1 Gp120 Promotes Lysosomal Exocytosis in Human Schwann Cells. Front. Cell. Neurosci. 2019, 13, 329. [Google Scholar] [CrossRef] [PubMed]
- Halcrow, P.W.; Lakpa, K.L.; Khan, N.; Afghah, Z.; Miller, N.; Datta, G.; Chen, X.; Geiger, J.D. HIV-1 Gp120-Induced Endolysosome de-Acidification Leads to Efflux of Endolysosome Iron, and Increases in Mitochondrial Iron and Reactive Oxygen Species. J. Neuroimmune Pharmacol. 2022, 17, 181. [Google Scholar] [CrossRef]
- Cinti, A.; le Sage, V.; Milev, M.P.; Valiente-Echeverría, F.; Crossie, C.; Miron, M.J.; Panté, N.; Olivier, M.; Mouland, A.J. HIV-1 Enhances MTORC1 Activity and Repositions Lysosomes to the Periphery by Co-Opting Rag GTPases. Sci. Rep. 2017, 7, 5515. [Google Scholar] [CrossRef]
- Scerra, G.; De Pasquale, V.; Scarcella, M.; Caporaso, M.G.; Pavone, L.M.; D’Agostino, M. Lysosomal Positioning Diseases: Beyond Substrate Storage. Open Biol. 2022, 12, 220155. [Google Scholar] [CrossRef] [PubMed]
- Machado, E.R.; Annunziata, I.; van de Vlekkert, D.; Grosveld, G.C.; d’Azzo, A. Lysosomes and Cancer Progression: A Malignant Liaison. Front. Cell Dev. Biol. 2021, 9, 642494. [Google Scholar] [CrossRef] [PubMed]
- Kundu, S.T.; Grzeskowiak, C.L.; Fradette, J.J.; Gibson, L.A.; Rodriguez, L.B.; Creighton, C.J.; Scott, K.L.; Gibbons, D.L. TMEM106B Drives Lung Cancer Metastasis by Inducing TFEB-Dependent Lysosome Synthesis and Secretion of Cathepsins. Nat. Commun. 2018, 9, 2731. [Google Scholar] [CrossRef]
- Hämälistö, S.; Jäättelä, M. Lysosomes in Cancer- Living on the Edge (of the Cell). Curr. Opin. Cell Biol. 2016, 39, 69. [Google Scholar] [CrossRef]
- Brix, D.M.; Rafn, B.; Bundgaard Clemmensen, K.; Andersen, S.H.; Ambartsumian, N.; Jäättelä, M.; Kallunki, T. Screening and Identification of Small Molecule Inhibitors of ErbB2-induced Invasion. Mol. Oncol. 2014, 8, 1703. [Google Scholar] [CrossRef]
- Medina, D.L.; Fraldi, A.; Bouche, V.; Annunziata, F.; Mansueto, G.; Spampanato, C.; Puri, C.; Pignata, A.; Martina, J.A.; Sardiello, M.; et al. Transcriptional Activation of Lysosomal Exocytosis Promotes Cellular Clearance. Dev. Cell 2011, 21, 421. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Webster, P.; Ortego, J.; Andrews, N.W. Lysosomes Behave as Ca2+-Regulated Exocytic Vesicles in Fibroblasts and Epithelial Cells. J. Cell Biol. 1997, 137, 93. [Google Scholar] [CrossRef] [PubMed]
- Purohit, V.; Rapaka, R.S.; Rutter, J. Cannabinoid Receptor-2 and HIV-Associated Neurocognitive Disorders. J. Neuroimmune Pharmacol. 2014, 9, 447–453. [Google Scholar] [CrossRef]
