Multiple Roles of Glycerate Kinase—From Photorespiration to Gluconeogenesis, C4 Metabolism, and Plant Immunity
Abstract
1. Introduction
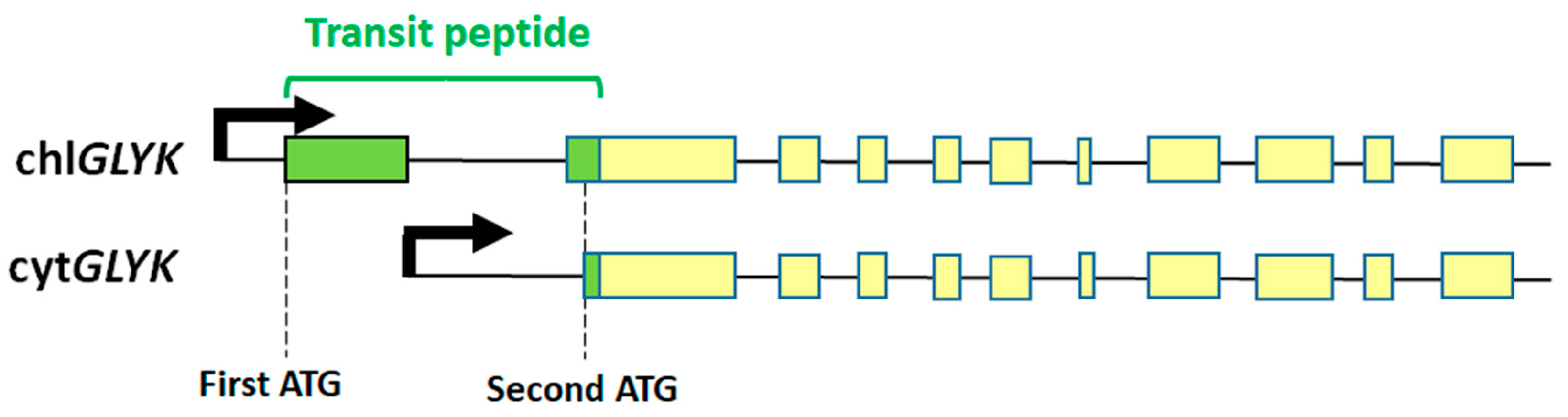
2. GK—Gene and Protein
3. GK Regulation and Subcellular Location
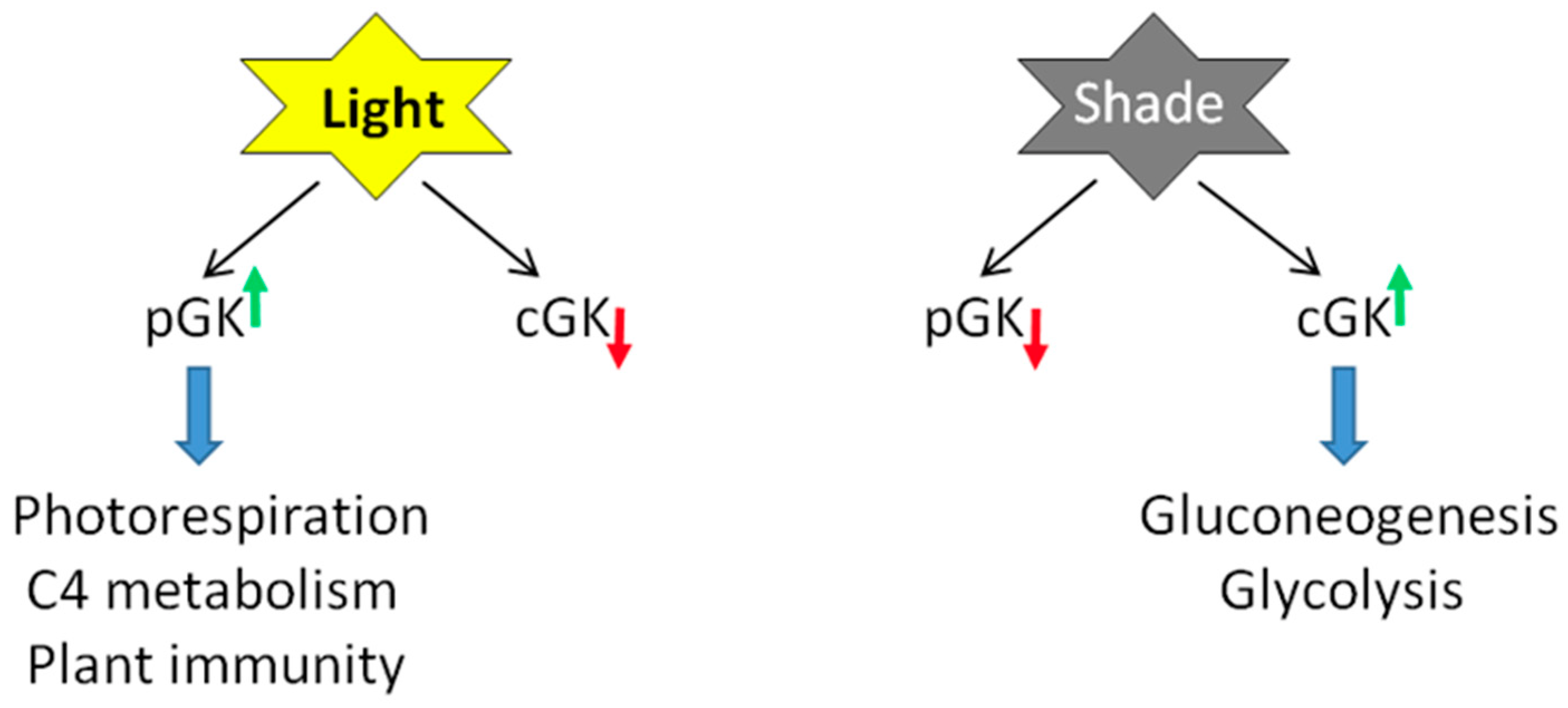
3.1. Transcriptional and Post-Transcriptional Regulation
3.2. Redox Regulation
3.3. Metabolite Regulation
4. Origins of D-Glycerate
4.1. HPR (and GR) Isozymes/Isoforms
4.2. 3PGA Phosphatase
4.3. Lactate Dehydrogenase
4.4. Tartronic Semialdehyde Reductase
5. Glycerate Transporters
5.1. Plastidial Glycerate Transporters
5.2. Vacuolar Glycerate Transporter
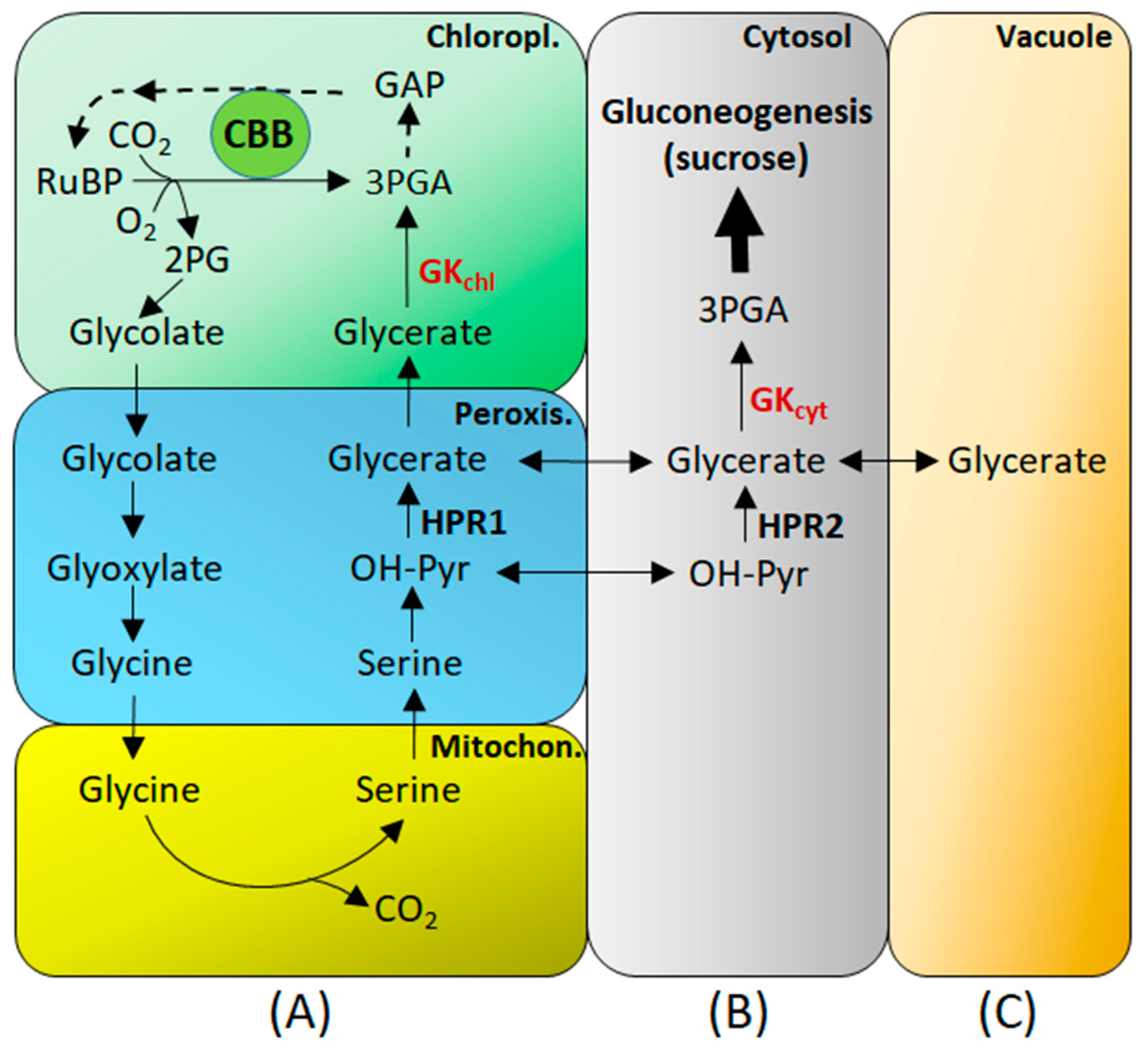
6. GK in Photorespiration
7. GK in Gluconeogenesis and Respiration
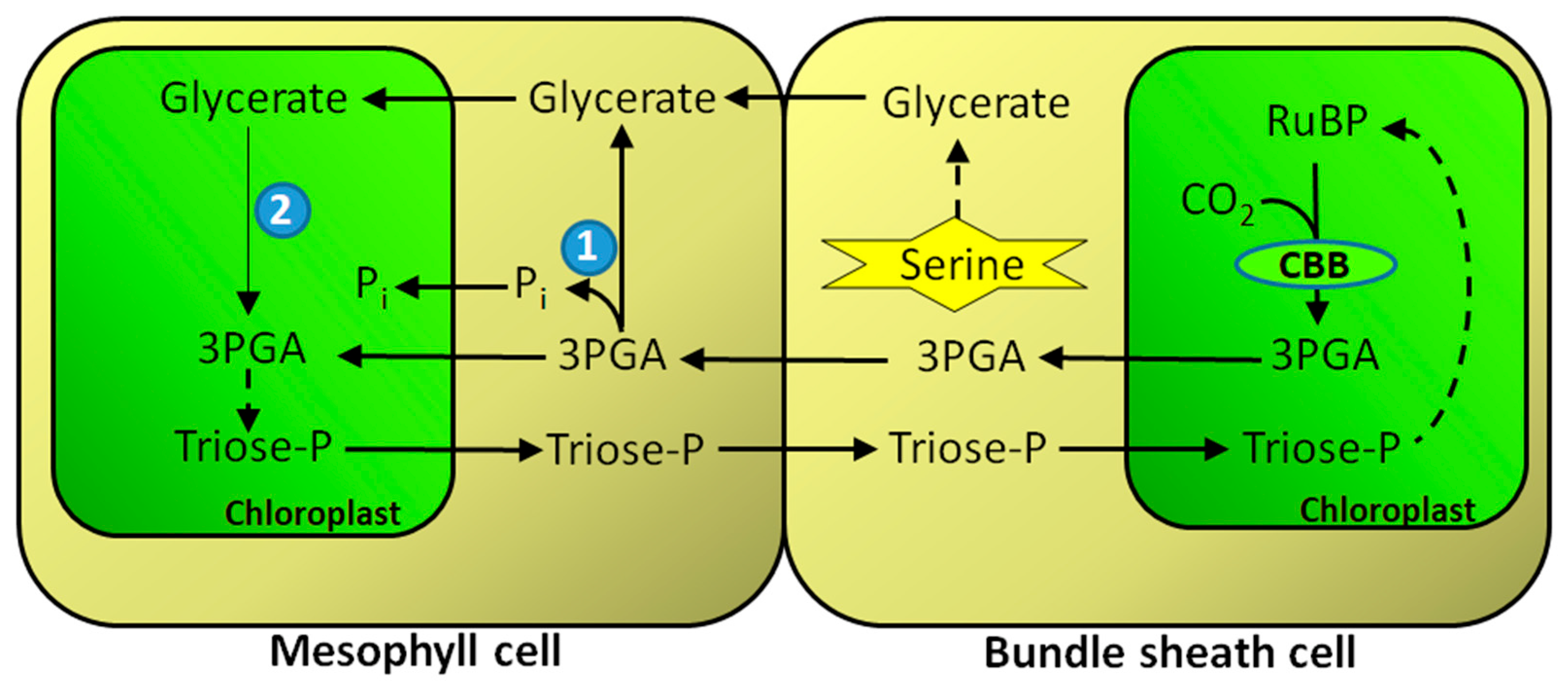
8. GK in C4 Metabolism
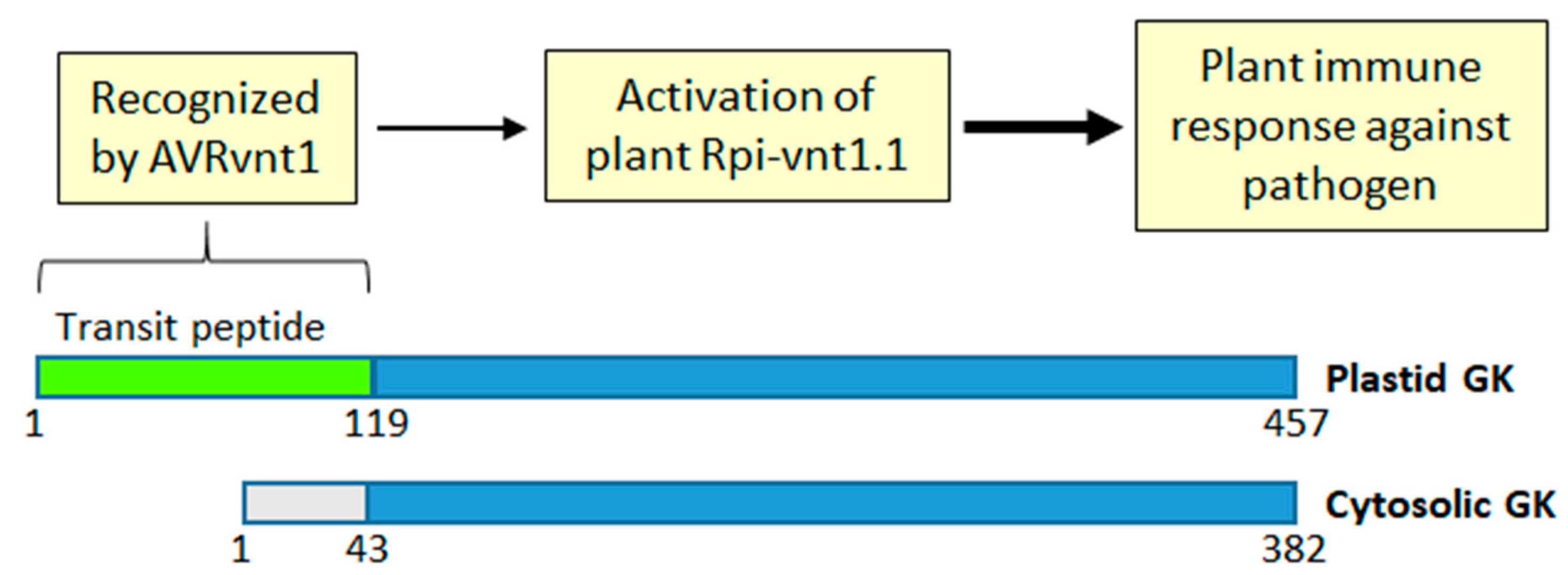
9. GK and Plant Immunity
10. Summary and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CBB cycle | Calvin–Benson–Bassham cycle |
| GABA | γ-aminobutyric acid |
| GHB | γ-hydroxybutyrate |
| GK | glycerate kinase |
| GR | glyoxylate reductase |
| HPR | hydroxypyruvate reductase |
| 2PG | 2-phosphoglycolate |
| 3PGA | 3-phosphoglycerate |
| PLGG1 | plastidial glycolate/glycerate transporter |
| RA | rosmarinic acid |
| SGAT | serine: glyoxylate aminotransferase |
| SSA | succinic semialdehyde |
| TCA cycle | tricarboxylic acid cycle |
| TSR | tartronic semialdehyde reductase |
| VGT | vacuolar glycerate transporter |
References
- Husic, D.W.; Husic, H.D.; Tolbert, N.E.; Black, C.C. The oxidative photosynthetic carbon cycle or C2 cycle. CRC Crit. Rev. Plant Sci. 1987, 5, 45–100. [Google Scholar] [CrossRef]
- Givan, C.V.; Joy, K.W.; Kleczkowski, L.A. A decade of photorespiratory nitrogen cycling. Trends Biochem. Sci. 1988, 13, 433–437. [Google Scholar] [CrossRef]
- Boldt, R.; Edner, C.; Kolukisaoglu, U.; Hagemann, M.; Weckwerth, W.; Wienkoop, S.; Morgenthal, K.; Bauwe, H. D-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 2005, 17, 2413–2420. [Google Scholar] [CrossRef] [PubMed]
- Flügel, F.; Timm, S.; Arrivault, S.; Florian, A.; Stitt, M.; Fernie, A.R.; Bauwe, H. The photorespiratory metabolite 2-phosphoglycolate regulates photosynthesis and starch accumulation in Arabidopsis. Plant Cell 2017, 29, 2537–2551. [Google Scholar] [CrossRef] [PubMed]
- Andrews, T.J.; Lorimer, G.H. Photorespiration—Still unavoidable? FEBS Lett. 1978, 90, 1–9. [Google Scholar] [CrossRef]
- Heber, U.; Krause, G.H. What is the physiological role of photorespiration? Trends Biochem. Sci. 1980, 5, 32–34. [Google Scholar] [CrossRef]
- Bartsch, O.; Hagemann, M.; Bauwe, H. Only plant-type (GLYK) glycerate kinases produce D-glycerate 3-phosphate. FEBS Lett. 2008, 582, 3025–3028. [Google Scholar] [CrossRef]
- Black, S.; Wright, N.G. Enzymatic formation of glyceryl and phosphoglyceryl methylthiol esters. J. Biol. Chem. 1956, 221, 171–180. [Google Scholar] [CrossRef]
- Kern, R.; Bauwe, H.; Hagemann, M. Evolution of enzymes involved in the photorespiratory 2-phosphoglycolate cycle from cyanobacteria via algae toward plants. Photosynth. Res. 2011, 109, 103–114. [Google Scholar] [CrossRef]
- Eisenhut, M.; Ruth, W.; Haimovich, M.; Bauwe, H.; Kaplan, A.; Hagemann, M. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. USA 2008, 105, 17199–17204. [Google Scholar] [CrossRef]
- Kroth, P.G.; Chiovitti, A.; Gruber, A.; Martin-Jezequel, V.; Mock, T.; Parker, M.S.; Stanley, M.S.; Kaplan, A.; Caron, L.; Weber, T.; et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 2008, 3, e1426. [Google Scholar] [CrossRef]
- Bauwe, H.; Hagemann, M.; Fernie, A.R. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010, 15, 330–336. [Google Scholar] [CrossRef]
- Bauwe, H.; Hagemann, M.; Kern, R.; Timm, S. Photorespiration has a dual origin and manifold links to central metabolism. Curr. Opin. Plant Biol. 2012, 15, 269–275. [Google Scholar] [CrossRef]
- Hagemann, M.; Fernie, A.R.; Espie, G.S.; Kern, R.; Eisenhut, M.; Reumann, S.; Weber, A.P.M. Evolution of the biochemistry of the photorespiratory C2 cycle. Plant Biol. 2013, 15, 639–647. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Lea, P.J. The role of peroxisomes in the integration of metabolism and evolutionary diversity of photosynthetic organisms. Phytochemistry 2002, 60, 651–674. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, Y.; Katayama, H.; Sugimoto, E. Identity of mitochondrial and cytosolic glycerate kinases in rat liver and regulation of their intracellular localization by dietary protein. Biochim. Biophys. Acta 1979, 582, 260–275. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Randall, D.D.; Zahler, W.L. The substrate specificity, kinetics, and mechanism of glycerate-3-kinase from spinach leaves. Arch. Biochem. Biophys. 1985, 236, 185–194. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Randall, D.D. Substrate stereospecificity of leaf glycerate kinase from C3 and C4 plants. Phytochemistry 1988, 27, 1269–1273. [Google Scholar] [CrossRef]