- Rom, S.; Persidsky, Y. Cannabinoid Receptor 2: Potential Role in Immunomodulation and Neuroinflammation. J. Neuroimmune Pharmacol. 2013, 8, 608–620. [Google Scholar] [CrossRef]
- Rizzo, M.D.; Henriquez, J.E.; Blevins, L.K.; Bach, A.; Crawford, R.B.; Kaminski, N.E. Targeting Cannabinoid Receptor 2 on Peripheral Leukocytes to Attenuate Inflammatory Mechanisms Implicated in HIV-Associated Neurocognitive Disorder. J. Neuroimmune Pharmacol. 2020, 15, 780–793. [Google Scholar] [CrossRef]
- Yadav-Samudrala, B.J.; Fitting, S. Mini-Review: The Therapeutic Role of Cannabinoids in NeuroHIV. Neurosci. Lett. 2021, 750, 135717. [Google Scholar] [CrossRef]
- Starr, A.; Jordan-Sciutto, K.L.; Mironets, E. Confound, Cause, or Cure: The Effect of Cannabinoids on HIV-Associated Neurological Sequelae. Viruses 2021, 13, 1242. [Google Scholar] [CrossRef]
- Kaminski, N.E.; Kaplan, B.L.F. Immunomodulation by Cannabinoids: Current Uses, Mechanisms, and Identification of Data Gaps to Be Addressed for Additional Therapeutic Application. Adv. Pharmacol. 2021, 91, 1–59. [Google Scholar] [CrossRef]
- Wang, L.; Zeng, Y.; Zhou, Y.; Yu, J.; Liang, M.; Qin, L.; Zhou, Y. Win55,212-2 Improves Neural Injury Induced by HIV-1 Glycoprotein 120 in Rats by Exciting CB2R. Brain Res. Bull. 2022, 182, 67–79. [Google Scholar] [CrossRef]
- Ribeiro, R.; Wen, J.; Li, S.; Zhang, Y. Involvement of ERK1/2, CPLA2 and NF-ΚB in Microglia Suppression by Cannabinoid Receptor Agonists and Antagonists. Prostaglandins Other Lipid Mediat. 2013, 100–101, 1–14. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Zhao, H.; Zheng, Q.; Xiao, L.; Zhao, M. CB2 Receptor Activation Ameliorates the Proinflammatory Activity in Acute Lung Injury Induced by Paraquat. Biomed Res. Int. 2014, 2014, 971750. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Yu, B.; Bai, J.; Wang, X.; Guo, X.; Liu, Y.; Lin, J.; Hu, S.; Zhang, W.; Tao, Y.; et al. Cannabinoid Receptor 2 Agonist Prevents Local and Systemic Inflammatory Bone Destruction in Rheumatoid Arthritis. J. Bone Miner. Res. 2019, 34, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ma, Y.; Liu, Y.; Wang, N.; Zhao, X.; Wen, D. CB2R Agonist JWH-133 Attenuates Chronic Inflammation by Restraining M1 Macrophage Polarization via Nrf2/HO-1 Pathway in Diet-Induced Obese Mice. Life Sci. 2020, 260, 118424. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Heo, D.R.; Kim, Y.A.; Lee, J.; Kim, N.S.; Bang, O.S. Coniferaldehyde Inhibits LPS-Induced Apoptosis through the PKC α/β II/Nrf-2/HO-1 Dependent Pathway in RAW264.7 Macrophage Cells. Environ. Toxicol. Pharmacol. 2016, 48, 85–93. [Google Scholar] [CrossRef]
- Kwak, M.K.; Wakabayashi, N.; Itoh, K.; Motohashi, H.; Yamamoto, M.; Kensler, T.W. Modulation of Gene Expression by Cancer Chemopreventive Dithiolethiones through the Keap1-Nrf2 Pathway. Identification of Novel Gene Clusters for Cell Survival. J. Biol. Chem. 2003, 278, 8135–8145. [Google Scholar] [CrossRef] [PubMed]
- Brand, S.R.; Kobayashi, R.; Mathews, M.B. The Tat Protein of Human Immunodeficiency Virus Type 1 Is a Substrate and Inhibitor of the Interferon-Induced, Virally Activated Protein Kinase, PKR. J. Biol. Chem. 1997, 272, 8388–8395. [Google Scholar] [CrossRef]
- Kang, K.W.; Lee, S.J.; Park, J.W.; Kim, S.G. Phosphatidylinositol 3-Kinase Regulates Nuclear Translocation of NF-E2-Related Factor 2 through Actin Rearrangement in Response to Oxidative Stress. Mol. Pharmacol. 2002, 62, 1001–1010. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Poulogiannis, G. Phosphoinositide 3-Kinase/Akt Signaling and Redox Metabolism in Cancer. Front. Oncol. 2018, 8, 160. [Google Scholar] [CrossRef] [PubMed]
- Bird, T.A.; Schooley, K.; Dower, S.K.; Hagen, H.; Virca, G.D. Activation of Nuclear Transcription Factor NF-κB by Interleukin-1 Is Accompanied by Casein Kinase II-Mediated Phosphorylation of the P65 Subunit. J. Biol. Chem. 1997, 272, 32606–32612. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Westerheide, S.D.; Hanson, J.L.; Baldwin, J. Tumor Necrosis Factor Alpha-Induced Phosphorylation of RelA/P65 on Ser529 Is Controlled by Casein Kinase II. J. Biol. Chem. 2000, 275, 32592–32597. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Pinna, L.A. One-Thousand-and-One Substrates of Protein Kinase CK2? FASEB J. 2003, 17, 349–368. [Google Scholar] [CrossRef]
- Martin, M.U.; Wesche, H. Summary and Comparison of the Signaling Mechanisms of the Toll/Interleukin-1 Receptor Family. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2002, 1592, 265–280. [Google Scholar] [CrossRef]
- Li, Q.; Verma, I.M. NF-κB Regulation in the Immune System. Nat. Rev. Immunol. 2002, 2, 725–734. [Google Scholar] [CrossRef]
- Baumann, B.; Weberjakob Troppmair, C.K.; Whiteside, S.; Israel, A.; Rapp, U.R.; Wirth, T. Raf Induces NF-κB by Membrane Shuttle Kinase MEKK1, a Signaling Pathway Critical for Transformation. Proc. Natl. Acad. Sci. USA 2000, 97, 4615–4620. [Google Scholar] [CrossRef]
- Mitchell, S.; Vargas, J.; Hoffmann, A. Signaling via the NFκB System. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016, 8, 227–241. [Google Scholar] [CrossRef]