- Zelcbuch, L.; Razo-Mejia, M.; Herz, E.; Yahav, S.; Antonovsky, N.; Kroytoro, H.; Milo, R.; Bar-Even, A. An in vivo metabolic approach for deciphering the product specificity of glycerate kinase proves that both E. coli’s glycerate kinases generate 2-phosphoglycerate. PLoS ONE 2015, 10, e0122957. [Google Scholar] [CrossRef] [PubMed]
- Hatch, M.D.; Slack, C.R. NADP-specific malate dehydrogenase and glycerate kinase in leaves and evidence for their location in chloroplasts. Biochem. Biophys. Res. Commun. 1969, 34, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Randall, D.D. Purification and partial characterization of spinach leaf glycerate kinase. FEBS Lett. 1983, 158, 313–316. [Google Scholar] [CrossRef]
- Schmitt, M.R.; Edwards, G.E. Glycerate kinase from leaves of C3 plants. Arch. Biochem. Biophys. 1983, 224, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Chaguturu, R. Glycerate kinase from spinach leaves: Partial purification, characterization and subcellular localization. Physiol. Plant. 1985, 63, 19–24. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Randall, D.D. Purification and characterization of D-glycerate-3-kinase from maize leaves. Planta 1988, 173, 221–229. [Google Scholar] [CrossRef]
- Bartsch, O.; Mikkat, S.; Hagemann, M.; Bauwe, H. An autoinhibitory domain confers redox regulation to maize glycerate kinase. Plant Physiol. 2010, 153, 832–840. [Google Scholar] [CrossRef]
- Kozaki, A.; Takeba, G. Photorespiration protects C3 plants from photooxidation. Nature 1996, 384, 557–560. [Google Scholar] [CrossRef]
- Foyer, C.H.; Bloom, A.J.; Queval, G.; Noctor, G. Photorespiratory metabolism: Genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 2009, 60, 455–484. [Google Scholar] [CrossRef] [PubMed]
- Bapatla, R.B.; Saini, D.; Aswani, V.; Rajsheel, P.; Sunil, B.; Timm, S.; Raghavendra, A.S. Modulation of photorespiratory enzymes by oxidative and photo-oxidative stress induced by menadione in leaves of pea (Pisum sativum). Plants 2021, 10, 987. [Google Scholar] [CrossRef]
- Saini, D.; Bapatla, R.B.; Vemula, C.K.; Gahir, S.; Bharath, P.; Gupta, K.J.; Raghavendra, A.S. Moderate modulation by S-nitrosoglutathione of photorespiratory enzymes in pea (Pisum sativum) leaves, compared to the strong effects of high light. Protoplasma 2024, 261, 43–51. [Google Scholar] [CrossRef]
- Timm, S.; Florian, A.; Wittmiss, M.; Jahnke, K.; Hagemann, M.; Fernie, A.R.; Bauwe, H. Serine acts as a metabolic signal for the transcriptional control of photorespiration-related genes in Arabidopsis. Plant Physiol. 2013, 162, 379–389. [Google Scholar] [CrossRef]
- Aroca, A.; García-Díaz, I.; García-Calderón, M.; Gotor, C.; Márquez, A.J.; Betti, M. Photorespiration: Regulation and new insights on the potential role of persulfidation. J. Exp. Bot. 2023, 74, 6023–6039. [Google Scholar] [CrossRef] [PubMed]
- Saini, D.; Bharath, P.; Gahir, S.; Raghavendra, A.S. Suppression of photorespiratory metabolism by low O2 and presence of aminooxyacetic acid induces oxidative stress in Arabidopsis thaliana leaves. Physiol. Mol. Biol. Plants 2023, 29, 1851–1861. [Google Scholar] [CrossRef]
- Laxa, M.; Fromm, S. Co-expression and regulation of photorespiratory genes in Arabidopsis thaliana: A bioinformatic approach. Curr. Plant Biol. 2018, 14, 2–18. [Google Scholar] [CrossRef]
- Modde, K.; Timm, S.; Florian, A.; Michl, K.; Fernie, A.R.; Bauwe, H. High serine:glyoxylate aminotransferase activity lowers leaf daytime serine levels, inducing the phosphoserine pathway in Arabidopsis. J. Exp. Bot. 2017, 68, 643–656. [Google Scholar] [CrossRef]
- Ushijima, T.; Hanada, K.; Gotoh, E.; Yamori, W.; Kodama, Y.; Tanaka, H.; Kusano, M.; Fukushima, A.; Tokizawa, M.; Yamamoto, Y.Y.; et al. Light controls protein localization through phytochrome-mediated alternative promoter selection. Cell 2017, 171, 1316–1325. [Google Scholar] [CrossRef]
- Usuda, H.; Edwards, G.E. Localization of glycerate kinase and some enzymes for sucrose synthesis in C3 and C4 plants. Plant Physiol. 1980, 65, 1017–1022. [Google Scholar] [CrossRef]
- Usuda, H.; Edwards, G.E. Photosynthetic formation of glycerate in isolated bundle sheath cells and its metabolism in mesophyll cells of the C4 plant Panicum capillare L. Funct. Plant Biol. 1980, 7, 655–662. [Google Scholar] [CrossRef]
- Oh, S.; Montgomery, B.L. Phytochromes: Where to Start? Cell 2017, 171, 1254–1256. [Google Scholar] [CrossRef]
- Gao, C.; Xu, H.; Huang, J.; Sun, B.; Zhang, F.; Savage, Z.; Duggan, C.; Yan, T.; Wu, C.H.; Wang, Y.; et al. Pathogen manipulation of chloroplast function triggers a light-dependent immune recognition. Proc. Natl. Acad. Sci. USA 2020, 117, 9613–9620. [Google Scholar] [CrossRef] [PubMed]