- Wang, P.; Wang, S.C.; Yang, H.; Lv, C.; Jia, S.; Liu, X.; Wang, X.; Meng, D.; Qin, D.; Zhu, H.; et al. Therapeutic Potential of Oxytocin in Atherosclerotic Cardiovascular Disease: Mechanisms and Signaling Pathways. Front. Neurosci. 2019, 13, 454. [Google Scholar] [CrossRef]
- Pype, S.; Declercq, W.; Ibrahimi, A.; Michiels, C.; van Rietschoten, J.G.I.; Dewulf, N.; de Boer, M.; Vandenabeele, P.; Huylebroeck, D.; Remacle, J.E. TTRAP, a Novel Protein That Associates with CD40, Tumor Necrosis Factor (TNF) Receptor-75 and TNF Receptor-Associated Factors (TRAFs), and That Inhibits Nuclear Factor-κB Activation. J. Biol. Chem. 2000, 275, 18586–18593. [Google Scholar] [CrossRef]
- di Paola, S.; Medina, D.L. TRPML1-/TFEB-Dependent Regulation of Lysosomal Exocytosis. Methods Mol. Biol. 2019, 1925, 143–144. [Google Scholar] [CrossRef]
- Tseng, H.H.L.; Vong, C.T.; Kwan, Y.W.; Lee, S.M.Y.; Hoi, M.P.M. Lysosomal Ca2+ Signaling Regulates High Glucose-Mediated Interleukin-1β Secretion via Transcription Factor EB in Human Monocytic Cells. Front. Immunol. 2017, 8, 1161. [Google Scholar] [CrossRef]
- Johnson, J.L.; Hong, H.; Monfregola, J.; Kiosses, W.B.; Catz, S.D. Munc13-4 Restricts Motility of Rab27a-Expressing Vesicles to Facilitate Lipopolysaccharide-Induced Priming of Exocytosis in Neutrophils. J. Biol. Chem. 2011, 286, 5647–5656. [Google Scholar] [CrossRef]
- Sharda, A.; Kim, S.H.; Jasuja, R.; Gopal, S.; Flaumenhaft, R.; Furie, B.C.; Furie, B. Defective PDI Release from Platelets and Endothelial Cells Impairs Thrombus Formation in Hermansky-Pudlak Syndrome. Blood 2015, 125, 1633–1642. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.S.Y.; Haydar, T.F.; Radoja, S. Protein Kinase C Delta Localizes to Secretory Lysosomes in CD8+ CTL and Directly Mediates TCR Signals Leading to Granule Exocytosis-Mediated Cytotoxicity. J. Immunol. 2008, 181, 4716–4722. [Google Scholar] [CrossRef] [PubMed]
- Lazure, F.; Blackburn, D.M.; Corchado, A.H.; Sahinyan, K.; Karam, N.; Sharanek, A.; Nguyen, D.; Lepper, C.; Najafabadi, H.S.; Perkins, T.J.; et al. Myf6/MRF4 Is a Myogenic Niche Regulator Required for the Maintenance of the Muscle Stem Cell Pool. EMBO Rep. 2020, 21, e49499. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Du, L.; Zhang, L.; Hu, Y.; Xia, W.; Wu, J.; Zhu, J.; Chen, L.; Zhu, F.; Li, C.; et al. Cathepsin B Contributes to Autophagy-Related 7 (Atg7)-Induced Nod-like Receptor 3 (NLRP3)-Dependent Proinflammatory Response and Aggravates Lipotoxicity in Rat Insulinoma Cell Line. J. Biol. Chem. 2013, 288, 30094. [Google Scholar] [CrossRef] [PubMed]
- Rafn, B.; Nielsen, C.F.; Andersen, S.H.; Szyniarowski, P.; Corcelle-Termeau, E.; Valo, E.; Fehrenbacher, N.; Olsen, C.J.; Daugaard, M.; Egebjerg, C.; et al. ErbB2-Driven Breast Cancer Cell Invasion Depends on a Complex Signaling Network Activating Myeloid Zinc Finger-1-Dependent Cathepsin B Expression. Mol. Cell 2012, 45, 764–776. [Google Scholar] [CrossRef]
- Lee, J.H.; Yu, W.H.; Kumar, A.; Lee, S.; Mohan, P.S.; Peterhoff, C.M.; Wolfe, D.M.; Martinez-Vicente, M.; Massey, A.C.; Sovak, G.; et al. Lysosomal Proteolysis and Autophagy Require Presenilin 1 and Are Disrupted by Alzheimer-Related PS1 Mutations. Cell 2010, 141, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Sardiello, M.; Palmieri, M.; di Ronza, A.; Medina, D.L.; Valenza, M.; Gennarino, V.A.; di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; et al. A Gene Network Regulating Lysosomal Biogenesis and Function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef]
- Zenón, F.; Cantres-Rosario, Y.; Adiga, R.; Gonzalez, M.; Rodriguez-Franco, E.; Langford, D.; Melendez, L.M. HIV-Infected Microglia Mediate Cathepsin B-Induced Neurotoxicity. J. Neurovirol. 2015, 21, 544–558. [Google Scholar] [CrossRef] [PubMed]
- ElZohary, L.; Weglicki, W.B.; Chmielinska, J.J.; Kramer, J.H.; Tong Mak, I. Mg-Supplementation Attenuated Lipogenic and Oxidative/Nitrosative Gene Expression Caused by Combination Antiretroviral Therapy (CART) in HIV-1-Transgenic Rats. PLoS ONE 2019, 14, e0210107. [Google Scholar] [CrossRef]
- Han, D.; Lu, X.; Yin, W.; Fu, H.; Zhang, X.; Cheng, L.; Liu, F.; Jin, C.; Tian, X.; Xie, Y.; et al. Activation of NRF2 Blocks HIV Replication and Apoptosis in Macrophages. Heliyon 2023, 9, e12575. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.A.; Cook, D.R.; Chi, A.W.S.; Vance, P.J.; Kolson, L.L.; Wong, B.J.; Jordan-Sciutto, K.L.; Kolson, D.L. Dimethyl Fumarate, an Immune Modulator and Inducer of the Antioxidant Response, Suppresses HIV Replication and Macrophage-Mediated Neurotoxicity: A Novel Candidate for HIV Neuroprotection. J. Immunol. 2011, 187, 5015–5025. [Google Scholar] [CrossRef]
- Gill, A.J.; Kovacsics, C.E.; Cross, S.A.; Vance, P.J.; Kolson, L.L.; Jordan-Sciutto, K.L.; Gelman, B.B.; Kolson, D.L. Heme Oxygenase-1 Deficiency Accompanies Neuropathogenesis of HIV-Associated Neurocognitive Disorders. J. Clin. Investig. 2014, 124, 4459. [Google Scholar] [CrossRef]