- Heber, U.; Kirk, M.R.; Gimmler, H.; Schafer, G. Uptake and reduction of glycerate by isolated chloroplasts. Planta 1974, 120, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Randall, D.D. Light and thiol activation of maize leaf glycerate kinase: The stimulating effect of reduced thioredoxins and ATP. Plant Physiol. 1985, 79, 274–277. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Randall, D.D. Thiol-dependent regulation of glycerate metabolism in leaf extracts: The role of glycerate kinase in C4 plants. Plant Physiol. 1986, 81, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Morisse, S.; Zaffagnini, M.; Gao, X.H.; Lemaire, S.D.; Marchand, C.H. Insight into protein S-nitrosylation in Chlamydomonas reinhardtii. Antioxid. Redox Signal. 2014, 21, 1271–1284. [Google Scholar] [CrossRef] [PubMed]
- Keech, O.; Gardeström, P.; Kleczkowski, L.A.; Rouhier, N. The redox control of photorespiration: From biochemical and physiological aspects to biotechnological considerations. Plant Cell Environ. 2017, 40, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Timm, S.; Nunes-Nesi, A.; Pärnik, T.; Morgenthal, K.; Wienkoop, S.; Keerberg, O.; Weckwerth, W.; Kleczkowski, L.A.; Fernie, A.R.; Bauwe, H. A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 2008, 20, 2848–2859. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Q.; Zhang, J.; Hou, Y.; Dai, Q.; Lv, H.; Cao, P.; Zhao, L. Hydroxypyruvate reductase gene family in Nicotiana benthamiana: Genome-wide identification and expression pattern profiling. Curr. Plant Biol. 2023, 35–36, 100305. [Google Scholar] [CrossRef]
- Zhang, Z.; Liang, X.; Lu, L.; Xu, Z.; Huang, J.; He, H.; Peng, X. Two glyoxylate reductase isoforms are functionally redundant but required under high photorespiration conditions in rice. BMC Plant Biol. 2020, 20, 357. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Randall, D.D. Purification and characterization of a novel NADPH (NADH)-dependent hydroxypyruvate reductase from spinach leaves. Comparison of immunological properties of leaf hydroxypyruvate reductases. Biochem. J. 1988, 250, 145–152. [Google Scholar] [CrossRef]
- Givan, C.V.; Kleczkowski, L.A. The enzymic reduction of glyoxylate and hydroxypyruvate in leaves of higher plants. Plant Physiol. 1992, 100, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Zelitch, I.; Gotto, A.M. Properties of a new glyoxylate reductase from leaves. Biochem. J. 1962, 84, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Kleczkowski, L.A. Glyoxylate metabolism during photorespiration: A cytosol connection. In Handbook of Photosynthesis; Pessarakli, M., Ed.; Marcel Dekker, Inc.: New York, NY, USA, 1997; pp. 269–279. [Google Scholar]
- Igamberdiev, A.U.; Bykova, N.V.; Kleczkowski, L.A. Origins and metabolism of formate in higher plants. Plant Physiol. Biochem. 1999, 37, 503–513. [Google Scholar] [CrossRef]
- Timm, S.; Florian, A.; Jahnke, K.; Nunes-Nesi, A.; Fernie, A.R.; Bauwe, H. The hydroxypyruvate-reducing system in Arabidopsis: Multiple enzymes for the same end. Plant Physiol. 2011, 155, 694–705. [Google Scholar] [CrossRef]
- Shi, M.; Zhao, L.; Wang, Y. Identification and characterization of genes encoding the hydroxypyruvate reductases in Chlamydomonas reveal their distinct roles in photorespiration. Front. Plant Sci. 2021, 12, 690296. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Givan, C.V.; Hodgson, J.M.; Randall, D.D. Subcellular location of NADPH-dependent hydroxypyruvate reductase activity in leaf protoplasts of Pisum sativum L. and its role in photorespiratory metabolism. Plant Physiol. 1988, 88, 1182–1185. [Google Scholar] [CrossRef] [PubMed]
- Mano, S.; Hayashi, M.; Nishimura, M. Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 1999, 17, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Mano, S.; Hayashi, M.; Nishimura, M. A leaf-peroxisomal protein, hydroxypyruvate reductase, is produced by light-regulated alternative splicing. Cell Biochem. Biophys. 2000, 32, 147–154. [Google Scholar] [CrossRef]
- Murray, A.J.; Blackwell, R.D.; Lea, P.J. Metabolism of hydroxypyruvate in a mutant of barley lacking NADH-dependent hydroxypyruvate reductase, an important photorespiratory enzyme activity. Plant Physiol. 1989, 91, 395–400. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Edwards, G.E.; Blackwell, R.D.; Lea, P.J.; Givan, C.V. Enzymology of the reduction of hydroxypyruvate and glyoxylate in a mutant of barley lacking peroxisomal hydroxypyruvate reductase. Plant Physiol. 1990, 94, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Yang, G.; Chen, Y.; Zhang, C.; Zhang, J.; Peng, X. Two hydroxypyruvate reductases encoded by OsHPR1 and OsHPR2 are involved in photorespiratory metabolism in rice. J. Integr. Plant Biol. 2014, 56, 170–180. [Google Scholar] [CrossRef]