- Ramirez, S.H.; Reichenbach, N.L.; Fan, S.; Rom, S.; Merkel, S.F.; Wang, X.; Ho, W.; Persidsky, Y. Attenuation of HIV-1 Replication in Macrophages by Cannabinoid Receptor 2 Agonists. J. Leukoc. Biol. 2013, 93, 801–810. [Google Scholar] [CrossRef]
- Asin, S.; Taylor, J.A.; Trushin, S.; Bren, G.; Paya, C.V. Ikappakappa Mediates NF-κB Activation in Human Immunodeficiency Virus-Infected Cells. J. Virol. 1999, 73, 3893–3903. [Google Scholar] [CrossRef]
- McElhinny, J.A.; MacMorran, W.S.; Bren, G.D.; Ten, R.M.; Israel, A.; Paya, C.V. Regulation of I Kappa B Alpha and P105 in Monocytes and Macrophages Persistently Infected with Human Immunodeficiency Virus. J. Virol. 1995, 69, 1500–1509. [Google Scholar] [CrossRef]
- Giacoppo, S.; Gugliandolo, A.; Trubiani, O.; Pollastro, F.; Grassi, G.; Bramanti, P.; Mazzon, E. Cannabinoid CB2 Receptors Are Involved in the Protection of RAW264.7 Macrophages against the Oxidative Stress: An in Vitro Study. Eur. J. Histochem. 2017, 61, 2749. [Google Scholar] [CrossRef]
- Luo, X.Q.; Li, A.; Yang, X.; Xiao, X.; Hu, R.; Wang, T.W.; Dou, X.Y.; Yang, D.J.; Dong, Z. Paeoniflorin Exerts Neuroprotective Effects by Modulating the M1/M2 Subset Polarization of Microglia/Macrophages in the Hippocampal CA1 Region of Vascular Dementia Rats via Cannabinoid Receptor 2. Chin. Med. 2018, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.C.; Appelberg, S.; Goldberger, B.A.; Klein, T.W.; Sleasman, J.W.; Goodenow, M.M. Δ9-Tetrahydrocannabinol Treatment during Human Monocyte Differentiation Reduces Macrophage Susceptibility to HIV-1 Infection. J. Neuroimmune Pharmacol. 2014, 9, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Cantres-Rosario, Y.M.; Acevedo-Mariani, F.M.; Pérez-Laspiur, J.; Haskins, W.E.; Plaud, M.; Cantres-Rosario, Y.M.; Skolasky, R.; Méndez-Bermúdez, I.; Wojna, V.; Meléndez, L.M.; et al. Microwave & Magnetic Proteomics of Macrophages from Patients with HIV-Associated Cognitive Impairment. PLoS ONE 2017, 12, e0181779. [Google Scholar] [CrossRef]
- Borges-Vélez, G.; Rosado-Philippi, J.; Cantres-Rosario, Y.M.; Carrasquillo-Carrion, K.; Roche-Lima, A.; Pérez-Vargas, J.; González-Martínez, A.; Correa-Rivas, M.S.; Meléndez, L.M. Zika Virus Infection of the Placenta Alters Extracellular Matrix Proteome. J. Mol. Histol. 2021, 53, 199–214. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Sharma, V.; Perez-Riverol, Y.; Carver, J.J.; Kundu, D.J.; García-Seisdedos, D.; Jarnuczak, A.F.; Hewapathirana, S.; Pullman, B.S.; et al. The ProteomeXchange Consortium in 2020: Enabling ‘Big Data’ Approaches in Proteomics. Nucleic Acids Res 2020, 48, D1145. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442. [Google Scholar] [CrossRef]