- Timm, S.; Nunes-Nesi, A.; Florian, A.; Eisenhut, M.; Morgenthal, K.; Wirtz, M.; Hell, R.; Weckwerth, W.; Hagemann, M.; Fernie, A.R.; et al. Metabolite profiling in Arabidopsis thaliana with moderately impaired photorespiration reveals novel metabolic links and compensatory mechanisms of photorespiration. Metabolites 2021, 11, 391. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Kleczkowski, L.A. Capacity for NADPH/NADP turnover in the cytosol of barley seed endosperm: The role of NADPH-dependent hydroxypyruvate reductase. Plant Physiol. Biochem. 2000, 38, 747–753. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Givan, C.V. Serine formation in leaves by mechanisms other than the glycolate pathway. J. Plant Physiol. 1988, 132, 641–652. [Google Scholar] [CrossRef]
- Ros, R.; Cascales-Miñana, B.; Segura, J.; Anoman, A.D.; Toujani, W.; Flores-Tornero, M.; Rosa-Tellez, S.; Muñoz-Bertomeu, J. Serine biosynthesis by photorespiratory and nonphotorespiratory pathways: An interesting interplay with unknown regulatory networks. Plant Biol. 2013, 15, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Ros, R.; Muñoz-Bertomeu, J.; Krueger, S. Serine in plants: Biosynthesis, metabolism, and functions. Trends Plant Sci. 2014, 19, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Kleczkowski, L.A. The glycerate and phosphorylated pathways of serine synthesis in plants: The branches of plant glycolysis linking carbon and nitrogen metabolism. Front. Plant Sci. 2018, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guérard, F.; Hodges, M.; Jossier, M. Phosphomimetic T335D mutation of hydroxypyruvate reductase 1 modifies cofactor specificity and impacts Arabidopsis growth in air. Plant Physiol. 2020, 183, 194–205. [Google Scholar] [CrossRef]
- Corpas, F.J.; Leterrier, M.; Begara-Morales, J.C.; Valderrama, R.; Chaki, M.; López-Jaramillo, J.; Luque, F.; Palma, J.M.; Padilla, M.N.; Sánchez-Calvo, B.; et al. Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochim. Biophys. Acta 2013, 1830, 4981–4989. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Edwards, G.E. Identification of hydroxypyruvate and glyoxylate reductases in maize leaves. Plant Physiol. 1989, 91, 278–286. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Randall, D.D.; Blevins, D.G. Inhibition of spinach leaf NADPH(NADH)-glyoxylate reductase by acetohydroxamate, aminooxyacetate, and glycidate. Plant Physiol. 1987, 84, 619–623. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Randall, D.D.; Edwards, G.E. Oxalate as a potent and selective inhibitor of spinach (Spinacia oleracea) leaf NADPH-dependent hydroxypyruvate reductase. Biochem. J. 1991, 276, 125–127. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Edwards, G.E.; Randall, D.D. Effects of oxalate on reduction of hydroxypyruvate and glyoxylate in leaves. Phytochemistry 1992, 31, 51–54. [Google Scholar] [CrossRef]
- Allan, W.L.; Clark, S.M.; Hoover, G.J.; Shelp, B.J. Role of plant glyoxylate reductases during stress: A hypothesis. Biochem. J. 2009, 423, 15–22. [Google Scholar] [CrossRef]
- Simpson, J.P.; Di Leo, R.; Dhanoa, P.K.; Allan, W.L.; Makhmoudova, A.; Clark, S.M.; Hoover, G.J.; Mullen, R.T.; Shelp, B.J. Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: Comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. J. Exp. Bot. 2008, 59, 2545–2554. [Google Scholar] [CrossRef]
- Zarei, A.; Brikis, C.J.; Bajwa, V.S.; Chiu, G.Z.; Simpson, J.P.; DeEll, J.R.; Bozzo, G.G.; Shelp, B.J. Plant glyoxylate/succinic semialdehyde reductases: Comparative biochemical properties, function during chilling stress, and subcellular localization. Front. Plant Sci. 2017, 8, 1399. [Google Scholar] [CrossRef]
- Givan, C.V.; Tsutakawa, S.; Hodgson, J.M.; David, N.; Randall, D.D. Glyoxylate reductase activity in pea leaf protoplasts: Nucleotide specificity and subcellular location. J. Plant Physiol. 1988, 132, 593–599. [Google Scholar] [CrossRef]
- Hücherig, S.; Petersen, M. RNAi suppression and overexpression studies of hydroxyphenylpyruvate reductase (HPPR) and rosmarinic acid synthase (RAS) genes related to rosmarinic acid biosynthesis in hairy root cultures of Coleus blumei. Plant Cell Tissue Organ Cult. 2013, 113, 375–385. [Google Scholar] [CrossRef]
- Xu, J.J.; Fang, X.; Li, C.Y.; Zhao, Q.; Martin, C.; Chen, X.Y.; Yang, L. Characterization of Arabidopsis thaliana hydroxyphenylpyruvate reductases in the tyrosine conversion pathway. Front. Plant Sci. 2018, 9, 1305. [Google Scholar] [CrossRef]
- Wang, L.; Qian, J.; Li, M.; Zheng, H.; Yang, X.; Zheng, M.; Hsu, Y.F. Arabidopsis PDE1 confers phosphate-deficiency tolerance in primary root growth. Plant Cell Rep. 2023, 43, 8. [Google Scholar] [CrossRef] [PubMed]
- Shelp, B.J.; Aghdam, M.S.; Flaherty, E.J. γ-Aminobutyrate (GABA) regulated plant defense: Mechanisms and opportunities. Plants 2021, 10, 1939. [Google Scholar] [CrossRef] [PubMed]
- Janiak, V.; Petersen, M.; Zentgraf, M.; Klebe, G.; Heine, A. Structure and substrate docking of a hydroxy(phenyl)pyruvate reductase from the higher plant Coleus blumei Benth. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Randall, D.D.; Tolbert, N.E. 3-Phosphoglycerate phosphatase in plants. I. Isolation and characterization from sugarcane leaves. J. Biol. Chem. 1971, 246, 5510–5517. [Google Scholar] [CrossRef] [PubMed]
- Randall, D.D.; Tolbert, N.E.; Gremel, D. 3-Phosphoglycerate phosphatase in plants. II. Distribution, physiological considerations, and comparison with P-glycolate phosphatase. Plant Physiol. 1971, 48, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Betsche, T. L-Lactate dehydrogenase from leaves of higher plants. Kinetics and regulation of the enzyme from lettuce (Lactuca sativa L. ). Biochem. J. 1981, 195, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, N.; Taniguchi, N. Evaluation of the role of lactate dehydrogenase in oxalate synthesis. Phytochemistry 1997, 44, 571–574. [Google Scholar] [CrossRef] [PubMed]
- South, P.F.; Cavanagh, A.P.; Liu, H.W.; Ort, D.R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 2019, 363, eaat9077. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.M.; Shen, B.R.; Li, B.D.; Zhang, C.L.; Lin, M.; Tong, P.P.; Cui, L.L.; Zhang, Z.S.; Peng, X.X. A synthetic photorespiratory shortcut enhances photosynthesis to boost biomass and grain yield in rice. Mol. Plant. 2020, 13, 1802–1815. [Google Scholar] [CrossRef] [PubMed]
- Howitz, K.T.; McCarty, R.E. D-Glycerate transport by the pea chloroplast glycolate carrier: Studies on [1-14C]D-glycerate uptake and D-glycerate dependent O2 evolution. Plant Physiol. 1986, 80, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Pick, T.R.; Bräutigam, A.; Schulz, M.A.; Obata, T.; Fernie, A.R.; Weber, A.P. PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc. Natl. Acad. Sci. USA 2013, 110, 3185–3190. [Google Scholar] [CrossRef]
- Shim, S.H.; Lee, S.K.; Lee, D.W.; Brilhaus, D.; Wu, G.; Ko, S.; Lee, C.H.; Weber, A.P.M.; Jeon, J.S. Loss of function of rice plastidic-glycolate/glycerate translocator 1 impairs photorespiration and plant growth. Front. Plant Sci. 2019, 10, 1726. [Google Scholar] [CrossRef]
- Cui, L.; Zhang, C.; Li, Z.; Xian, T.; Wang, L.; Zhang, Z.; Zhu, G.; Peng, X. Two plastidic glycolate/ glycerate translocator 1 isoforms function together to transport photorespiratory glycolate and glycerate in rice chloroplasts. J. Exp. Bot. 2021, 72, 2584–2599. [Google Scholar] [CrossRef]
- South, P.F.; Walker, B.J.; Cavanagh, A.P.; Rolland, V.; Badger, M.; Ort, D.R. Bile acid sodium symporter BASS6 can transport glycolate and is involved in photorespiratory metabolism in Arabidopsis thaliana. Plant Cell 2017, 29, 808–823. [Google Scholar] [CrossRef]
- Lin, Y.C.; Tsay, Y.F. Study of vacuole glycerate transporter NPF8.4 reveals a new role of photorespiration in C/N balance. Nat. Plants 2023, 9, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Keller, R.; Ziegler, C.; Schneider, D. When two turn into one: Evolution of membrane transporters from half modules. Biol. Chem. 2014, 395, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
- Timm, S.; Eisenhut, M. Four plus one: Vacuoles serve in photorespiration. Trends Plant Sci. 2023, 28, 1340–1343. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lai, Z.; Wang, W.; Zhong, Q.; Wu, F.; Yang, S.; Xie, B.; Li, Y.; Sun, W.; Peng, X.; et al. The characterization of Arabidopsis photorespiration D-glycerate 3-kinase mutants generated by CRISPR/Cas9 and identification of its interacting Proteins. J. Plant Growth Regul. 2023, 42, 2458–2473. [Google Scholar] [CrossRef]
- Dai, Z.; Locasale, J.W. Thermodynamic constraints on the regulation of metabolic fluxes. J. Biol. Chem. 2018, 293, 19725–19739. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Gregory, L.M.; Weise, S.E.; Walker, B.J. Integrated flux and pool size analysis in plant central metabolism reveals unique roles of glycine and serine during photorespiration. Nat. Plants 2023, 9, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Bykova, N.V. Mitochondria in photosynthetic cells: Coordinating redox control and energy balance. Plant Physiol. 2023, 191, 2104–2119. [Google Scholar] [CrossRef]
- Shameer, S.; Ratcliffe, R.G.; Sweetlove, L.J. Leaf energy balance requires mitochondrial respiration and export of chloroplast NADPH in the light. Plant Physiol. 2019, 180, 1947–1961. [Google Scholar] [CrossRef]
- Dahal, K.; Martyn, G.D.; Alber, N.A.; Vanlerberghe, G.C. Coordinated regulation of photosynthetic and respiratory components is necessary to maintain chloroplast energy balance in varied growth conditions. J. Exp. Bot. 2017, 68, 657–671. [Google Scholar] [CrossRef]