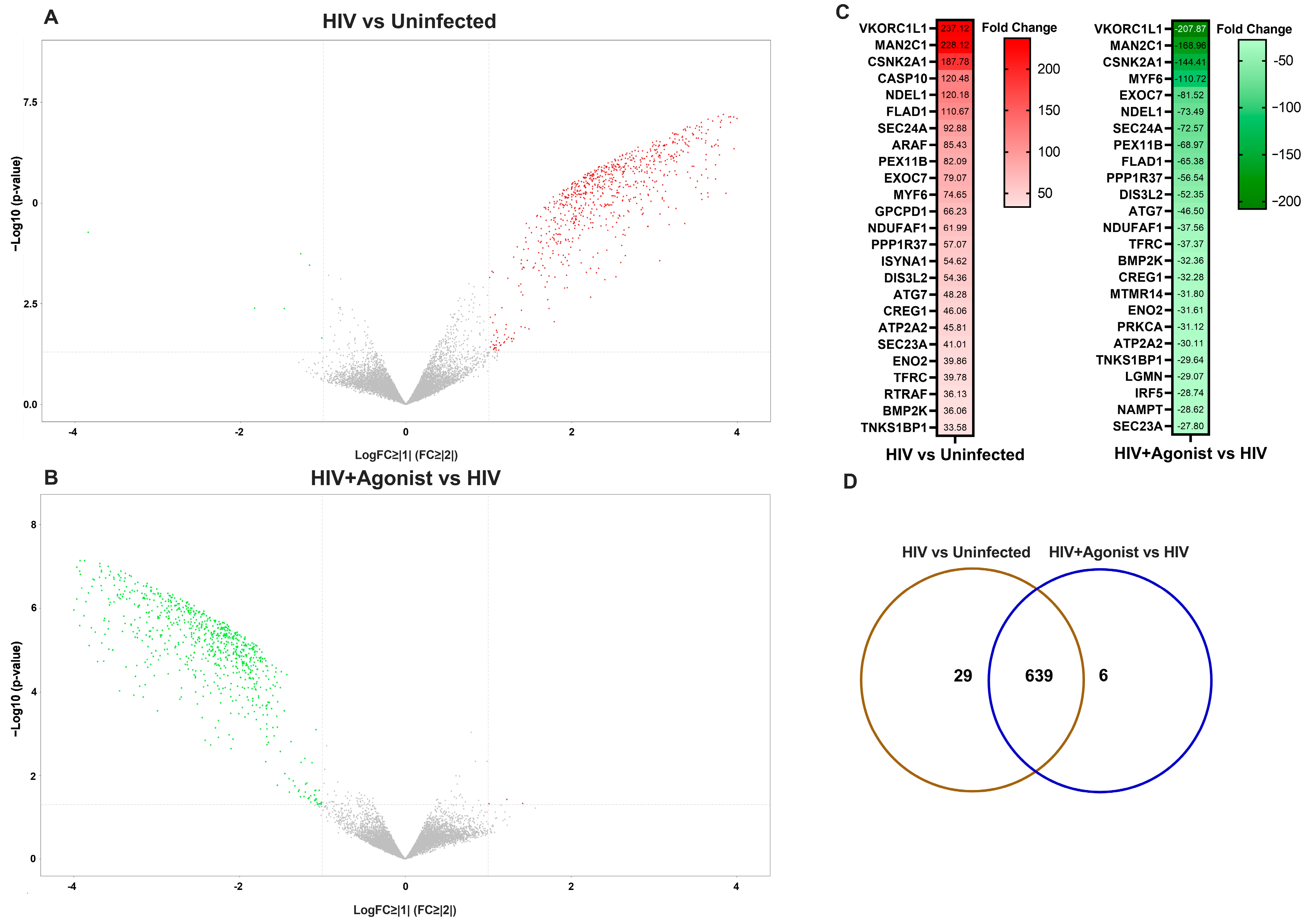
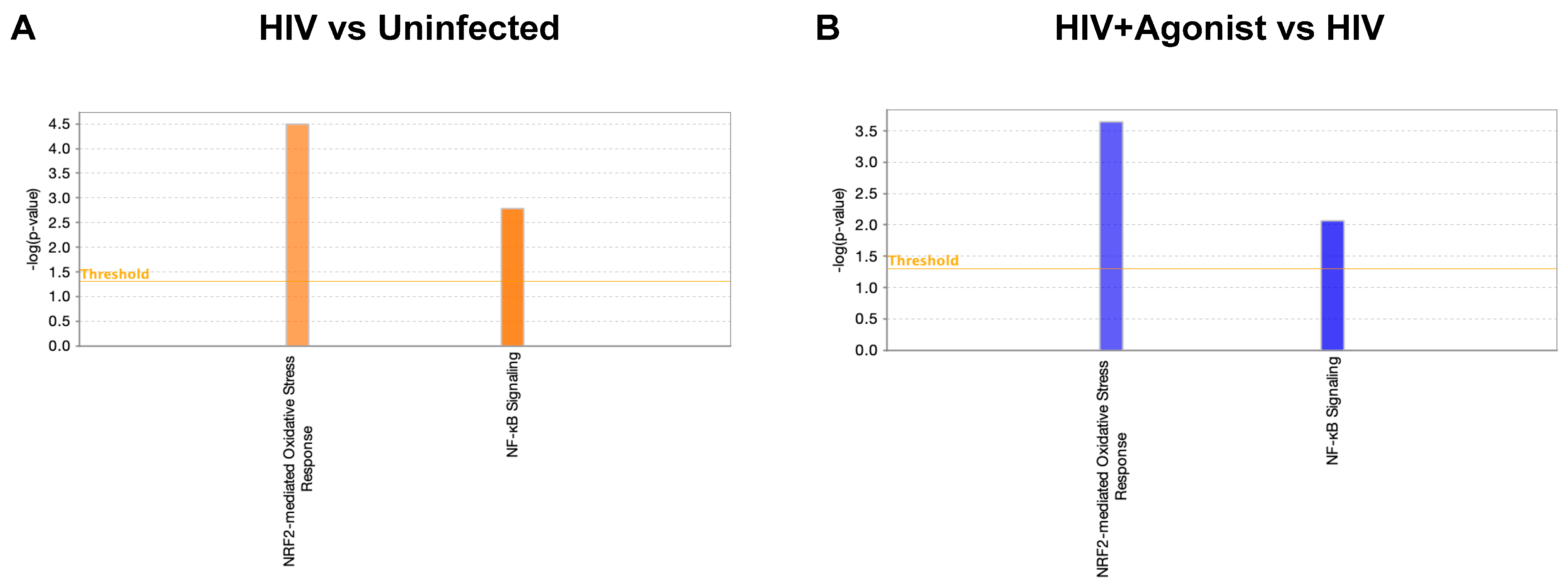
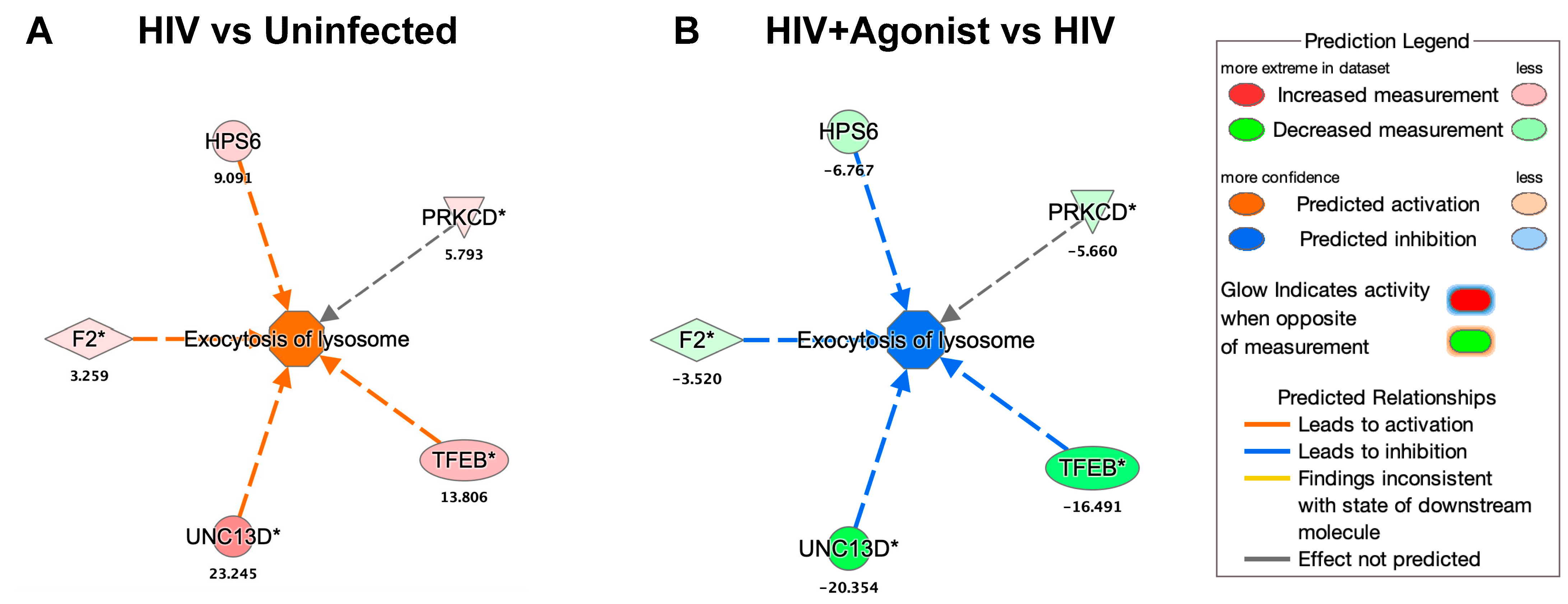
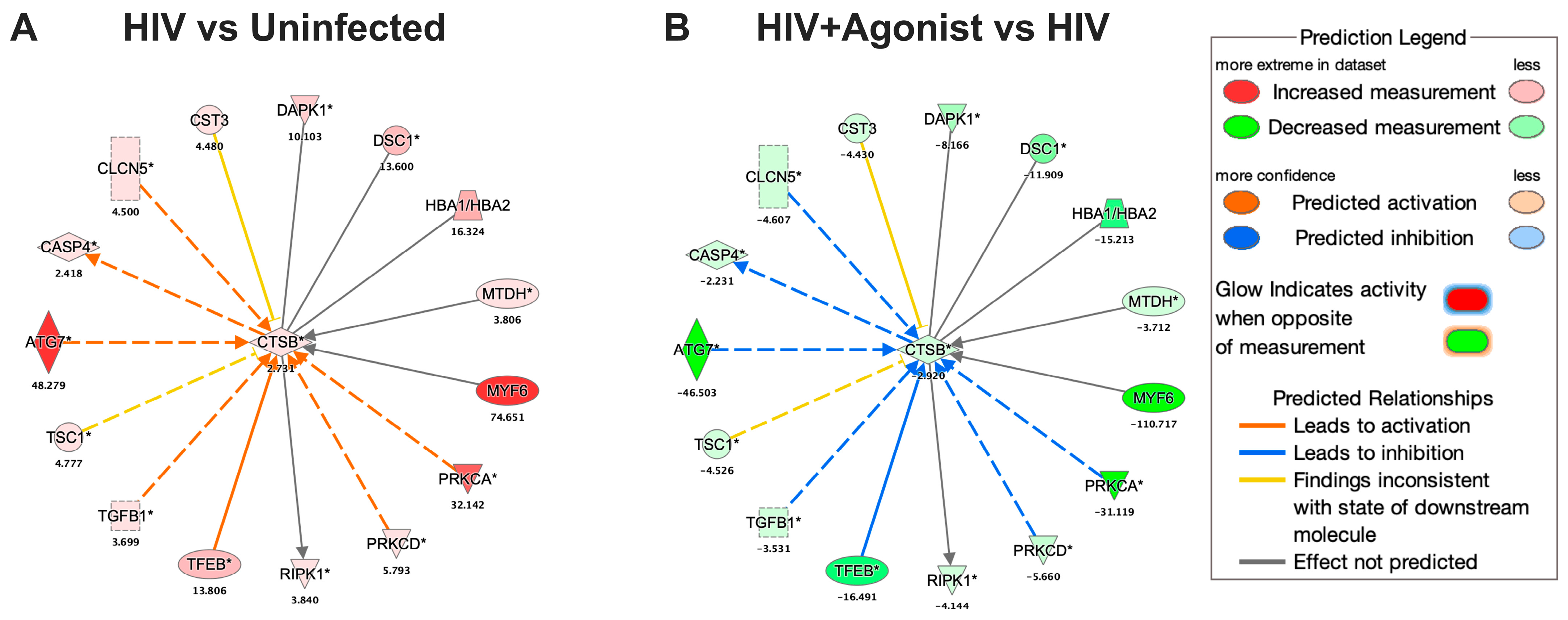
| HIV vs. Uninfected | HIV + Agonist vs. HIV | ||||
|---|---|---|---|---|---|
| Symbol | Entrez Gene Name | Expr Fold Change | Expr p-Value | Expr Fold Change | Expr p-Value |
| PRKCA | protein kinase C alpha | 32.142 | 4.63 × 10−8 | −31.119 | 3.94 × 10−6 |
| HACD3 | 3-hydroxyacyl-CoA dehydratase 3 | 25.127 | 1.51 × 10−7 | −23.856 | 1.07 × 10−7 |
| CUL3 | cullin 3 | 9.871 | 6.53 × 10−7 | −9.554 | 4.99 × 10−6 |
| PIK3R2 | phosphoinositide-3-kinase regulatory subunit 2 | 9.386 | 7.64 × 10−7 | −9.018 | 5.89 × 10−6 |
| DNAJC7 | DnaJ heat shock protein family (Hsp40) member C7 | 7.967 | 1.12 × 10−6 | −8.220 | 1.40 × 10−5 |
| DNAJC11 | DnaJ heat shock protein family (Hsp40) member C11 | 6.470 | 1.67 × 10−5 | −6.012 | 1.01 × 10−6 |
| PRKCD | protein kinase C delta | 5.793 | 1.29 × 10−6 | −5.660 | 5.75 × 10−6 |
| HMOX1 | heme oxygenase 1 | 5.773 | 2.78 × 10−6 | −5.579 | 2.41 × 10−6 |
| STIP1 | stress induced phosphoprotein 1 | 5.593 | 4.72 × 10−6 | −5.338 | 2.06 × 10−6 |
| FTH1 | ferritin heavy chain 1 | 4.999 | 2.25 × 10−6 | −5.082 | 1.07 × 10−4 |
| ACTG2 | actin gamma 2, smooth muscle | 4.746 | 1.42 × 10−5 | −4.700 | 4.70 × 10−5 |
| DNAJC8 | DnaJ heat shock protein family (Hsp40) member C8 | 4.268 | 1.81 × 10−5 | −4.202 | 1.20 × 10−5 |
| DNAJB12 | DnaJ heat shock protein family (Hsp40) member B12 | 4.025 | 9.19 × 10−5 | −3.563 | 8.55 × 10−6 |
| PIK3CB | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta | 3.723 | 5.97 × 10−6 | −4.084 | 6.96 × 10−6 |
| SQSTM1 | sequestosome 1 | 3.639 | 6.16 × 10−5 | −3.879 | 6.59 × 10−6 |
| GCLC | glutamate-cysteine ligase catalytic subunit | 3.280 | 2.52 × 10−3 | −3.711 | 3.33 × 10−4 |
| HIV vs. Uninfected | HIV + Agonist vs. HIV | ||||
|---|---|---|---|---|---|
| Symbol | Entrez Gene Name | Expr Fold Change | Expr p-Value | Expr Fold Change | Expr p-Value |
| CSNK2A1 | casein kinase 2 alpha 1 | 187.779 | 1.01 × 10−8 | −144.412 | 1.16 × 10−7 |
| ARAF | A-Raf proto-oncogene, serine/threonine kinase | 85.425 | 2.27 × 10−8 | −13.087 | 1.90 × 10−5 |
| MAP4K4 | mitogen-activated protein kinase kinase kinase kinase 4 | 22.938 | 2.13 × 10−7 | −15.934 | 1.12 × 10−6 |
| PIK3R2 | phosphoinositide-3-kinase regulatory subunit 2 | 9.386 | 7.64 × 10−7 | −9.018 | 5.89 × 10−6 |
| TDP2 | tyrosyl-DNA phosphodiesterase 2 | 6.952 | 3.23 × 10−6 | −7.308 | 9.59 × 10−6 |