- Anoman, A.D.; Flores-Tornero, M.; Rosa-Telléz, S.; Muñoz-Bertomeu, J.; Segura, J.; Ros, R. The specific role of plastidial glycolysis in photosynthetic and heterotrophic cells under scrutiny through the study of glyceraldehyde-3-phosphate dehydrogenase. Plant Signal. Behav. 2016, 11, e1128614. [Google Scholar] [CrossRef] [PubMed]
- Andriotis, V.M.; Kruger, N.J.; Pike, M.J.; Smith, A.M. Plastidial glycolysis in developing Arabidopsis embryos. New Phytol. 2010, 185, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E. Compartmentation in plant metabolism. J. Exp. Bot. 2007, 58, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Selinski, J.; Scheibe, R. Malate valves: Old shuttles with new perspectives. Plant Biol. 2019, 21, 21–30. [Google Scholar] [CrossRef]
- Lim, S.L.; Voon, C.P.; Guan, X.; Yang, Y.; Gardeström, P.; Lim, B.L. In planta study of photosynthesis and photorespiration using NADPH and NADH/NAD+ fluorescent protein sensors. Nat. Commun. 2020, 11, 3238. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Romanowska, E.; Gardeström, P. Photorespiratory flux and mitochondrial contribution to energy and redox balance of barley leaf protoplasts in the light and during light-dark transitions. J. Plant Physiol. 2001, 158, 1325–1332. [Google Scholar] [CrossRef]
- Gardeström, P.; Igamberdiev, A.U. The origin of cytosolic ATP in photosynthetic cells. Physiol. Plant. 2016, 157, 367–379. [Google Scholar] [CrossRef]
- Voon, C.P.; Guan, X.; Sun, Y.; Sahu, A.; Chan, M.N.; Gardeström, P.; Wagner, S.; Fuchs, P.; Nietzel, T.; Versaw, W.K.; et al. ATP compartmentation in plastids and cytosol of Arabidopsis thaliana revealed by fluorescent protein sensing. Proc. Natl. Acad. Sci. USA 2018, 115, E10778–E10787. [Google Scholar] [CrossRef]
- Kleczkowski, L.A.; Randall, D.D.; Blevins, D.G. Purification and characterization of a novel NADPH(NADH)-dependent glyoxylate reductase from spinach leaves. Comparison of immunological properties of leaf glyoxylate reductase and hydroxypyruvate reductase. Biochem. J. 1986, 239, 653–659. [Google Scholar] [CrossRef]
- Harley, P.C.; Sharkey, T.D. An improved model of C3 photosynthesis at high CO2: Reversed O2 sensitivity explained by lack of glycerate reentry into the chloroplast. Photosynth. Res. 1991, 27, 169–178. [Google Scholar] [CrossRef]
- Kondracka, A.; Rychter, A.M. The role of Pi recycling processes during photosynthesis in phosphate-deficient bean plants. J. Exp. Bot. 1997, 48, 1461–1468. [Google Scholar] [CrossRef]
- Sharkey, T.D. O2-insensitive photosynthesis in C3 plants: Its occurrence and a possible explanation. Plant Physiol. 1985, 78, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, T.D.; Vanderveer, P.J. Stromal phosphate concentration is low during feedback limited photosynthesis. Plant Physiol. 1989, 91, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Pieters, A.J.; Paul, M.J.; Lawlor, D.W. Low sink demand limits photosynthesis under Pi deficiency. J. Exp. Bot. 2001, 52, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Edwards, G.E.; Walker, D.A. C3, C4: Mechanisms and Cellular and Environmental Regulation of Photosynthesis; Wiley/Blackwell: Hoboken, NJ, USA, 1983; ISBN 10: 0520050185. [Google Scholar]
- Hatch, M.D. C4 photosynthesis: A unique blend of modified biochemistry, anatomy and ultrastructure. Biochim. Biophys. Acta 1987, 895, 81–106. [Google Scholar] [CrossRef]
- Truszkiewicz, W.; Paszkowski, A. Serine:glyoxylate aminotransferases from maize and wheat leaves: Purification and properties. Photosynth. Res. 2004, 82, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, S.E.; Benstein, R.M.; Flores-Tornero, M.; Blau, S.; Anoman, A.D.; Rosa-Téllez, S.; Gerlich, S.C.; Salem, M.A.; Alseekh, S.; Kopriva, S.; et al. The phosphorylated pathway of serine biosynthesis links plant growth with nitrogen metabolism. Plant Physiol. 2021, 186, 1487–1506. [Google Scholar] [CrossRef]
- Wang, L.; Kuang, Y.; Zheng, S.; Tong, Y.; Zhu, Y.; Wang, Y. Overexpression of the phosphoserine phosphatase-encoding gene (AtPSP1) promotes starch accumulation in Lemna turionifera 5511 under sulfur deficiency. Plants 2023, 12, 1012. [Google Scholar] [CrossRef]
- García-Calderón, M.; Pérez-Delgado, C.M.; Credali, A.; Vega, J.M.; Betti, M.; Márquez, A.J. Genes for asparagine metabolism in Lotus japonicus: Differential expression and interconnection with photorespiration. BMC Genom. 2017, 18, 781. [Google Scholar] [CrossRef]
- Murray, A.J.; Blackwell, R.D.; Joy, K.W.; Lea, P.J. Photorespiratory N donors, aminotransferase specificity and photosynthesis in a mutant of barley deficient in serine: Glyoxylate aminotransferase activity. Planta 1987, 172, 106–113. [Google Scholar] [CrossRef]
- Ohnishi, J.; Yamazaki, M.; Kanai, R. Differentiation of photorespiratory activity between mesophyll and bundle sheath cells of C4 Plants II. Peroxisomes of Panicum miliaceum L. Plant Cell Physiol. 1985, 26, 797–803. [Google Scholar] [CrossRef]
- Ohnishi, J.; Kanai, R. Glycerate uptake into mesophyll and bundle sheath chloroplasts of a C4 plant, Panicum miliaceum. J. Plant Physiol. 1988, 133, 119–121. [Google Scholar] [CrossRef]
- Leegood, R.C. Strategies for engineering C4 photosynthesis. J. Plant Physiol. 2013, 170, 378–388. [Google Scholar] [CrossRef]
- Ludwig, M. The roles of organic acids in C4 photosynthesis. Front. Plant Sci. 2016, 7, 647. [Google Scholar] [CrossRef]
- Chen, T.M.; Dittrich, P.; Campbell, W.H.; Black, C.C. Metabolism of epidermal tissues, mesophyll cells, and bundle sheath strands resolved from mature nutsedge leaves. Arch. Biochem. Biophys. 1974, 163, 246–262. [Google Scholar] [CrossRef] [PubMed]
- Downton, W.J.S.; Berry, J.A.; Tregunna, E.B. C4-photosynthesis: Noncyclic electron flow and grana development in bundle sheath chloroplasts. Z. Pflanzenphysiol. 1970, 63, 194–198. [Google Scholar] [CrossRef]
- Chapman, K.S.; Berry, J.A.; Hatch, M.D. Photosynthetic metabolism in bundle sheath cells of the C4 species Zea mays: Sources of ATP and NADPH and the contribution of photosystem II. Arch. Biochem. Biophys. 1980, 202, 330–341. [Google Scholar] [CrossRef]
- Meierhoff, K.; Westhoff, P. Differential biogenesis of photosystem II in mesophyll and bundle-sheath cells of monocotyledonous NADP-malic enzyme-type C4 plants: The non-stoichiometric abundance of the subunits of photosystem II in the bundle-sheath chloroplasts and the translational activity of the plastome-encoded genes. Planta 1993, 191, 23–33. [Google Scholar] [CrossRef]
- Mayne, B.C.; Dee, A.M.; Edwards, G.E. Photosynthesis in mesophyll protoplasts and bundle sheath cells of various type of C4 plants. III. Fluorescence emission spectra, delayed light emission, and P700 content. Z. Pflanzenphysiol. 1974, 74, 275–291. [Google Scholar] [CrossRef]
- Sowiński, P.; Szczepanik, J.; Minchin, P.E. On the mechanism of C4 photosynthesis intermediate exchange between Kranz mesophyll and bundle sheath cells in grasses. J. Exp. Bot. 2008, 59, 1137–1147. [Google Scholar] [CrossRef]
- Danila, F.R.; Quick, W.P.; White, R.G.; Furbank, R.T.; von Caemmerer, S. The metabolite pathway between bundle sheath and mesophyll: Quantification of plasmodesmata in leaves of C3 and C4 monocots. Plant Cell 2016, 28, 1461–1471. [Google Scholar] [CrossRef]
- Leegood, R.C. The intercellular compartmentation of metabolites in leaves of Zea mays L. Planta 1985, 164, 163–171. [Google Scholar] [CrossRef]
- Monson, R.K.; Edwards, G.E.; Ku, M.S.B. C3-C4 intermediate photosynthesis in plants. Bioscience 1984, 34, 563–574. [Google Scholar] [CrossRef]
- Sage, R.F. Russ Monson and the evolution of C4 photosynthesis. Oecologia 2021, 197, 823–840. [Google Scholar] [CrossRef]
- Alonso-Cantabrana, H.; von Caemmerer, S. Carbon isotope discrimination as a diagnostic tool for C4 photosynthesis in C3-C4 intermediate species. J Exp. Bot. 2016, 67, 3109–3121. [Google Scholar] [CrossRef]
- Blätke, M.A.; Bräutigam, A. Evolution of C4 photosynthesis predicted by constraint-based modelling. eLife 2019, 8, e49305. [Google Scholar] [CrossRef]
- Bauwe, H. Photorespiration—Rubisco’s repair crew. J. Plant Physiol. 2023, 280, 153899. [Google Scholar] [CrossRef]
- Medeiros, D.B.; Ishihara, H.; Guenther, M.; Rosado de Souza, L.; Fernie, A.R.; Stitt, M.; Arrivault, S. 13CO2 labeling kinetics in maize reveal impaired efficiency of C4 photosynthesis under low irradiance. Plant Physiol. 2022, 190, 280–304. [Google Scholar] [CrossRef] [PubMed]
- Furbank, R.T.; Kelly, S. Finding the C4 sweet spot: Cellular compartmentation of carbohydrate metabolism in C4 photosynthesis. J. Exp. Bot. 2021, 72, 6018–6026. [Google Scholar] [CrossRef] [PubMed]
- Downton, W.J.S.; Hawker, J.S. Enzymes of starch and sucrose metabolism in Zea mays leaves. Phytochemistry 1973, 12, 1551–1556. [Google Scholar] [CrossRef]
- Smith, A.M.; Zeeman, S.C. Starch: A flexible, adaptable carbon store coupled to plant growth. Annu. Rev. Plant Biol. 2020, 71, 217–245. [Google Scholar] [CrossRef] [PubMed]
- Borghi, G.L.; Arrivault, S.; Günther, M.; Barbosa Medeiros, D.; Dell’Aversana, E.; Fusco, G.M.; Carillo, P.; Ludwig, M.; Fernie, A.R.; Lunn, J.E.; et al. Metabolic profiles in C3, C3-C4 intermediate, C4-like, and C4 species in the genus Flaveria. J. Exp. Bot. 2022, 73, 1581–1601. [Google Scholar] [CrossRef]
- Bleau, J.R.; Spoel, S.H. Selective redox signaling shapes plant–pathogen interactions. Plant Physiol. 2021, 186, 53–65. [Google Scholar] [CrossRef]
- Neill, S.; Desikan, R.; Hancock, J. Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 2002, 5, 388–395. [Google Scholar] [CrossRef]
- Williams, A.; Pétriacq, P.; Schwarzenbacher, R.E.; Beerling, D.J.; Ton, J. Mechanisms of glacial-to-future atmospheric CO2 effects on plant immunity. New Phytol. 2018, 218, 752–761. [Google Scholar] [CrossRef]