| EIF2AK2 | eukaryotic translation initiation factor 2 alpha kinase 2 | 6.464 | 8.41 × 10−6 | −6.394 | 6.53 × 10−6 |
| TRADD | TNFRSF1A associated via death domain | 6.013 | 1.54 × 10−6 | −4.772 | 8.51 × 10−6 |
| IGF2R | insulin like growth factor 2 receptor | 5.983 | 3.18 × 10−6 | −5.753 | 3.25 × 10−6 |
| PRKACA | protein kinase cAMP-activated catalytic subunit alpha | 4.901 | 2.52 × 10−6 | −4.533 | 2.30 × 10−6 |
| RIPK1 | receptor interacting serine/threonine kinase 1 | 3.84 | 9.41 × 10−6 | −4.144 | 3.88 × 10−6 |
| PIK3CB | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta | 3.723 | 5.97 × 10−6 | −4.084 | 6.96 × 10−6 |
| MYD88 | MYD88 innate immune signal transduction adaptor | 2.785 | 1.36 × 10−4 | −2.638 | 1.20 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosario-Rodríguez, L.J.; Cantres-Rosario, Y.M.; Carrasquillo-Carrión, K.; Rodríguez-De Jesús, A.E.; Cartagena-Isern, L.J.; García-Requena, L.A.; Roche-Lima, A.; Meléndez, L.M. Quantitative Proteomics Reveal That CB2R Agonist JWH-133 Downregulates NF-κB Activation, Oxidative Stress, and Lysosomal Exocytosis from HIV-Infected Macrophages. Int. J. Mol. Sci. 2024, 25, 3246. https://doi.org/10.3390/ijms25063246
Rosario-Rodríguez LJ, Cantres-Rosario YM, Carrasquillo-Carrión K, Rodríguez-De Jesús AE, Cartagena-Isern LJ, García-Requena LA, Roche-Lima A, Meléndez LM. Quantitative Proteomics Reveal That CB2R Agonist JWH-133 Downregulates NF-κB Activation, Oxidative Stress, and Lysosomal Exocytosis from HIV-Infected Macrophages. International Journal of Molecular Sciences. 2024; 25(6):3246. https://doi.org/10.3390/ijms25063246
Chicago/Turabian StyleRosario-Rodríguez, Lester J., Yadira M. Cantres-Rosario, Kelvin Carrasquillo-Carrión, Ana E. Rodríguez-De Jesús, Luz J. Cartagena-Isern, Luis A. García-Requena, Abiel Roche-Lima, and Loyda M. Meléndez. 2024. "Quantitative Proteomics Reveal That CB2R Agonist JWH-133 Downregulates NF-κB Activation, Oxidative Stress, and Lysosomal Exocytosis from HIV-Infected Macrophages" International Journal of Molecular Sciences 25, no. 6: 3246. https://doi.org/10.3390/ijms25063246
APA StyleRosario-Rodríguez, L. J., Cantres-Rosario, Y. M., Carrasquillo-Carrión, K., Rodríguez-De Jesús, A. E., Cartagena-Isern, L. J., García-Requena, L. A., Roche-Lima, A., & Meléndez, L. M. (2024). Quantitative Proteomics Reveal That CB2R Agonist JWH-133 Downregulates NF-κB Activation, Oxidative Stress, and Lysosomal Exocytosis from HIV-Infected Macrophages. International Journal of Molecular Sciences, 25(6), 3246. https://doi.org/10.3390/ijms25063246