- Jiang, X.; Walker, B.J.; He, S.Y.; Hu, J. The role of photorespiration in plant immunity. Front. Plant Sci. 2023, 14, 1125945. [Google Scholar] [CrossRef]
- Trémulot, L.; Macadré, C.; Gal, J.; Garmier, M.; Launay-Avon, A.; Paysant-Le Roux, C.; Ratet, P.; Noctor, G.; Dufresne, M. Impact of high atmospheric carbon dioxide on the biotic stress response of the model cereal species Brachypodium distachyon. Front. Plant Sci. 2023, 14, 1237054. [Google Scholar] [CrossRef]
- Eisenhut, M.; Roell, M.S.; Weber, A.P.M. Mechanistic understanding of photorespiration paves the way to a new green revolution. New Phytol. 2019, 223, 1762–1769. [Google Scholar] [CrossRef]
- Kacar, B.; Hanson-Smith, V.; Adam, Z.R.; Boekelheide, N. Constraining the timing of the Great Oxidation Event within the Rubisco phylogenetic tree. Geobiology 2017, 15, 628–640. [Google Scholar] [CrossRef]
- Garcia, A.K.; Kaçar, B. How to resurrect ancestral proteins as proxies for ancient biogeochemistry. Free Radic. Biol. Med. 2019, 140, 260–269. [Google Scholar] [CrossRef]
- von Bismarck, T.; Wendering, P.; Perez de Souza, L.; Ruß, J.; Strandberg, L.; Heyneke, E.; Walker, B.J.; Schöttler, M.A.; Fernie, A.R.; Nikoloski, Z.; et al. Growth in fluctuating light buffers plants against photorespiratory perturbations. Nat. Commun. 2023, 14, 7052. [Google Scholar] [CrossRef]
- Li de La Sierra-Gallay, I.; Collinet, B.; Graille, M.; Quevillon-Cheruel, S.; Liger, D.; Minard, P.; Blondeau, K.; Henckes, G.; Aufrère, R.; Leulliot, N.; et al. Crystal structure of the YGR205w protein from Saccharomyces cerevisiae: Close structural resemblance to E. coli pantothenate kinase. Proteins 2004, 54, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A. Inhibitors of photosynthetic enzymes/ carriers and metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1994, 45, 339–367. [Google Scholar] [CrossRef]
- Kuo, Y.M.; Henry, R.A.; Andrews, A.J. Measuring specificity in multi-substrate/product systems as a tool to investigate selectivity in vivo. Biochim. Biophys. Acta 2016, 1864, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Jiang, J.; Zhang, C.; Jiang, L.; Ye, N.; Lu, Y.; Yang, G.; Liu, E.; Peng, C.; He, Z.; et al. Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J. Exp. Bot. 2010, 61, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Segel, I.H. Enzyme Kinetics. Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems; John Wiley & Sons: Hoboken, NJ, USA, 1975. [Google Scholar]
- Cornish-Bowden, A. A Simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J. 1974, 137, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Eastmond, P.J.; Graham, I.A. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 2001, 6, 72–78. [Google Scholar] [CrossRef]
- Graham, I.A. Seed storage oil mobilization. Annu. Rev. Plant Biol. 2008, 59, 115–142. [Google Scholar] [CrossRef]
| Enzyme | Reaction | Location | Inhibitors |
|---|---|---|---|
| HPR1 | OH-pyruvate + NAD(P)H → D-Glycerate + NAD(P)+ Glyoxylate + NAD(P)H → Glycolate + NAD(P)+ | Per | None |
| HPR2 | OH-pyruvate + NAD(P)H → D-Glycerate + NAD(P)+ Glyoxylate + NAD(P)H → Glycolate + NAD(P)+ OH-phenylpyruvate + NAD(P)H → 4-OH-phenyllactic acid + NAD(P)+ | Cyt | Oxalate, Phosphohydroxypyruvate, Tartronate |
| HPR3 | OH-pyruvate + NAD(P)H → D-Glycerate + NAD(P)+ Glyoxylate + NAD(P)H → Glycolate + NAD(P)+ | Chl | Oxalate |
| GR1 | Glyoxylate + NAD(P)H → Glycolate + NAD(P)+ SSA + NAD(P)H → γ-OH-butyrate + NAD(P)+ | Cyt | AHA, AOA, Glycidate, Oxalate? |
| GR2 | Glyoxylate + NAD(P)H → Glycolate + NAD(P)+ | Chl, Mit | Oxalate? |
| PGA-Pase | 3PGA → D-Glycerate + Pi | Cyt | None |
| LDH | Pyruvate + NAD(P)H ⇌ Lactate + NAD(P)+ OH-pyruvate + NAD(P)H ⇌ L-Glycerate + NAD(P)+ | Cyt, Mit, Chl | None |
| TSR | TSA + NAD(P)H ⇌ D-Glycerate + NAD(P)+ | Not in plants |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleczkowski, L.A.; Igamberdiev, A.U. Multiple Roles of Glycerate Kinase—From Photorespiration to Gluconeogenesis, C4 Metabolism, and Plant Immunity. Int. J. Mol. Sci. 2024, 25, 3258. https://doi.org/10.3390/ijms25063258
Kleczkowski LA, Igamberdiev AU. Multiple Roles of Glycerate Kinase—From Photorespiration to Gluconeogenesis, C4 Metabolism, and Plant Immunity. International Journal of Molecular Sciences. 2024; 25(6):3258. https://doi.org/10.3390/ijms25063258
Chicago/Turabian StyleKleczkowski, Leszek A., and Abir U. Igamberdiev. 2024. "Multiple Roles of Glycerate Kinase—From Photorespiration to Gluconeogenesis, C4 Metabolism, and Plant Immunity" International Journal of Molecular Sciences 25, no. 6: 3258. https://doi.org/10.3390/ijms25063258
APA StyleKleczkowski, L. A., & Igamberdiev, A. U. (2024). Multiple Roles of Glycerate Kinase—From Photorespiration to Gluconeogenesis, C4 Metabolism, and Plant Immunity. International Journal of Molecular Sciences, 25(6), 3258. https://doi.org/10.3390/ijms25063258







